Abstract
Pak choi (Brassica rapa ssp. chinensis Makino) is a typical seed vernalization vegetable. Because premature bolting in spring causes significant economic losses, it is very important to clarify the flower formation mechanism to prevent this. Cytokinins are important plant hormones involved in regulating plant growth and development. To understand the relationship between cytokinin metabolism and flowering of pak choi, in this study, we determined the cytokinin trans-zeatin content in shoot apices of pak choi at different developmental stages by enzyme-linked immunosorbent assay. The results showed that cytokinin levels increased significantly after low temperature treatment at 4 °C, and continued to increase with vegetative growth after transplanting, reaching a peak at the critical period which is immediately prior to flower buds’ differentiation (S0), then decreased thereafter. The levels for low temperature treatment were consistently higher than controls. To explore the molecular mechanism underpinning the cytokinin changes, expression of homologous genes encoding cytokinin metabolic enzymes was analysed by transcriptome sequencing. The expression levels of nine genes (Bra004037, Bra023701, Bra002204, Bra014968, Bra028326, Bra028182, Bra034022, Bra009143 and Bra005869) were consistent with the changes in cytokinin content. The correlation between differentially expressed genes and cytokinin content in different developmental stages of apexes was analysed, and six closely related genes (Bra023701, Bra002204, Bra014968, Bra028182, Bra005869 and Bra009143) were identified. The results help to illuminate the molecular mechanisms controlling flowering of pak choi.
Introduction
Pak choi (Brassica rapa ssp. chinensis Makino) is a cruciferous vegetable that requires vernalization at low temperature before flowering under high temperature and long sunshine conditions. Weak winterness cultivars are more prone to bolting, and premature bolting in spring causes significant economic losses [Citation1]. Therefore, it is of great significance to clarify the flowering mechanism to prevent premature bolting.
Phytohormones play an important role in the flowering process of plants. Among them, cytokinins play a key role in long-distance signalling from root to shoot. This regulates root and shoot growth, photomorphogenesis, flowering time, senescence and seed development [Citation2]. High levels of cytokinins can increase the activity of shoot apical meristem (SAM) [Citation3] and promote the growth of axillary buds [Citation4]. Cytokinins can also promote cell division in Chinese cabbage sprouts [Citation5]. In Arabidopsis thaliana, the SAM at the early stage of floral transition was found to contain more cytokinins [Citation6], the external application of cytokinin can promote the transition of flower formation [Citation7, Citation8], increase the number of female flower and inhibit stamen development [Citation9]. Treatment of Brassica napus with cytokinins increases the ovule and seed numbers [Citation10]. Some scholars found that cytokinins can induce cabbage tip callus to form shoots [Citation11]. In B. napus, cytokinins increase the trans-zeatin content during flower bud differentiation [Citation12], and overexpression of the isopentenyl-transferase (IPT) gene increases the inflorescence branching in plants [Citation13]. In A. thaliana, cytochrome P450 monooxygenase, family 735, subfamily A (CYP735As) has been shown to catalyse the synthesis of trans-zeatin [Citation14], overexpression of uridine diphosphate glucose glycosyltransferase 76Cs (UGT76Cs) reduces cytokinin content [Citation15]. In addition, cytokinin can also delay the senescence of Chinese cabbage leaves during storage [Citation16], and the shedding of Chinese cabbage petals will lead to the reduction of cytokinins [Citation17], drought stress conditions increase the cytokinin content of rapeseed [Citation18]. These findings indicated that cytokinins play a role in different stages of plant growth and development, but the role in the process of flower bud differentiation is particularly important. However, for pak choi, the process of cytokinin metabolism during flower formation is poorly understood.
At present, more than 30 kinds of cytokinins have been reported, including isopentenyl adenine (ip), dihydro-zeatin (dZ), cis-zeatin (cZ) and trans-zeatin (tZ), among which tZ is considered to be the most important cytokinin [Citation19]. Therefore, in the present study, the content of cytokinin in the apexes of pak choi were measured at different developmental stages, and expression levels of associated genes were analysed by transcriptome sequencing to explore the molecular mechanism of cytokinins regulating the flowering process of pak choi.
Materials and methods
Material handling and sampling
The pak choi inbred line 75# was used in the experiment, and seeds with full grains were soaked in water and germinated at 24 °C in an incubator. At 3 days after seed germination, seedlings were placed at low temperature (4 °C) for 20 days and traditional management was carried out. Among them, 800 plants were treated with low temperature, and 500 plants were control.
On days 0 and 10 after transplanting, the shoot apexes of plants subjected to low temperature treatment and control (CK) plants were sampled and recorded as DAV0, CK-DAV0 and DAV10, CK-DAV10, respectively. Flower bud differentiation was observed under a stereo microscope. The shoot apex was sampled when it is at flower buds differentiated stage 0 (S0), CK-S0 and stage 1 (S1), according to Song et al. [Citation20]. A 0.2 g sample was used to determine the cytokinin content, and RNA extraction was performed from 0.1 g samples. Three biological replicates were included. After sampling, samples were snap-frozen in liquid nitrogen and stored at −80 °C for later use.
Cytokinin content determination
The content of cytokinin in the shoot apexes of pak choi at different developmental stages (DAV0, CK-DAV0, DAV10, CK-DAV10, S0, CK-S0 and S1) was determined by enzyme-linked immunosorbent assay at China Agricultural University. The determined component was tZ, and the method was performed as described previously [Citation21].
Transcriptome analysis
Total RNA was extracted using an RNA Prep Pure Plant Kit (Tiangen, DP432) according to the manufacturer’s instructions. Extracted RNA was stored at −80 °C. Transcriptome sequencing was performed on samples DAV10, S0 and S1 by Biomarker Technologies Co. Ltd (Beijing, China), and sequencing steps, expression analysis and functional annotation were carried out as described previously [Citation22].
The transcriptome sequencing data of this study were uploaded to the NCBI SRA database and can be obtained through the accession number PRJNA821155. The corresponding data sequences refer to SRR18534521 (for DAV10-1), SRR18534520 (for DAV10-2), SRR18534519 (for DAV10-3), SRR18534518 (for S0–1), SRR18534517 (for S0–2), SRR18534516 (for S0–3), SRR18534515 (for S1–1), SRR18534514 (for S1–2) and SRR18534513 (for S1–3).
Gene expression and functional annotation analysis
In order to avoid the influence of gene length and sequencing depth on the expression level, the method of Fragments Per Kilobasesper Millionmapped Reads (FPKM) was used to normalize the sequencing data. Differentially expressed genes (DEGs) at different developmental stages were identified using DESeq2 software. With FDR < 0.01 and Fold Change (FC) ≥ 2 as the threshold, pairwise comparisons were made in DAV10 vs. S0 and S0 vs. S1, and all DEGs were annotated by Gene Ontology (GO).
Real-time quantitative PCR validation
To validate the transcriptome sequencing result, GO annotations to cytokinin-related genes were screened in the self-tested transcriptome data, and six DEGs that related to cytokinin were randomly selected and used specific primers for real-time quantitative PCR (RT-qPCR). According to the general principles of primer design, Primer-BLAST in NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was used to design and select specific primers with amplified fragments of about 200 bp (), with ACTIN as an internal reference gene. RT-qPCR amplifications were performed on a Quant Studio 3 real-time PCR machine with an initial denaturation step at 94 °C for 5 min, followed by 40 cycles at 94 °C for 30 s, 49.5 °C for 30 s and 72 °C for 30 s, with three biological replicates. Relative expression levels were calculated using the 2–ΔΔCT method to normalize against the internal reference gene [Citation23], and the specific method was applied described previously [Citation24].
Table 1. Real-time quantitative PCR primers.
Results
Comparison of cytokinin content in shoot apexes at different developmental stages
The cytokinin(tZ)content of the shoot apexes at different developmental stages was measured for pak choi subjected to low temperature treatment and untreated controls (). The results showed that in the low temperature treatment group, the cytokinin contents were increased during the transition from the vegetative growth phase to the reproductive growth phase, and the levels reached a peak value of 6.92 ng/gFW when the flower bud differentiation was at stage 0 (S0). That is to say, once flower bud differentiation started, the cytokinin content decreased. Our results are consistent with these of prior observations, indicating that increased cytokinin content could promote flower bud differentiation in pak choi. The content of cytokinin in the control group showed the same trend, but the levels were consistently lower than in the low temperature treatment group throughout the growth period. This indicated that, within a certain range, higher accumulation of cytokinin contributes to flower bud differentiation of pak choi.
Figure 1. Comparison of cytokinin content in the shoot apexes of pak choi at different developmental stages. DAV0, 0 days after transplanting; DAV10, 10 days after transplanting; S0, Stage immediately prior to flower bud differentiation; S1, flower bud differentiation stage 1.
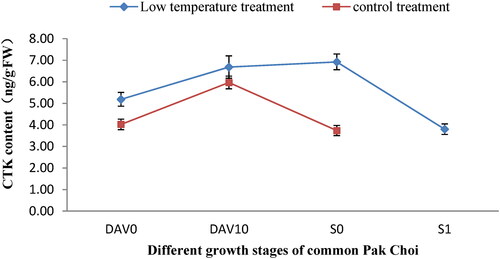
The cytokinin content of the low temperature treatment and control groups at 0 days after transplanting were compared, and the cytokinin levels of the low temperature treatment group were higher, which indicates that low temperature treatment can increase the accumulation of cytokinin and thereby promote the differentiation of flower buds.
Transcriptome analysis
Quality assessment of transcriptome sequencing
In order to analyse the gene expression of shoot apexes at different developmental stages in pak choi, nine libraries (DAV10-1, DAV10-2, DAV10-3, S0–1, S0–2, S0–3, S1–1, S1–2 and S1–3) were constructed (). We, respectively, obtained 53,756,300, 46,233,658, 41,857,180, 46,995,194, 45,981,760, 39,083,622, 40,207,374, 56,949,456 and 51,518,858 reads. Mapped reads, respectively, accounted for 87.05%, 86.67%, 86.74%, 87.61%, 85.95%, 85.74%, 86.43%, 86.16% and 86.09% of total reads, >85% in all cases. The number of unique mapped reads was between 83.36% and 85.44%, and the comparison efficiency was high, indicating that the sequencing quality was good, and could meet the needs of subsequent analysis.
Table 2. Illumina DNA sequencing reads from nine libraries and mapping results.
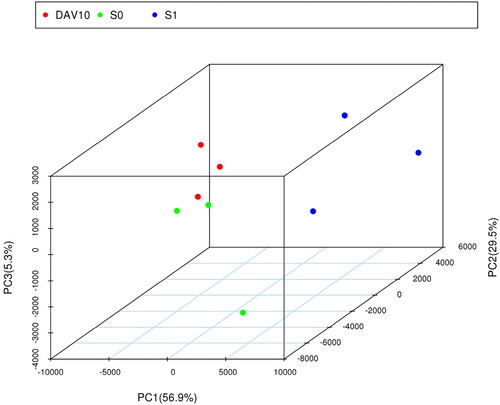
Principal component analysis of samples
Principal component analysis (PCA) was performed on nine samples of shoot apexes at different developmental stages, and the contribution rate of the first principal component (PC1) was 56.9%, compared with 29.5% for PC2 and 5.3% for PC3 (). For PC1, 2 and 3, samples with the same colour were clustered together, and samples with different colours were separate, indicating that the repeatability within the group was good, and there were differences between the groups, hence subsequent gene mining could be carried out.
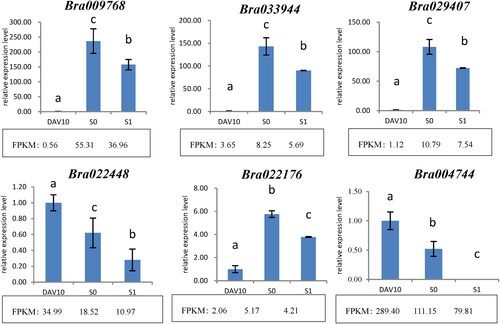
Verification of transcriptome results by RT-qPCR
In order to verify the reliability of transcriptome data, six genes were randomly selected for RT-qPCR verification, and the results were consistent with the sequencing results, confirming that the transcriptome data were accurate and reliable ().
FPKM, fragments per kilobase of transcript per million mapped reads. Different letters indicate a significant difference (p < 0.05).
Expression analysis of genes encoding cytokinin metabolism-related enzymes
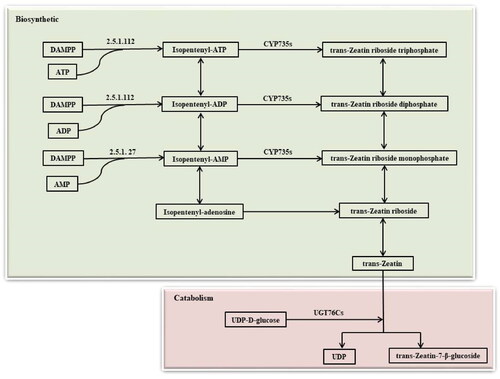
The major sites of cytokinin synthesis in cells are the plastid and the cytoplasmic matrix [Citation25], and tZ is the most important cytokinin [Citation19]. There are two pathways: de novo synthesis and tRNA hydrolysis. De novo synthesis mainly results from the deoxy-xylulose phosphate pathway [Citation26]. This is the main pathway for trans-zeatin synthesis, which is carried out in the plastid [Citation27]. The first step is catalysed by IPT (EC 2.5.1.122; EC 2.5.1.27) that utilizes ATP, ADP and AMP as dimethylallyl diphosphate (DAMPP) receptors to form isopentenyl ATP, isopentenyl ADP and isopentenyl AMP, respectively [Citation28]. Under the action of CYP735As, trans-zeatin riboside triphosphate, trans-zeatin riboside diphosphate and trans-zeatin riboside monophosphate are formed, and tZ is finally formed [Citation14]. tZ is degraded into trans-zeatin 7-β-d-glucoside and UDP by UGT76Cs involving UDP-d-glucose [Citation29], and degradation reactions are irreversible ().
In order to further analyse the reasons for the changes in cytokinin content in different developmental stages of pak choi, genes encoding key enzymes involved in cytokinin metabolic processes were statistically analysed (). The results revealed 13 genes that encode IPT enzymes and CYP735As enzymes, which are involved in cytokinin biosynthesis. Three genes encode UGT76Cs enzymes, which are involved in cytokinin catabolism.
Table 3. Genes and enzymes related to cytokinin metabolism.
The results of transcriptome sequencing were used to analyse the expression changes of genes encoding enzymes related to cytokinin metabolism. Bra028182 and Bra034022 encoding CYP735A2 were upregulated in DAV10 vs. S0 and downregulated in S0 vs. S1; Bra009143 encoding UGT76C1 and Bra005869 encoding UGT76C4 in catabolism were downregulated in DAV10 vs. S0 and S0 vs. S1. The results of hormone content analysis showed that the cytokinin levels in S0 were higher than in DAV10 and S1, and the changes in gene expression were consistent with the changes in hormone content, indicating that these nine genes may be related to the rate and extent of cytokinin synthesis.
Identification of cytokinin metabolism-related genes
According to FDR < 0.01 and Fold Change (FC) ≥ 2, DAV10 vs. S0, S0 vs. S1 were compared and screened for DEGs (). Based on the GO functional annotation of DEGs, seven genes (Bra023701, Bra002204, Bra014968, Bra028182, Bra034021, Bra009143 and Bra005869) and three genes (Bra002204, Bra014968 and Bra009142) related to cytokinin metabolism were found in DAV10 vs. S0 and S0 vs. S1, respectively. Among them, the expression levels of six DEGs (Bra023701, Bra002204, Bra014968, Bra028182, Bra005869 and Bra009143) were consistent with the changing trend of cytokinin content.
Table 4. Analysis of DEGs related to cytokinin metabolism in different developmental stages of pak choi.
Bra023701, Bra002204 and Bra014968 encode the IPT enzyme, which catalyses the first and rate-limiting step in cytokinin biosynthesis, using ATP, ADP and AMP as DAMPP receptors to ultimately form isopentenyl adenine (iP) and tZ ( and ). Bra028182 and Bra034022 encode the enzyme CYP735As, which utilizes isopentenyl adenine nucleotides to generate tZ nucleotides, In DAV10 vs. S0, the expression of the above genes was increased; we thought that this is beneficial to the synthesis of cytokinin, which promotes the flowering of pak choi. Upregulation of CYP735A1 in pak choi can increase the content of cytokinin, but the expression of Bra034021, which encodes CYP735A2, was downregulated ( and ). The reasons for this need to be further analysed. Bra005869 and Bra009143 are involved in the degradation of cytokinins and were downregulated in DAV10 vs. S0 ( and ). Their homologous genes in Arabidopsis are UGT76C4 and UGT76C1, and downregulation of these two genes at this stage reduces cytokinin decomposition, which promotes flower bud differentiation of pak choi.
Figure 5. Differentially expressed genes closely related to cytokinin metabolism at different developmental stages of pak choi. DAV10, 10 days after transplanting; S0, stage immediately prior to flower bud differentiation; S1, flower bud differentiation stage 1; green, blue and red indicate gene expression from low to high. Genes in purple font are closely related to cytokinin content.
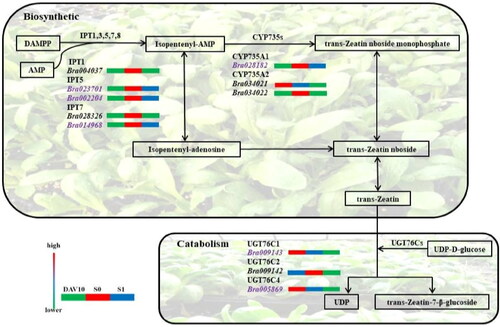
In S0 vs. S1 cytokinin anabolism, Bra002204 encoding IPT5 and Bra014968 encoding IPT7 were downregulated, and the transcript levels of AtIPT1, AtIPT5 and AtIPT7 are negatively regulated by CTK in Arabidopsis [Citation38]. Thus, higher cytokinin content inhibits IPT transcription to reduce cytokinin content, consistent with our results. In cytokinin catabolism, Bra009142, which encodes UGT76C2, was downregulated, which may be related to the initiation of flower bud differentiation.
Discussion
The relationship between cytokinins and flower bud differentiation and development
Cytokinins play an important role in flower bud differentiation for plant flower formation. Previous studies have shown that bud activation and growth depend on local cytokinin synthesis in axillary buds or stems [Citation30], and cytokinins move toward the shoot end to promote Arabidopsis bud growth [Citation31]. During lychee flower bud differentiation, cytokinin activity in flower buds increases [Citation32]. In rapeseed, cytokinins can regulate stem and shoot development [Citation33], increase in cytokinin content is linked with the start of flower bud differentiation [Citation34]. Our results were consistent with theirs, suggesting that increased cytokinin content can promote flower bud differentiation.
Expression of some important genes involved in cytokinin metabolism
In order to further elucidate the reasons for the change of cytokinin content in different stages of flower bud differentiation in pak choi, the metabolic pathway of cytokinin was analysed. The results of RNA sequencing revealed that the expression levels of nine genes correlated with changes in cytokinin content. Among them, the expression of six genes reached a significant level of difference, including Bra004037 encoding IPT1, Bra002204 encoding IPT5, Bra014968 encoding IPT7, Bra028182 encoding CYP735A1, Bra009143 encoding UGT76C1 and Bra005869 of UGT76C4, which we believe is a DEG closely related to cytokinin metabolism. Studies have shown that overexpression of the IPT gene can make plants grow faster, have longer flowering periods [Citation35], increase flowers [Citation36] and increase the number of seeds [Citation37]. In Arabidopsis, the transcript levels of AtIPT1, AtIPT5 and AtIPT7 are negatively regulated by CTK [Citation38], suggesting that higher cytokinin content inhibits IPT transcription to reduce cytokinin content, which is also confirmed by our results. CYP735As utilize isopentenyl adenine nucleotides to generate trans-zeatin nucleotides [Citation14] and promote the transcriptional levels of trans-zeatin, which are lower during dormancy and increase during flowering [Citation39]. Our results verified this, which indicates that the flower bud differentiation of pak choi requires a higher content of cytokinins. UGT76C4 and UGT76C1 can catalyse the glycosylation at the 7-, 9-position of the free-state cytokinin ring to generate N-glycosides [Citation29], inactivating cytokinin to reduce cytokinin content, and in our results, the expression of UGT76Cs decreased, especially the expression change of UGT76C1 reached a significant level, which would reduce the decomposition of cytokinin, and then promote the flower bud differentiation of pak choi. However, further studies are needed to confirm the targeting effect of genes in cytokinin metabolism on flower bud differentiation of pak choi.
Conclusions
In this study, the cytokinin contents in the shoot apexes in pak choi at different developmental stages were measured. The results showed that low temperature treatment increased the content of cytokinin, and higher cytokinin levels could promote flower bud differentiation of pak choi. To elucidate the molecular mechanism of changes in cytokinin content, the expression levels of the genes encoding cytokinin metabolic enzymes were analysed based on RNA sequencing results. The results showed that the expression patterns of most genes were consistent with the changes in cytokinin content, especially Bra004037, Bra023701, Bra002204, Bra014968 and Bra028326 encoding IPT enzymes, Bra028182 and Bra034022 encoding CYP735As enzymes, Bra009143 and Bra005869 encoding UGT76Cs enzymes. By comparing the content of tZ and expression of DEGs between different developmental stages in pak choi, six genes (Bra023701, Bra002204, Bra014968, Bra028182, Bra005869 and Bra009143) whose expression levels were consistent with changes in cytokinin content were identified. Among them, Bra023701, Bra002204, Bra014968 and Bra028182 involved in cytokinin synthesis were upregulated in DAV10 vs. S0 and downregulated in S0 vs. S1, while Bra005869 and Bra009143 involved in cytokinin breakdown, were downregulated in DAV10 vs. S0. These result in accelerating synthesis and slowing down degradation of cytokinin at a specific stage. Because the trends in expression changes were consistent with the cytokinin content in different developmental stages, six genes were predicted as most closely related to the flowering process of pak choi. The findings can help us to understand the molecular mechanism underlying cytokinin regulation of flower bud differentiation in pak choi.
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/sra/PRJNA821155
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Liu RY, Hou LP, Wang L, et al. Effect analysis of low temperature promoting flowering in chinese cabbage. Acta Agricul Boreali-Sin. 2009;24(06):193–197.
- Davies P, Krikorian AD. Plant hormones: physiology, biochemistry and molecular biology. Sci Hortic. 1996;66(3):267–270.
- Hirose N, Takei K, Kuroha T, et al. Regulation of cytokinin biosynthesis, compartmentaliz-ation and translocation. J Exp Bot. 2008;59(1):75–83.
- Cao XW, Cui HM, Yao Y, et al. Effects of endogenous hormones on variation of shoot branching in a variety of non-heading Chinese cabbage and related gene expression. J Plant Biol. 2017;60(4):343–351.
- Lan M, Li G, Hu J, et al. iTRAQ-based quantitative analysis reveals proteomic changes in Chinese cabbage (brassica rapa L.) in response to plasmodiophora brassicae infection. Sci Rep. 2019;9(1):12058–12058.
- Corbesier L, Prinsen E, Jacqmard A, et al. Cytokinin levels in leaves, leaf exudate and shoot apical meristem of Arabidopsis thaliana during floral transition. J Exp Bot. 2003;54(392):2511–2517.
- Besnard WC. Effectiveness of gibberellins and 6-benzyladenine on flowering of Arabidopsis thaliana. Physiol Plant. 1981;53(3):205–212.
- Michniewicz M, Kamieńska A. Flower formation induced by kinetin and vitamin E treatment in long-day plant (Arabidopsis thaliana) grown in short day. Naturwissenschaften. 1965;52(22):623–623.
- Nibau C, Stilio V, Wu HM, et al. Arabidopsis and tobacco SUPERMAN regulate hormone signalling and mediate cell proliferation and differentiation. J Exp Bot. 2011;62(3):949–961.
- Zuñiga-Mayo VM, Baños-Bayardo CR, Díaz-Ramírez D, et al. Conserved and novel responses to cytokinin treatments during flower and fruit development in brassica napus and Arabidopsis thaliana. Sci Rep. 2018;8(1):6836. May 1
- Kamaliah T, Rashid AA, Aziz MA, et al. Effects of cytokinin on multiple shoot formation from shoot tips of cabbage (Brassica oleracea subsp. capitata cv. tropicana). 2001.
- Tarkowská D, Filek M, Krekule J, et al. The dynamics of cytokinin changes after grafting of vegetative apices on flowering rapeseed plants. Plants (Basel, Switzerland). 2019;8(4):78.
- Roeckel P, Oancia T, Drevet J. Effects of seed-specific expression of a cytokinin biosynthetic gene on canola and tobacco phenotypes. Transgenic Res. 1997;6(2):133–141.
- Takei K, Yamaya T, Sakakibara H. Arabidopsis CYP735A1 and CYP735A2 encode cytokinin hydroxylases that catalyze the biosynthesis of trans-zeatin. J Biol Chem. 2004;279(40):41866–41872.
- Wang J, Ma X-M, Kojima M, et al. N-Glucosyltransferase UGT76C2 is involved in cytokinin homeostasis and cytokinin response in Arabidopsis thaliana. Plant Cell Physiol. 2011;52(12):2200–2213.,.
- Li F, Huang H, Ding X, et al. Effect of CPPU on postharvest attributes of Chinese flowering cabbage during storage. Postharvest Biol Technol. 2021;174(15):111438.
- Meng C, Gu AX, Zhao JX, et al. Analysis of gene expression and changes of endogenous hormone levels in ABCDE model of missing Chinese cabbage. Hortic Plant J. 2017;3(4):133–140. 000
- Zhu ZH, Sami A, Xu QQ, et al. Effects of seed priming treatments on the germination and development of two rapeseed (brassica napus L.) varieties under the co-influence of low temperature and drought. PLoS One. 2021;16(9):e0257236.
- Schmitz RY, Skoog F, Playtis AJ, et al. Cytokinins: synthesis and biological activity of geometric and position isomers of zeatin. Plant Physiol. 1972;50(6):702–705.
- Song HX, Fu C, Hou LP, et al. Screening of paraffin section staining methods and morphological identification of flower bud differentiation of pak choi. Jiangsu Agric Sci. 2018;46(2):88–91.
- Deng A, Tan W, He S, et al. Monoclonal antibody-based enzyme linked immunosorbent assay for the analysis of Jasmonates in plants. J Integr Plant Biol. 2008;50(8):1046–1052.
- Sun M, Qi X, Hou L, et al. Gene expression analysis of pak choi in response to vernalization. PLoS One. 2015;10(10):e0141446.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta CT) method. Methods. 2001;25(4):402–408.
- Feng XJ, Ma JJ, Liu ZQ, et al. Analysis of glucosinolate content and metabolism related genes in different parts of Chinese flowering cabbage. Front Plant Sci. 2022;12:767898.
- Kasahara H, Takei K, Ueda N, et al. Distinct iso-prenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. J Biol Chem. 2004;279(14):14049–14054.
- Hecht S, Eisenreich W, Adam P, et al. Studies on the nonmevalonate pathway to terpenes: the role of the GcpE (IspG) protein. Proc Natl Acad Sci U S A. 2001;98(26):14837–14842.
- Rohmer M, Knani M, Simonin P, et al. Isoprenoid biosynthesis in bacteria: a novel pathway for the early steps leading to isopentenyl diphosphate. Biochem J. 1993;295(2)::517–524. 1)
- Sakakibara H, Kasahara H, Ueda N, et al. Agrobacterium tumefaciens increases cytokinin production in plastids by modifying the biosynthetic pathway in the host plant. Proc Natl Acad Sci U S A. 2005;102(28):9972–9977.
- Hou B, Lim EK, Higgins GS, et al. N-glucosylation of cytokinins by glycosyltransferases of Arabidopsis thaliana. J Biol Chem. 2004;279(46):47822–47832.
- Li GF, Tan M, Cheng F, et al. Molecular role of cytokinin in bud activation and outgrowth in apple branching based on transcriptomic analysis. Plant Mol Biol. 2018;98(3):261–274.
- Ongaro V, Leyser O. Hormonal control of shoot branching. J Exp Bot. 2008;59(1):67–74.
- Chen WS. Changes in cytokinins before and during early flower bud differentiation in lychee (Litchi chinensis sonn.). Plant Physiol. 1991;96(4):1203–1206.
- Zhao W, Chao H, Zhang L, et al. Integration of QTL mapping and gene fishing techniques to dissect the multi-main stem trait in rapeseed (Brassica napus L.). Front Plant Sci. 2019;10(10):1152.
- Bouille P, Sotta B, Miginiac E, et al. Hormones and pod development in oilseed rape (Brassica napus). Plant Physiol. 1989;90(3):876–880.
- Guo JC, Duan RJ, Hu XW, et al. Isopentenyl transferase gene (ipt) downstream transcriptionally fused with gene expression improves the growth of transgenic plants. Transgenic Res. 2010;19(2):197–209.
- Khodakovskaya M, Zhao D, Smith W, et al. Expression of ipt gene controlled by an ethylene and auxin responsive fragment of the LEACO1 promoter increases flower number in transgenic Nicotiana tabacum. Plant Cell Rep. 2006;25(11):1181–1192.
- Ashikari M, Sakakibara H, Lin S, et al. Cytokinin oxidase regulates rice grain production. Science. 2005;309(5735):741–745.
- Miyawaki K, Miho MK, Kakimoto T. Expression of cytokinin biosynthetic isopentenyltransferase genes in Arabidopsis: tissue specificity and regulation by auxin, cytokinin, and nitrate. Plant J. 2004;37(1):128–138.
- Ito A, Tuan PA, Saito T, et al. Changes in phytohormone content and associated gene expression throughout the stages of pear (Pyrus pyrifolia nakai) dormancy. Tree Physiol. 2021;41(4):529–543.