Abstract
The ladder-shape melting temperature isothermal amplification (LMTIA) is a newly developed technique with advantages of simple mechanism, easy primer design, quick turn-around time, high specificity, high sensitivity and short target sequence compared to the loop-mediated isothermal amplification (LAMP) technique. The objective of this research was to establish the LMTIA assay for detection of chicken in beef. The LMTIA primers targeting the prolactin receptor gene of Gallus gallus were designed, the LMTIA reaction system was optimized, the specificity and the sensitivity of the LMTIA assay were determined. Our results showed that the LMTIA assay was able to specifically detect 100 ng genomic DNA of Gallus gallus, without detecting the 100 ng genomic DNA of Anas platyrhynchos, Felis catus, Sus scrofa, Canis lupus familiaris, Bos taurus or Capra hircus. The sensitivity of the LMTIA assay was 100 pg genomic DNA of G. gallus. Furthermore, the LMTIA assay was able to detect the chicken adulterated in beef with ≥0.1% (w/w) detection limit (LOD). This study holds a promise for facilitation of the surveillance of the commercial adulteration of chicken in beef.
Introduction
High-value foods have recently become the objects of food fraud; the main types of fraud include origin mislabeling [Citation1] and species adulteration [Citation1–3], e.g. chicken has been commonly adulterated into beef [Citation4]. The research on food authentication has already become the hot spot. The advanced technologies including nuclear magnetic resonance spectroscopy [Citation5], isotopic mass spectrometry [Citation6], high resolution mass spectrometry [Citation7], immunoassay [Citation8, Citation9], infrared spectroscopy [Citation10, Citation11], electrochemical genosensor [Citation12], chemometrics [Citation13], nucleic acid detection have been explored for detection of food adulteration, among which the nucleic acid detection is suitable for species identification for its specification, sensitivity and accuracy [Citation14].
Many of the existing nucleic acids amplification methods have been improved or modified for detection of meat adulteration, e.g. recombinase polymerase amplification (RPA) [Citation15, Citation16], polmyerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) [Citation17, Citation18], quantitative polmyerase chain reaction (qPCR) [Citation19], multiplex real-time PCR assay [Citation20], digital PCR [Citation21–23], saltatory rolling circle amplification (SRCA) [Citation24], clustered regularly interspaced short palindromic repeats (CRISPR)/cas12a-mediated liposome-amplified strategy [Citation25] and loop-mediated isothermal amplification (LAMP) [Citation26]. The current available assays are very specific and sensitive but time-consuming and require highly sophisticated equipment and qualified personnel [Citation27], thus, a new nucleic acids assay for detection of meat adulteration is urgently needed.
Ladder-shape melting temperature isothernmal amplification (LMTIA) is a recently developed nucleic acids amplification technique. The LMTIA provides single-strand DNA template via the melting temperature (Tm) difference between the primer and its target sequence rather than thermal denaturation or enzyme catalysis. LMTIA has the following advantages over the previously established nucleic acids amplification techniques: simple mechanism, easy primer design, quick turn-around time, high specificity, high sensitivity, short target sequence, wide application range of single-stranded DNA and double-stranded DNA or RNA (for reverse transcriptase activity of Bst DNA Polymerase) as its target [Citation28]. To verify the reported excellent performance [Citation28], we previously used the LMTIA technique to detect the adulterant chicken adulteration in beef and to detect African swine fever virus (ASFV) [Citation29]. The objective of this study was to develop the LMTIA assay for detection of chicken in beef, which would provide a promising solution for facilitating the surveillance of the commercial adulteration of chicken in beef.
Materials and methods
Target sequence selection and LMTIA primer design
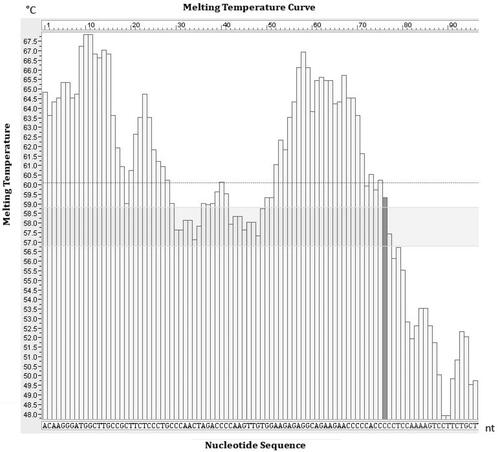
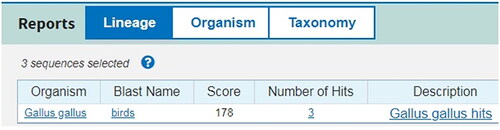
According to the principle for the target sequence selection of LMTIA, the sequence with ladder-shape melting temperature curve was selected as target from the prolactin receptor gene of Gallus gallus (GenBank accession No. KU553470.1) using software Oligo 7 (Molecular Biology Insights, Inc. Colorado Springs, CO, USA), the length of the selected target sequence was 96 nt with GC content of 53.4%. The melting temperature curve of the selected sequence was ladder-shape is shown in , and the 96-nt sequence was blasted in Genbank, only the DNA sequence from Gallus gallus was aligned, as shown in , so the selected sequence was highly specific to G. gallus. The LMTIA primers were designed with the online software Primer3Plus (https://dev.primer3plus.com/index.html), with the selected 96-nt sequence as the target; the parameters of primers were set as described in a previous report [Citation28], and the primer sequences are listed in .
Table 1. LMTIA primers targeting the prolactin receptor gene of Gallus gallus.
DNA preparation
The meat samples of Gallus gallus, Anas platyrhynchos, Felis catus, Sus scrofa, Canis lupus familiaris, Bos taurus and Capra hircus were provided by Henan Tuodao Biotechnology Co., Ltd. Pieces of chicken and beef meat were minced, then 1.0 g, 10.0 g, 50.0 g 100.0 g and 200.0 g of minced chicken were mixed with 999.0 g, 990.0 g, 950.0 g, 900.0 g and 200.0 g of minced beef; respectively, the samples of beef adulterated with 0.1%, 1%, 5%, 10% and 20% chicken were prepared. The genomic DNA was extracted from these samples using an Animal Tissue DNA Extraction Kit (Tiangen Biotech (Beijing) Co., Ltd) and stored at −20 °C until use.
Optimization of LMTIA reaction temperature
The LMTIA reaction temperature was optimized using 10 μL reaction mixture containing 1.3 μmol each of the left primer L and the right primer R, 1× General LMTIA Reaction Mix (1.0 mmol/L deoxyribonucleoside triphosphates, 20 mmol/L Tris-HCl [pH 8.8], 10 mmol/L KCl, 10 mmol/L (NH4)2SO4, 6 mmol/L MgSO4, 0.1% Triton™ X-100, 1 × EvaGreen, 0.32 U/μL Bst DNA polymerase) (Merit Biotech [Shandong] Co., Ltd, Heze, Shandong Province, China), and 100 ng genomic DNA of Gallus gallus as template. The reaction mixture was overlaid with 20 μL of liquid paraffin to avoid aerosol contamination [Citation28]. The reaction mixture was heated at 52 °C, 54 °C, 56 °C and 58 °C for 60 min (1.5 min per cycle) in a StepOne™ System Real-Time PCR System (Applied Biosystems, CA, USA), respectively.
Specificity determination of the LMTIA assay
The specificity of the newly developed LMTIA assay was determined using 100 ng genomic DNA of Gallus gallus, Anas platyrhynchos, Felis catus, Sus scrofa, Canis lupus familiaris, Bos taurus and Capra hircus, and the reaction mixtures were heated at the optimized temperature for 60 min (1.5 min per cycle) in a StepOne™ System Real-Time PCR System (Applied Biosystems, CA, USA).
Sensitivity determination of the LMTIA assay
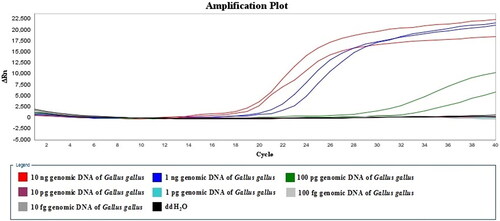
The sensitivity of the newly developed LMTIA assay was determined using the genomic DNA of Gallus gallus ranging from 10 ng to 10 fg, and the reaction mixtures were heated at the selected temperature for 60 min (1.5 min per cycle) in a StepOne™ System Real-Time PCR System (Applied Biosystems, CA, USA).
Detection limit determination of the LMTIA assay for chicken adulterated in beef
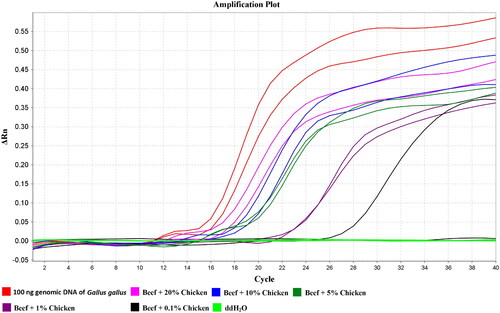
The detection limit (LOD) of the newly developed LMTIA assay was determined using the genomic DNA extracted from the beef, which was adulterated with 0.1%, 1%, 5%, 10% and 20% chicken, and the reaction mixtures were heated at the optimized temperature for 60 min (1.5 min per cycle) in an Archimed time resolution real-time fluorescence quantitative PCR system (RocGene (Beijing) Technology Co., Ltd, Beijing, China).
Results
Selection of the LMTIA reaction temperature
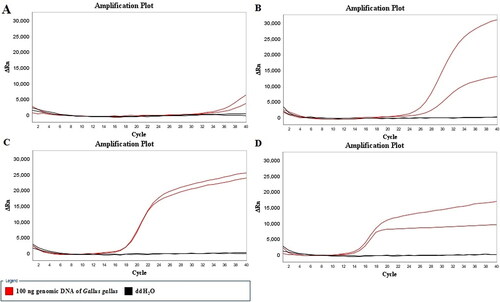
All positive controls (with 100 ng genomic DNA of Gallus gallus as template) tested positiveand all negative controls (DNA template substituted by ddH2O) tested negative at 52 °C, 54 °C, 56 °C and 58 °C (). The amplification efficiency of the LMTIA reaction was the highest at 58 °C. Although the amplification efficiency at 56 °C was slightly lower than that at 58 °C, the repeatability at 56 °C was better; therefore, the temperature of 56 °C was selected as the reaction temperature for the subsequent experiment.
LMTIA is highly specific to chicken
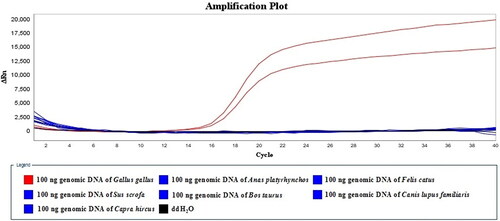
The specificity of LMTIA assay was tested using 100 ng genomic DNA of Gallus gallus, Anas platyrhynchos, Felis catus, Sus scrofa, Canis lupus familiaris, Bos taurus and Capra hircus. As shown in , only G. gallus was successfully detected. The results of the two repeated experiments were consistent, and there was no detectable false-positive response of the negative controls. Cytochrome b gene [Citation17, Citation26], ligand dependent nuclear receptor corepressor (LcoR) gene [Citation19] as well as interleukin 2 (IL2) gene [Citation21] had been previously selected for target sequences, while the 96-nt sequence on the prolactin receptor gene of Gallus gallus was selected as the target of the developed LMTIA assay. The sequence was highly specific to Gallus gallus by sequence and LMTIA specificity assay.
Sensitivity of the LMTIA assay
Sensitivity of the LMTIA assay was tested using the genomic DNA of Gallus gallus ranging from 10 ng to 10 fg at 56 °C for 60 min. The sensitivity was 100 pg genomic DNA of G. gallus, as shown in .
Detection limit of the LMTIA assay for chicken adulterated in beef
Genomic DNA extracted from the artificial samples of beef adulterated with 0.1%, 1%, 5%, 10% and 20% chicken were used for determination of the detection limit of the LMTIA assay. As shown in , one sample of the beef adulterated with 0.1% chicken was positive while the other was negative. Therefore, the LMTIA assay can detect the chicken adulterated in beef with detection limit (LOD) of 0.1% (w/w).
Discussion
The LMTIA technique is a recently reported modification of LAMP [Citation28]. LMTIA technique achieves isothermal amplification in PCR via Tm difference between primers and their target sequences, and it shortens the reaction time from more than 1 h to less than 25 min using the primer design method of LAMP as reference. Compared to LAMP, LMTIA has the advantages of clear single-strand template generation mechanism, rapidity, high specificity and sensitivity as well as being suitable for short target sequence amplification. In this study, the LMTIA primers were designed to target a 96-nt sequence on the prolactin receptor gene of G. gallus. The LMTIA assay for detection of chicken adulteration in beef was established, so the experiment demonstrated that the reaction mechanism of LMTIA is accurate and the technique is reproducible. In order to prove the reproducibility of LMTIA, we have used the LMTIA technique to detect adulterant pork in beef, adulterant duck in beef, adulterant chicken in beef, adulterant cassava starch in sweet potato noodles, adulterant corn starch in sweet potato noodles, adulterant rape honey in Robinia honey, and to detect African swine fever virus (ASFV) [Citation29]. All of these results confirmed the advantages of the LMTIA technique.
In this study, the 96-nt sequence on the prolactin receptor gene of Gallus gallus was selected as the target sequence of the LMTIA assay., The alignment in GenBank indicates that the target sequence is of high specificity to Gallus gallus, and it is also confirmed by the specificity determination that the developed LMTIA assay is highly specific to chicken.
The developed LMTIA assay is able to detect the chicken adulterated in beef with detection limit (LOD) of 0.1% (w/w), while the LOD of single primer-triggered isothermal amplification (SAMP) [Citation30], multiplex PCR assay [Citation31], direct lysis-multiplex PCR assay [Citation32] and LAMP [Citation33] are 2% (w/w), 0.05% (w/w), 0.1% (w/w) and 0.1% (w/w), respectively. The LOD of the developed LMTIA is almost the same as other nucleic acids amplification methods, and the research has provided a new sensitive method for detection of chicken adulterated in beef.
In the future of LMTIA assay, as recently reported [Citation29], the Bst DNA polymerase (large fragment) should be modified to improve the catalytic efficiency and shorten the amplification time to less than 10 min, and the Bst DNA polymerase (large fragment) should be made compatible with the UNG-dUTP system to reduce the product pollution. Moreover, a fluorescent probe suitable for the LMTIA technique should be designed to overcome the shortcomings of absence of internal amplification controls (IACs) as well as limitations of quantitative assessments and multiplexing [Citation34].
Conclusions
In this study, the LMTIA assay was developed to specifically detect chicken adulterated in beef. The sensitivity was 100 ng genomic DNA, and the LOD was 0.1%. The experiment verified that the reaction mechanism of LMTIA is correct and the technique is reproducible and provided a reference for authentication of other species meat using the LMTIA technique.
Acknowledgments
The authors wish to thank all members from Key Laboratory of Biomarker Based Rapid-detection Technology for Food Safety of Henan Province, and Dr. Yanhong Liu from US Department of Agriculture for critical reading of the manuscript.
Data availability statement
The data that support this study are available from the corresponding author upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Francois G, Fabrice V, Didier M. Traceability of fruits and vegetables. Phytochemistry. 2020;173:112291.
- Brigante FI, Mas AL, Pigni NB, et al. Targeted metabolomics to assess the authenticity of bakery products containing chia, sesame and flax seeds. Food Chem. 2020;312:126059.
- Yang QQ, Qian ZX, Ye ZH, et al. Widespread mislabeling of nonnative apple snails (Ampullariidae Pomacea) as native field snails (Viviparidae: Cipangopaludina) on the Chinese food markets. Aquaculture. 2021;530:735756.
- Fengou LC, Tsakanikas P, Nychas G. Rapid detection of minced pork and chicken adulteration in fresh, stored and cooked ground meat. Food Control. 2021;125(6):108002.
- Gallo V, Ragone R, Musio B, et al. A contribution to the harmonization of non-targeted NMR methods for data-driven food authenticity assessment. Food Anal Methods. 2020;13(2):530–541.
- Martinelli LA, Nardoto GB, Perez MAZ, et al. Carbon and nitrogen isotope ratios of food and beverage in Brazil. Molecules. 2020;25(6):1457.
- Li Y, Zhang Y, Li H, et al. Simultaneous determination of heat stable peptides for eight animal and plant species in meat products using UPLC-MS/MS method. Food Chem. 2018;245:125–131.
- Mandli J, El Fatimi I, Seddaoui N, et al. Enzyme immunoassay (ELISA/immunosensor) for a sensitive detection of pork adulteration in meat. Food Chem. 2018;255:380–389.
- Seddaoui N, Amine A. Smartphone-based competitive immunoassay for quantitative on-site detection of meat adulteration. Talanta. 2021;230:122346.
- Ling Y, Wu T, Liu Y, et al. Rapid identification of pork adulterated in the beef and mutton by infrared spectroscopy. J Spectrosc. 2018;2018:2413874.
- Rady A, Adedeji A. Assessing different processed meats for adulterants using visible-near-infrared spectroscopy. Meat Sci. 2018;136:59–67.
- Flauzino JMR, Nguyen EP, Yang Q, et al. Label-free and reagentless electrochemical genosensor based on graphene acid for meat adulteration detection. Biosens Bioelectron. 2022;195:113628.
- Song W, Yun YH, Wang H, et al. Smartphone detection of minced beef adulteration. Microchem J. 2021;164:106088.
- Amaral JS. Target and non-target approaches for food authenticity and traceability. Foods. 2021;10(1):172.
- Cao Y, Zheng K, Jiang J, et al. A novel method to detect meat adulteration by recombinase polymerase amplification and SYBR green I. Food Chem. 2018;266:73–78.
- Lin L, Zheng Y, Huang H, et al. A visual method to detect meat adulteration by recombinase polymerase amplification combined with lateral flow dipstick. Food Chem. 2021;354:129526.
- Sunutcha S, Lawan C, Montri S. Identification of sea snake meat adulteration in meat products using PCR-RFLP of mitochondrial DNA. Food Sci Hum Wellness. 2018;7(2):170–174.
- Vaithiyanathan S, Vishnuraj MR, Reddy GN, et al. Authentication of camel meat using species-specific PCR and PCR-RFLP. J Food Sci Technol. 2021;58(10):3882–3889.
- Wang W, Fu M, Zhang Q, et al. A novel quantitative real-time PCR method for the detection of mammalian and poultry species based on a shared single-copy nuclear DNA sequence. Food Chem. 2021;341(Pt 2):128170.
- Zhu T, Zhou X, Zhang W, et al. Multiplex and real-time PCR for qualitative and quantitative donkey meat adulteration. J Food Meas Charact. 2021;15(2):1–8.
- Chen X, Ji Y, Li K, et al. Development of a duck genomic reference material by digital PCR platforms for the detection of meat adulteration. Foods. 2021;10(8):1890.
- Temisak S, Thangsunan P, Boonnil J, et al. Accurate determination of meat mass fractions using DNA measurements for quantifying meat adulteration by digital PCR. Int J of Food Sci Tech. 2021;56(12):6345–6358.
- Yu N, Ren J, Huang W, et al. An effective analytical droplet digital PCR approach for identification and quantification of fur-bearing animal meat in raw and processed food. Food Chem. 2021;355:129525.
- Hu X, Xu H, Zhang Y, et al. Saltatory rolling circle amplification (SRCA) for sensitive visual detection of horsemeat adulteration in beef products. Eur Food Res Technol. 2021;247(11):2667–2676.
- Liu J, Chen J, Wu D, et al. CRISPR-/Cas12a-mediated liposome-amplified strategy for the surface-enhanced Raman scattering and naked-eye detection of nucleic acid and application to food authenticity screening. Anal Chem. 2021;93(29):10167–10174.
- Wang Y, Zhu K, Wang D. Visual detection of donkey-derived ingredients by loop-mediated isothermal amplification with 4-(2-pyridylazo)-resorcinol sodium salt. CYTA-J Food. 2020;18(1):240–244.
- Wang Z, Yu W, Xie R, et al. A strip of lateral flow gene assay using gold nanoparticles for point-of-care diagnosis of African swine fever virus in limited environment. Anal Bioanal Chem. 2021;413(18):4665–4672.
- Wang D, Wang Y, Zhang M, et al. Ladder-shape melting temperature isothermal amplification of nucleic acids. BioTechniques. 2021;71(1):358–369.
- Wang Y, Wang B, Xu D, et al. Development of a ladder-shape melting temperature isothermal amplification (LMTIA) assay for detection of African swine fever virus (ASFV). J Vet Sci. 2022;23:e51.
- Chen J, Zhang P, Wang H, et al. Identification for adulteration of beef with chicken based on single primer-triggered isothermal amplification. Int J Food Eng. 2021;17(5):337–344.
- Qin P, Qu W, Xu J, et al. A sensitive multiplex PCR protocol for simultaneous detection of chicken, duck, and pork in beef samples. J Food Sci Technol. 2019;56(3):1266–1274.
- Zhao G, Shen X, Liu Y, et al. Direct lysis-multiplex polymerase chain reaction assay for beef fraud substitution with chicken, pork and duck. Food Control. 2021;129:108252.
- Sul SY, Kim MJ, Kim HY. Development of a direct loop-mediated isothermal amplification (LAMP) assay for rapid and simple on-site detection of chicken in processed meat products. Food Control. 2019;98:194–199.
- Kokkinos PA, Ziros PG, Bellou M, et al. Loop-Mediated isothermal amplification (LAMP) for the detection of Salmonella in food. Food Anal. Methods. 2014;7(2):512–526.