Abstract
The genus Leea L. includes some species that are known to have ethnomedicinal properties. They have been used as cures against common ailments such as skin infections and arthritis. To augment the morphological identification of this taxon, four candidate chloroplast DNA barcodes (matK, rbcL, trnH-psbA and trnL-F) were tested for their efficiency as single-locus barcodes for four Leea species. Genomic DNA from silica-dried leaf samples were isolated and used as template for generating DNA barcodes. Pairwise sequence divergence using the Kimura two-parameter model was used to analyse interspecific and intraspecific variations among the barcodes whereas BLAST and neighbour-joining analyses were employed to examine discrimination success. The results show that matK is the most efficient single-locus barcode for Leea by yielding the highest rate of universality as well as the best discriminatory and authentication power among the barcodes examined.
Introduction
Leea L. is recognized by several taxonomists in its own monogeneric family Leeaceae with 34 recognized species worldwide (Ridsdale Citation1974, Citation1976; Shetty and Singh Citation2000; Ren et al. Citation2002; Wen Citation2007). Recently, the genus was classified under the grape family Vitaceae (APG Citation2009). Leea can be recognized by a combination of its habit (herbs to trees with barbed raphides), leaves (toothed edges, small glandular apex with one lateral vein lining the course above its tooth), stipules (borne along the petiole margin), inflorescences (cymose terminal, up to 18 cm across), and flowers (monoecious or hermaphrodite, calyx cupulate, corolla spreading, flower disc annular, stigmas minute) (Soltis et al. Citation2000).
In the Philippines, there are 15 recorded Leea species of which six are endemic (Pelser, Barcelona, and Nickrent Citation2011). Some of them have ethnomedicinal importance in the Philippines. The leaves and roots of Leea indica (Burm. f.) Merr. for instance, are used to treat skin complaints and relieve dizziness; leaves of Leea guineensis G. Don are used for arthritis and pounded roots are applied for healing wounds; and Leea macrophylla Roxb. ex Hornem. and Leea aequata L. both aid in wounds and sores (Uji Citation2001). Ethnomedicinal plants are used as a practical source of medicine for common health disorders (Maramba et al. Citation1982) but the threats of adulteration and misidentification are always present for these important plants. In particular, L. guineensis and L. indica can be distinguished by flower colour, having red to reddish orange and greenish-white flowers, respectively. In the absence of inflorescence however, L. guineensis can be misidentified as L. indica, and vice-versa, because they have overlapping vegetative forms (Ridsdale Citation1974, Citation1976). Life-threatening cases (Bilia Citation2013; Roy, Mallick, and Kaur Citation2013; Viljoen and Vermaak Citation2013) and deceptive attempts to raise the market value of the products (Wallace et al. Citation2012) have been reported when adulterations and misidentifications occur.
The emergence of nucleic acid analysis has provided more dependable techniques for revealing plant-based adulterants compared with conventional analytical methods (i.e. morphology, spectrometry, thin layer chromatography) (Sasikumar et al. 2004; Ng et al. Citation2005; Lum and Hirsch Citation2006; Aida et al. Citation2007). One such technique is DNA barcoding, which promises fast and accurate species identification by focusing the analysis on a short standardized segment of the genome (Hebert et al. Citation2003). Unfortunately, the DNA barcode involving the standard mitochondrial cytochrome c oxidase subunit 1 for animals evolves too slowly in plants (Fazekas et al. Citation2008). Selecting a standard DNA barcode for plants has been difficult because the barcoding efficiency varies for different taxa. This has led to a continuous search of the standard DNA barcode for plants with emphasis on nuclear ribosomal DNA (nrDNA) and plastid DNA (cpDNA). The CBOL-PWG (Citation2009) recommended having a core barcode consisting of two portions of plastid-coding regions, rbcL and matK, to be supplemented with additional markers as required. Other markers including trnH-psbA and trnL-F showed reasonable levels of species discrimination in floristic and taxonomic studies (Kress et al. Citation2005; Fazekas et al. Citation2012; Song et al. Citation2009; Yao et al. Citation2009).
In this study, four candidate cpDNA barcodes namely matK, rbcL, trnH-psbA and trnL-F, were tested for their efficiency as a single-locus barcode for selected Leea species. The candidate barcodes were assessed by their universality, discriminatory power and resolution (Kress et al. Citation2005; CBOL-PWG Citation2009; Hollingsworth, Graham, and Little Citation2011).
Material and methods
Sample collection and preservation
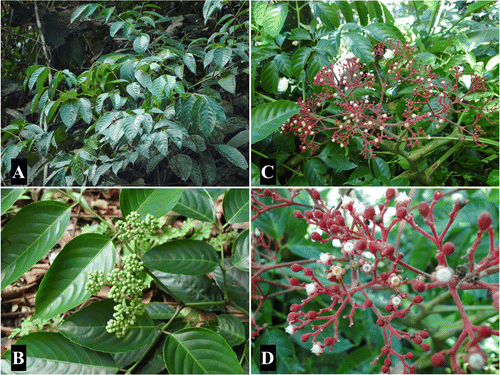
A total of eight specimens representing four Leea species, Leea aculeata Blume ex Spreng., Leea congesta Elmer, L. guineensis and Leea philippinensis Merr., were collected from six Philippine provinces (Table ). Leaf samples for genomic DNA extraction were placed in re-sealable bags with silica gel (Chase and Hills Citation1991). Herbarium vouchers were prepared for each specimen and deposited in the University of Santo Tomas Herbarium (USTH) for accessions. Voucher specimens and some field photographs (Figure ) of the plants were submitted to the Philippine National Herbarium (PNH) to confirm the identification
Table 1. List of Leea species collected in different Philippine provinces showing USTH voucher numbers.
Generation of DNA Barcodes
DNA of silica gel-dried leaf tissues was extracted following the protocols of DNeasy Plant Minikit (Qiagen, Hilden, Germany). Four candidate barcodes (matK, rbcL, trnH-psbA, trnL-F) were used to investigate the application of DNA barcoding for molecular authentication. The universal primer pairs (Table ) for matK (CBOL-PWG Citation2009), rbcL (Kress and Erickson Citation2007) trnH-psbA (Kress et al. Citation2005) and trnL-F (Taberlet et al. Citation1991) were amplified using a KapaTaq polymerase chain reaction (PCR) Kit (Kapa Biosystems, Wilmington, MA, USA) in Biometra® T-Gradient thermo cycler. The PCR cocktail for all markers contained (25 μl reaction): 17.35 μl water, 2.5 μl 10× buffer, 1.0 μl of 25 mm MgCl2, 2 μl 2 mm of dNTP, 1.0 μl of 10 μm forward and reverse primers, 0.15 μl Taq DNA polymerase and 0.5 μl DNA template. PCR conditions were set as follows: initial denaturation of 90 s at 97°C, followed by 35 cycles of 30 s at 95°C; 20 s at 50°C (for matK and rbcL) or 55°C (for trnH-psbA) or 20 s at 72°C (for trnL-F); 60 s at 72°C; finishing with 10 min at 72°C (Li et al. Citation2012). Agarose gel electrophoresis and QIA-quick PCR Purification Kit (Qiagen, Germany) were employed to resolve the amplicons and purify specific DNA fragments, respectively. Purified DNA was sent to Macrogen Inc. (Seoul, South Korea) for bidirectional DNA sequencing. All DNA sequences were assembled and edited using CodonCode Aligner v.4.1.1 (Codoncode Co., Centerville, MA, USA).
Table 2. Universal primers of the four candidate barcodes used in the study.
Sequence analyses
The new sequences together with sixteen GenBank accessions were automatically and manually aligned per marker in SeaView v.4 (Guoy, Guindon, and Gascuel Citation2010). Pairwise sequence divergence was calculated using the Kimura two-parameter (Kimura Citation1980) model in MEGA 6.0 (Tamura et al. Citation2013) to evaluate the mean interspecific and mean intraspecific (Chen et al. Citation2010; Yao et al. Citation2010) divergences. The Wilcoxon two-sample tests for interspecific and intraspecific divergences were conducted using SPSS 15.0 software (SPSS Inc, Chicago, IL, USA). The Basic Local Alignment Search Tool (BLAST, NCBI-GenBank) analysis of acquired sequences was conducted as previously described (Chen et al. Citation2010). A neighbour-joining method using Kimura-two-parameter distances (1000 bootstrap replicates) was conducted to test phylogenetic relationships in MEGA 6.0.
Results
A total of 29 new sequences were generated from four markers (Appendix 1.). After the multiple sequence alignments, trnL-F produced the longest mean length of 893.9 base pairs (bp) followed by matK, rbcL and trnH-psbA. Interestingly, trnH-psbA has the shortest mean length of 513.9 bp but possesses the highest number of parsimony informative characters with 37 from 72 variable sites (Table ). To measure the primers’ universality, amplification and sequencing success were assessed (Figure ). Only matK has 100% amplification and sequencing success whereas rbcL, trnH-psbA, and trnL-F have at least 75% success rates.
Table 3. Sequence characteristics from multiple sequence alignment of the four candidate barcodes used in this study.
Figure 2. Efficiency of polymerase chain reaction amplification and sequencing success rates of the four candidate barcodes for the selected Leea sp.
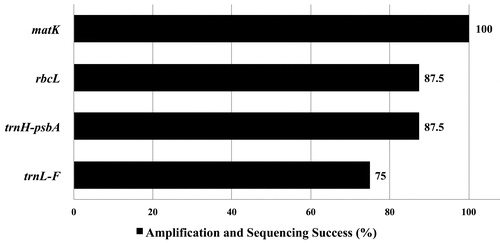
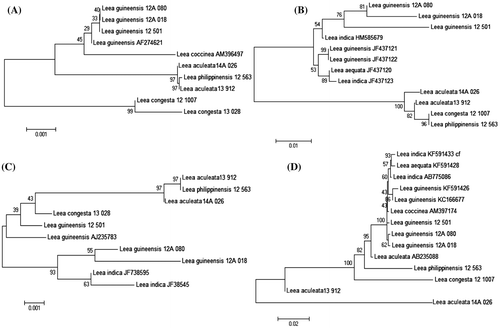
Table summarizes the pairwise sequence divergence analysis for each candidate barcode. The results show trnL-F as the barcode with the highest interspecific divergence of 0.055 ± 0.071 and matK has the lowest with 0.007 ± 0.003. The intraspecific divergences give similar results with trnL-F as the barcode with the highest value of 0.032 ± 0.607 whereas matK has the lowest with 0.000 ± 0.001. All barcodes were able to identify all of the specimens under the genus Leea using BLAST but gave indistinct resolutions in neighbour-joining except that for matK. In the rbcL tree for example, the four isolates of L. guineensis did not form into a monophyletic clade (Figure B).
Table 4. Calculated mean interspecific and intraspecific divergences of the four candidate barcodes using Kimura two-parameter.
Discussion
Universality
An ideal barcode should have a relatively short length to facilitate easy DNA extraction, amplification and sequencing such that it can be tractable across a wide range of species (Kress et al. Citation2005). The results shows that only matK has all samples (100%) successfully amplified and sequenced with minimum editing. Even after several attempts to amplify all samples using pure and diluted (1/10 and 1/100) DNA extracts for rbcL, trnH-psbA and trnL-F, they still gave lower success. Based on the results, matK is the most universal among the barcodes used. This is in contrast to the findings of Fazekas et al. (Citation2008) with 56% universality for 92 species from 32 genera and Newmaster et al. (Citation2008) with 49% for the family Myristicaceae, although both used substantially larger data sets.
Discrimination
Mean interspecific and intraspecific divergences were used to characterize the pairwise divergences of the samples per barcode. It is crucial for a barcode to have significantly higher interspecific than intraspecific divergence to better distinguish one species from another but not individuals of the same species (Lahaye et al. Citation2008; Galimberti et al. Citation2013). The Wilcoxon two-sample test shows that there are significant differences between the interspecific and intraspecific divergences of the candidate DNA regions, except for rbcL, which compromises its discriminatory power. The interspecific divergences of matK, trnH-psbA and trnL-F are significantly higher than their respective intraspecific variations (p < 0.05, Table ). In consequence matK, trnH-psbA and trnL-F can effectively discriminate one species from another.
Table 5. Wilcoxon two-sample test for interspecific and intraspecific divergences of the four candidate barcodes.
By comparison on the interspecific divergences, trnH-psbA and trnL-F have relatively higher mean interspecific distances compared with matK. Even with its relatively short length (approximately 450 bp), trnH-psbA is considered as the most variable plastid region in angiosperms and is easily amplified across a broad range of land plants with the potential to discriminate among the largest number of plant species for barcoding purposes (Kress et al. Citation2005; Song et al. Citation2009; Yao et al. Citation2009). However, trnH-psbA and trnL-F still cannot be better than matK because the latter has a very minimal intraspecific divergence.
Authentication
To be an efficient tool for molecular authentication, a barcode should identify a species up to the specific epithet. Unfortunately, BLAST analysis only identified all new sequences conclusively at the genus level. One sequence for example, is identified as different species with the same query cover and identity. In particular, the trnL-F sequences of L. guineensis isolates 12A-080 and 12-501 are identified as L. macrophylla (GenBank Acc. no. KF591434; 99% query cover and identity) and L. guineensis (GenBank Acc. no. KF501426; 99% query cover and identity). In the case of matK, there are only nine available accessions for Leea, which limit the accurate identification. It should be considered that BLAST analysis is primarily dependent on the availability and accuracy of published accessions. Hence, BLAST alone will not suffice as the basis for molecular authentication.
This is where phylogenetic analysis, mostly neighbour-joining, comes in handy. Unfortunately, because of the limited number of samples and the fact that these are congeneric species, the taxonomic groupings per species are mostly unresolved except for matK. Given that Leea coccinea is a synonym of L. guineensis, matK has successfully resolved all Leea species with multiple isolates (Figure A).
Conclusion
Compared with the other barcodes examined in this study, matK proved to be the most universal, having the best discriminatory and authentication power. This study serves as good baseline information regarding the application of DNA barcoding in the genus Leea that includes important ethnomedicinal species. It is vital to include more Leea species and isolates per species in future studies.
Notes on contributors
Vincent Louie D. Cabelin is an instructor at Notre Dame of Dadiangas University. He is currently completing his PhD in Biological Science at the Graduate School (GS) of University of Santo Tomas (UST). Contribution: V.L.D. Cabelin carried out the experiment, contributed most on manuscript writing, and read and approved the final version of the manuscript.
Propa Joy S. Santor is an M.Sc. in Biological Science student of the GS of UST. Contribution: P.J.R. Santor carried out the experiment, contributed most on manuscript writing, read and approved the final version of the manuscript.
Grecebio Jonathan D. Alejandro is a Full Professor and currently the Director of the Office for Graduate Research of the GS of UST. Dr Alejandro is recognized for his pioneering research on Plant Molecular Phylogenetics in the Philippines and established the Thomasian Angiosperm Phylogeny and Barcoding Group of UST. Contribution: G.J.D. Alejandro provided valuable opinions and suggestions in the earlier version of the manuscript, and read and approved the final version of the manuscript.
Acknowledgements
The authors thank the Department of Science and Technology Science (DOST)-Philippine Council for Health Research and Development (PCHRD) for the research fund. V.L.D. Cabelin is indebted to DOST-Science Educational Institute under the Accelerated S & T Human Resource Development Programme for his scholarship.
References
- APG (Angiosperm Phylogeny Group). 2009. “An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III.” Botanical Journal of the Linnean Society 141 (4): 399–436.
- Aida, A. A., C. Man, B. Yaakob, A. R. Raha, and R. Son. 2007. “Detection of pig derivatives in food products for halal authentication by polymerase chain reaction–restriction fragment length polymorphism.” Journal of the Science of Food and Agriculture 87 (4): 569–572.
- Bilia, A. R. 2013. “NMR as integrative or alternative analytical tool for the quality control of herbal drugs and their preparations.” Planta Medica 79 (5): OP18.
- CBOL-PWG (Consortium for the Barcode of Life – Plant Working Group). 2009. “A DNA barcode for land plants.” Proceedings of the National Academy of Sciences of the United States of America 106: 12794–12797.
- Chase, M. W., and H. H. Hills. 1991. “Silica gel: an ideal material for field preservation of leaf samples for DNA studies.” Taxon 40 (2): 215–220.
- Chen, S., H. Yao, J. Han, C. Liu, J. Song, L. Shi, Y. Zhu, et al. 2010. “Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species.” PloS One 5 (1): e8613.
- Fazekas, A. J., K. S. Burgess, P. R. Kesanakurti, S. W. Graham, S. G. Newmaster, B. C. Husband, D. M. Percy, M. Hajibabaei, and S. C. Barrett. 2008. “Multiple multilocus DNA barcodes from the plastid genome discriminate plant species equally well.” PLoS One 3 (7): e2802.
- Fazekas, A. J., M. L. Kuzminia, S. G. Newmaster, and M. Hollingsworth. 2012. In DNA Barcodes: Methods and Protocols, Methods in Molecular Biology, edited by Kress W. J., and D. L. Erickson, 858: 223–252. Berlin Heidelberg: Springer.
- Galimberti, A., F. De Mattia, A. Losa, I. Bruni, S. Federici, M. Casiraghi, S. Martellos, and M. Labra. 2013. “DNA barcoding as a new tool for food traceability.” Food Research International 50 (1): 55–63.
- Gouy, M., S. Guindon, and O. Gascuel. 2010. “SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building.” Molecular Biology and Evolution 27 (2): 221–224.
- Hebert, P. D., A. Cywinska, S. L. Ball, and J. R. deWaard. 2003. “Biological identifications through DNA barcodes.” Proceedings of the Royal Society of London B: Biological Sciences 270 (1512): 313–321.
- Hollingsworth, P. M., S. W. Graham, and D. P. Little. 2011. “Choosing and using a plant DNA barcode.” PloS One 6 (5): e19254.
- Kimura, M. 1980. “A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences.” Journal of Molecular Evolution 16 (2): 111–120.
- Kress, W. J., and D. L. Erickson. 2007. “A two-locus global DNA barcode for land plants: the coding rbcL gene complements the non-coding trnH-psbA spacer region.” PLoS One 2 (6): e508.
- Kress, W. J., K. J. Wurdack, E. A. Zimmer, L. A. Weigt, and D. H. Janzen. 2005. “Use of DNA barcodes to identify flowering plants.” Proceedings of the National Academy of Sciences of the United States of America 102 (23): 8369–8374.
- Lahaye, R., M. Van der Bank, D. Bogarin, J. Warner, F. Pupulin, G. Gigot, O. Maurin, S. Duthoit, T. G. Barraclough, and V. Savolainen. 2008. “DNA barcoding the floras of biodiversity hotspots.” Proceedings of the National Academy of Sciences of the United States of America 105 (8): 2923–2928.
- Li, M., K. Y. Au, H. Lam, L. Cheng, R. W. Jiang, P. P. H. But, and P. C. Shaw. 2012. “Identification of Baiying (Herba Solani Lyrati) commodity and its toxic substitute Xungufeng (Herba Aristolochiae Mollissimae) using DNA barcoding and chemical profiling techniques.” Food Chemistry 135 (3): 1653–1658.
- Lum, M. R., and A. M. Hirsch. 2006. “Molecular methods for the authentication of botanicals and detection of potential contaminants and adulterants.” Acta Horticulturae 720: 50–72.
- Maramba, N. P. C., J. D. Saludez, I. C. Sia, O. Y. Alegre, G. A. Solis-de Asis, L. B. Bagnaes, R. U. Macalagay, and A. R. Bajao. 1982. Guidebook on the Proper Use of Medicinal Plants. Quezon City, Philippines: Katha Publishing Co., Inc.
- Newmaster, S. G., A. J. Fazekas, R. A. D. Steeves, and J. Janovec. 2008. “Testing candidate plant barcode regions in the Myristicaceae.” Molecular Ecology Resources 8 (3): 480–490.
- Ng, C. C., C. Y. Lin, W. S. Tzeng, C. C. Chang, and Y. T. Shyu. 2005. “Establishment of an internal transcribed spacer (ITS) sequence-based differentiation identification procedure for mei (Prunus mume) and plum (Prunus salicina) and its use to detect adulteration in preserved fruits.” Food Research International 38 (1): 95–101.
- Pelser, P. B., J. F. Barcelona, and D. L. Nickrent (eds.). 2011 onwards. Co's Digital Flora of the Philippines. Accessed March 21, 2015. http://www.philippineplants.org/Families/Vitaceae.html
- Ren, H., K. Pan, Z. Chen, and R. Wang. 2002. “Structural characters of leaf epidermis and their systematic significance in Vitaceae.” Acta Phytotaxonomica Sinica 41 (6): 531–544.
- Ridsdale, C. E. 1974. “A revision of the family Leeaceae.” Blumea-Biodiversity, Evolution and Biogeography of Plants 22 (1): 57–100.
- Ridsdale, C. E. 1976. “Leeaceae.” Flora Malesiana-Series 1, Spermatophyta 7 (1), 755–782.
- Roy, A., A. Mallick, and A. Kaur. 2013. “Adulteration and substitution in Indian medicinal plants.” International Journal of Pharmaceutical Research and Bio-Science 2 (1): 208–18.
- Sasikumar, B., S. Syamkumar, R. Remya, and T. John Zachariah. 2005. “PCR based detection of adulteration in the market samples of turmeric powder.” Food Biotechnology 18 (3): 299–306.
- Shetty, B.V., and P. Singh. 2000. “Vitaceae.” In Flora of India, edited by Singh, N. P., J. N. Vohra, P. K. Hajra, D. K. Singh, 5: 246–324. Calcutta: Botanical Survey of India.
- Soltis, D. E., P. S. Soltis, M. W. Chase, M. E. Mort, D. C. Albach, M. Zanis, V. Savolainen, et al. 2000. “Angiosperm phylogeny inferred from 18S rDNA, rbcL and atpB sequences.” Botanical Journal of the Linnean Society 133 (4): 381–461.
- Song, J., H. Yao, Y. Li, X. Li, Y. Lin, C. Liu, J. Han, C. Xie, and S. Chen. 2009. “Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique.” Journal of Ethnopharmacology 124 (3): 434–439.
- Taberlet, P., L. Gielly, G. Pautou, and J. Bouvet. 1991. “Universal primers for amplification of three non-coding regions of chloroplast DNA.” Plant Molecular Biology 17 (5): 1105–1109.
- Tamura, K., G. Stecher, D. Peterson, A. Filipski, and S. Kumar. 2013. “MEGA6: molecular evolutionary genetics analysis version 6.0.” Molecular Biology and Evolution 30 (12): 2725–2729.
- Uji, T., 2001. “Leea van Royen ex L.” In PROSEA: Plant Resources of South-East Asia 12, (2) medicinal and poisonous plants 2, edited by van Valkenburg, J. L. C. H. and N. Bunyapraphatsara, 327–331. Leiden, the Netherlands: Backhuys Publishers.
- Viljoen, A. M., and I. Vermaak. 2013. “Standardization and quality control of herbal medicinal products – Does vibrational spectroscopy offer the solution?” Planta Medica 79(05): OP20.
- Wallace, L. J., S. M. Boilard, S. H. Eagle, J. L. Spall, S. Shokralla, and M. Hajibabaei. 2012. “DNA barcodes for everyday life: Routine authentication of natural health products.” Food Research International 49 (1): 446–452.
- Wen, J. 2007. “Leeaceae.” In The Families and Genera of Vascular Plants, Volume IX: Flowering Plants, Eudicots, edited by K. Kubitzki, 221–225. Berlin Heidelberg: Springer.
- Yao, H., J. Song, C. Liu, K. Luo, J. Han, Y. Li, X. Pang, et al. 2010. “Use of ITS2 region as the universal DNA barcode for plants and animals.” PloS One 5 (10): e13102.
- Yao, H., J. Y. Song, X. Y. Ma, C. Liu, Y. Li, H. X. Xu, J. P. Han, L. S. Dun, and S. L. Chen. 2009. “Identification of Dendrobium species by a candidate DNA barcode sequence: the chloroplast psbA-trnH intergenic region.” Planta Medica 75 (6): 667–669.