ABSTRACT
Introduction: Prostate cancer (PCa) is one of the most common malignancies in men and a major cause of cancer deaths among men worldwide. Prostate specific antigen (PSA) monitoring and histopathological examination of tumor biopsies remain gold standards in PCa diagnostics. These clinical parameters are not well suited for patient stratification, predicting and monitoring treatment response. On the other hand, liquid biopsies offer a unique opportunity to easily isolate tumor-derived material for longitudinal clinical assessment.
Areas covered: In this review we focus on the clinical application of novel liquid biomarkers that have the potential to monitor and stratify patients in order to achieve better therapeutic effects and improve clinical outcomes. Enumeration and characterization of circulating tumor cells (CTCs), tumor-educated platelets, exosomes, and cell-free nucleic acids have been studied for their clinical utility in PCa diagnostics, prognostics, monitoring treatment response and guiding treatment choice.
Expert opinion: Liquid biomarkers have high potential to be used for prognosis, monitoring treatment response and guiding treatment selection. Although there is a remarkable progress in PCa biomarker discovery, their clinical validation is very limited. Research should be focused on biomarker validation and the incorporation of these biomarkers in clinical practice.
1. Introduction
Prostate cancer (PCa) is the most common malignancy in men in Western countries, and a leading cause of male cancer-related deaths. Although serum prostate-specific antigen (sPSA) is widely used for diagnostics, prognostics, and therapeutic monitoring purposes, its limitations are well known. The specificity of sPSA is poor and its use can lead to overdiagnosis and overtreatment. Tumor biopsies therefore remain the gold standard for cancer diagnosis. However, taking biopsies from the primary tumor is an invasive procedure with complications such as bleeding, urinary retention, infection and sepsis. Furthermore, PCa predominantly metastasizes to the bones and bone biopsies are hard to perform and often limited by the low yield of tumor tissue. For metastasized PCa that progressed under androgen deprivation therapy (ADT), termed metastatic castration-resistant PCa (mCRPC), many treatment options are available nowadays, including next generation androgen receptor targeting agents (ARTAs) (e.g. abiraterone and enzalutamide), taxane-based chemotherapy (docetaxel and cabazitaxel) and other agents such as radium-223 and sipuleucel-T [Citation1–Citation8]. De novo resistance to the ARTAs is observed in almost a quarter of the mCRPC patients. Moreover, cross-resistance is common for these drugs. However, and unfortunately, sPSA is not an adequate marker for evaluation of treatment response and biomarkers predicting treatment resistance are needed.
In addition to traditional PCa therapies, the use of poly ADP-ribose polymerase (PARP) and AKT pathway inhibitors for patients with DNA-repair gene mutations and phosphatase and tensin homolog (PTEN) loss, respectively, are promising [Citation9,Citation10]. For these new types of therapy (e.g. PARP-inhibitors), innovative ways to select in advance those patients that have gene aberrations and are therefore most likely to benefit most from the therapy are required. Due to genomic evolution, a tissue biopsy taken from a single metastatic site may underestimate the heterogeneous character of the tumor. Therefore, less invasive biomarkers that are able to reflect tumor heterogeneity in one sample are required for guiding treatment selection.
Thus, there is a clinical unmet need for predictive biomarkers to help steer PCa treatment strategies. Biomarker tests should be none or minimally invasive. Liquid biopsy serve as a valuable source of biomarkers (hereinafter referred to as liquid biomarkers) and are being extensively investigated for clinical use. Liquid biopsy approaches can use blood, urine, stool, liquor, saliva, pleural fluid, ascites and seminal fluid as potential sources of tumor-derived material. Liquid biomarkers can be obtained using non- or minimally invasive procedures and therefore also allow disease and treatment response monitoring longitudinally (i.e. at several time points). Furthermore, liquid biomarkers may provide insights into individual tumor drivers, that potentially can be used to guide personalized medicine. Biomarkers are shed into the bloodstream or urine by the tumor or tumor metastasis and include circulating tumor cells (CTC), tumor-educated platelets (containing tumor RNA), exosomes (containing e.g. tumor-derived RNAs) and cell-free nucleic acids (incl. DNA, messenger RNA (mRNA), microRNA (miRNA) and long non-coding RNA (lncRNA)). In this review, we provide an overview of the most promising biomarker tests in blood and urine for PCa, and focus on those that are (almost) being used in the clinic.
2. Urinary liquid biomarkers
Urine is a promising source for PCa biomarkers. The prostatic fluid may contain products derived from prostate tumor cells that can be excreted via urine, which can be obtained non-invasively. Digital rectal examination (DRE) can enhance excretion of prostatic fluid. Until now, urine-derived biomarkers have proven their clinical use as diagnostic and prognostic tools. Four urinary liquid biomarker tests are commercially and clinically available: Progensa PCA3, SelectMDx, Michigan Prostate Score (MiPS) and ExoDx Prostate (IntelliScore). Studies have shown that those urine biomarkers outperform PSA as a diagnostic and, more importantly, prognostic biomarker. All tests have a high negative predictive value (NPV) and can be used to prevent unnecessary biopsies (). The SelectMDx, MiPS and IntelliScore tests have a high sensitivity and NPV for high grade (Gleason score ≥7) PCa, and hence the most useful tests for clinical application. The clinical utility and comparison of the four urinary liquid biomarkers recently have been reviewed in detail by Hendriks and colleagues [Citation11]. For clinically advanced patients these tests have limited value, since they often lack the primary tumor.
Table 1. Clinically (commercially) available urinary liquid biomarker tests for PCa.
3. Biomarkers in blood
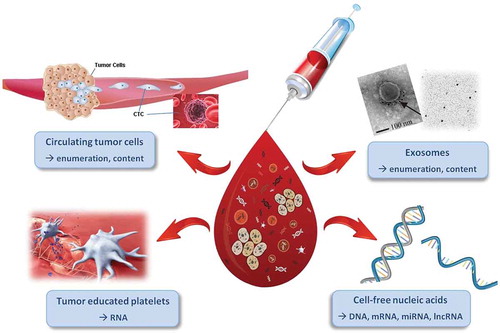
Biomarkers in blood are rapidly evolving and represent a valuable tool for diagnostic, prognostic and predictive purposes in several cancers. Furthermore, liquid biomarkers can be used to evaluate treatment response and guide precision medicine. Circulating tumor cells (CTC), tumor-educated platelets (containing tumor RNA), exosomes (containing e.g. non-coding RNAs) and cell-free nucleic acids (incl. DNA, mRNA, miRNA and lncRNA) are released from tumors into the circulation (see ). These cells and nucleic acids are promising biomarkers, some of which are now close to clinical implementation. Certainly, proteins and metabolites are also potentially very suitable biomarkers for prostate cancer, but the protein markers identified so far are still quite far away from clinical implementation [Citation21]. Here, we outline different blood-based biomarkers and describe their clinical relevance and utility.
Figure 1. Liquid biopsies contain a variety of clinically informative components. Blood samples can be analyzed for circulating tumor cells (CTCs (§3.1), tumor educated platelets (§3.2), exosomes (§3.3) and cell-free nucleic acids (§3.4) with each of them providing valuable numerical and molecular information. Adapted with permission from: Mader & Pantel. Oncol Res Treat 2017; 40: 404–8. S. Karger AG, Basel.
3.1. Circulating tumor cells
Circulating tumor cells form new metastatic sites by gaining plasticity and motility through, for example, epithelial-mesenchymal transition (EMT). In turn, CTCs are able to migrate, intravasate and extravasate from the bloodstream into tissues. The presence of CTCs can be a surrogate for disease stage and disease progression. CTCs can be identified for almost all solid malignancies, especially in advanced stages of the disease. However, isolation of CTCs is very challenging due to their low abundance and the presence of vast amounts of other cells in blood like erythrocytes and leukocytes. Several techniques are being developed for CTC enrichment and detection.
3.1.1. CTC enumeration
The enumeration of CTCs has emerged as a powerful biomarker for the assessment of prognosis and evaluation of treatment response. A decrease in sPSA can be seen in patients using next generation ARTAs without necessarily inducing apoptosis of the tumor cells. Therefore, the value of using sPSA as a biomarker for treatment response in patients using androgen inhibitors is limited. In contrast to sPSA, CTC counts are not directly modulated by androgens and therefore a much better predictive biomarker for treatment response. At this moment, CellSearch® is the only FDA approved system for CTC enumeration. The CellSearch® system enables standardized enumeration of CTCs based on immunomagnetic enrichment of epithelial cell adhesion molecule (EpCAM)-positive tumor cells. The Prostate Cancer Working Group 3 (PCWG3) has incorporated CTC enumeration by the CellSearch® system as a clinical endpoint in trials for treatment response [Citation22].
It has been shown that CTC enumeration can be used to assess treatment efficacy in mCRPC patients [Citation23,Citation24]. In a phase II trial with abiraterone acetate in mCRPC patients, CTC counts combined with lactate dehydrogenase (LDH) level was shown to be a marker for survival at the individual-patient level [Citation24]. Heller et al. explored CTC enumeration compared with PSA as a response measure of prolonged survival for mCRPC patients across five randomized phase III trials. A drop to zero in CTC count and conversion (≥ 5 CTCs at baseline followed by ≤ 4 at 13 weeks) had the highest discriminatory power for overall survival (OS) [Citation23]. The use of CTC counts to reflect treatment response has been investigated as an endpoint in clinical trials. However, it is important to highlight that in many patients less than 5 CTCs are detected. In that case, monitoring of CTCs can only inform on disease progression. Despite promising results in CTC enumeration trials, the clinical benefit of changing treatment based on unfavorable CTC counts has not been studied yet. CTC counts can also be used as a prognostic biomarker. The prognostic value of CTC enumeration using CellSearch® in PCa, especially in mCRPC, has been established in clinical trials and multiple large phase II and phase III trials [Citation25–Citation28]. A baseline level of ≥5 CTCs detected per 7.5mL blood is associated with significantly shorter, ~50% reduced OS [Citation25–Citation28].
Although the prognostic value of CTCs has been established in mCRPC, for other stages of PCa there is less evidence. Nonetheless, increasing CTC counts are indicative of shorter OS and time to progression in metastatic hormone-sensitive prostate cancer (mHSPC) [Citation29]. Furthermore, CTC counts are predictive for treatment response in mHSPC [Citation30,Citation31]. However, CTC counts are typically low in the early disease setting, resulting in limited patient inclusion in early disease CTC studies. Finally, to form distant metastases, tumor cells must gain plasticity and motility. This process involves EMT during which EpCAM may be down regulated, and hence CellSearch will underestimate the number of CTCs. CTC enrichment techniques targeting a wider scope of non-EpCAM CTC phenotypes (e.g. mesenchymal) are currently being developed. Unfortunately, CTC samples usually have to be processed within a short period of time, which limits applicability in the routine clinical setting. As a result, and in combination with the relatively high costs for sample collection and processing, CTC enumeration has not yet become standard care.
3.1.2. CTC PHENOTYPING: AR-v7 status
Besides the enumeration of CTCs also (single cell) molecular characterization of CTCs has been proposed to yield valuable predictive biomarkers. PCa is androgen driven and therefore androgen receptor (AR) signaling plays an important role. Elevated AR expression, enhanced AR activity and development of AR mutations are frequently observed in CRPC. These findings have given the impetus to the development of next generation ARTAs (e.g. abiraterone, enzalutamide, apalutamide, darolutamide). Since their registration, patient outcomes have significantly improved. Nonetheless, primary and acquired resistance to ARTAs is common. This might be due to the expression of AR splice variants in CRPC cells. AR splice variants lack the ligand-binding C-terminal domain which can lead to constitutive ligand-independent activation of the AR. Splice variant AR-v7 is regarded as the most common and most biologically and clinically relevant splice variant [Citation32].
In 2014, Antonarakis et al. showed that detection of AR-v7 mRNA in CTCs is related to lower PSA response rates and shorter progression-free survival (PFS) in mCRPC patients treated with enzalutamide or abiraterone [Citation33]. These findings were striking, as all patients with detectable AR-v7 did not show PSA responses. Additional studies have associated AR-v7 with resistance to abiraterone and enzalutamide in mCRPC patients, while AR-v7 positive patients are likely still sensitive to taxane-based chemotherapy [Citation34–Citation36]. Furthermore, combining CTC and AR-v7 status provides strong prognostic information. CTC-negative patients have the most favorable outcome, followed by CTC-positive/AR-v7-negative patients and CTC-positive/AR-v7-positive patients have the poorest prognosis [Citation37]. Thus, CTC AR-v7-status has the potential to serve as a predictive biomarker for abiraterone and enzalutamide resistance.
Bernemann et al. reported a study that not fully supported the use of AR-v7 as a predictive biomarker for enzalutamide and abiraterone response, as AR-v7 positive patients responded to enzalutamide or abiraterone therapy [Citation38]. The first study of Antonarakis et al. showing AR-v7 positive patients as non-responders to abiraterone or enzalutamide had almost all received one of these drugs before [Citation33]. AR-v7 status is likely most relevant for second-line next generation ARTAs, since AR-v7 levels might increase under anti-androgen treatment. Moreover, AR-v7 negative men may also display resistance to next generation ARTAs. Other resistance mechanisms such as alternative AR splice variants, AR mutations, AR amplifications, AR copy number gain or androgen overproduction may underlie this problem. Non-AR-related escape mechanisms might also contribute to AR upregulation by cross-talk between growth factor signaling pathways [Citation39,Citation40].
Beside AR-v7 detection in CTCs, AR-v7 can also be detected in exosomes or even whole blood. Del Re et al. extracted RNA from plasma-derived exosomes of CRPC patients and determined the presence of AR-v7 by digital droplet PCR. AR-v7 positive status was associated with a significantly shorter OS [Citation41]. Detection of AR-v7 mRNA in whole blood revealed similar results [Citation42–Citation44]. There are two CLIA-certified LDT tests for detecting the splice variant AR-v7; the AdnaTest and Oncotype DX (Epic Sciences platform). The AdnaTest allows analysis of gene expression in immunomagnetically enriched CTCs by RT-PCR. The Epic Sciences platform identifies AR-v7 protein in the nucleus of CTCs. Remarkably, the AdnaTest was already used in clinical practice while prospective clinical trial were still recruiting [Citation45]. Recently, the PROPHECY study validated the detection of AR-v7 in CTCs by AdnaTest and Epic Sciences platform in a prospective multicenter study. This study found AR-v7 detection in CTCs to be independently associated with shorter PFS and OS in men with mCRPC treated with abiraterone and enzalutamide regardless of line of treatment, and concluded that these patients should be offered alternative treatments [Citation46]. Although relevant, this study has some important limitations. First, the number of AR-v7 positive patients was relatively small and no alternative treatments were tested. Second, there were discrepancies in AR-v7 positive results between both tests. Eleven patients tested positive for AR-v7 with the Epic Sciences platform, while 28 patients were AR-v7 positive tested using the AdnaTest.
Taken together, AR-v7 detection is a promising predictive biomarker. Detection of AR-v7 in CTCs might be predictive for response to enzalutamide and abiraterone treatment, and it seems that for these patients taxane treatment should be preferred. However, large prospective clinical trials are needed to validate the AdnaTest and Epic Sciences platform to confirm their clinical utility for mCRPC patients.
3.2. Tumor educated platelets
It has been demonstrated that platelets can store tumor-derived RNA molecules. Blood contains about 150,000 to 350,000 platelets per microliter, and hence platelets are a highly available source of tumor-derived biomarkers. Furthermore, blood platelet isolation is relatively simple and common practice is clinical laboratories. However, only one study has shown the potential of using tumor-educated platelets for biomarker purposes in PCa [Citation47]. In CRPC patients receiving docetaxel or abiraterone treatment, the detection of tumor-derived biomarkers (KLK3, FOLH1, NPY) in platelets was associated with resistance to abiraterone [Citation47]. Further studies are ongoing to establish the value of platelets as biomarkers for PCa.
3.3. Exosomes
Exosomes are small (30–150 nm) extracellular vesicles that are produced in the endosomal compartment of most eukaryotic cells. Exosomes are enriched with DNA, RNA or protein, and they are highly representative for their cell of origin. Exosomes are present in tissues and can also be found in biological fluids including blood and urine. By characterizing exosomes, insight in the tumor biology can be obtained. Liquid biopsies could provide a platform to screen for tumoral changes along treatment regimes. Isolating exosomes essentially enriches for tumor-derived material as hematopoietic cells are removed in this process. Finally, exosomes protect biomarkers from potential degradation by nucleases, proteases or other environmental stressors present in plasma and urine. However, the capture of tumor-derived exosomes is challenging, and a critical step to developing exosome-based diagnostic tests. Exosome biogenesis, isolation and characterization methods have been reviewed by Gurunathan et al. [Citation48], and will not be further discussed in this review.
3.4. Circulating nucleic acids
3.4.1. RNAs
3.4.1.1. Messenger RNAs
Messenger RNA (mRNA) encompasses a large family of RNA molecules which transfer genetic information from genes to ribosomes for translation into protein products. Dysregulation of oncogenic and tumor suppressive mRNAs is central to the progression of all cancers. Detecting cancer-specific mRNA transcripts in body fluids is a direct and relatively simple method to characterize tumors. The established mRNA markers KLK3 and TMPRSS2-ERG are important for many urine-based diagnostic tests (SelectMDx, MiPS, IntelliScore). Recent studies have explored the prognostic value of these markers in blood. In a study on post-chemotherapy treated CRPC patients, 37% of peripheral blood mononuclear cell (PBMC) fractions were positive for TMPRSS2-ERG [Citation49]. When treated with abiraterone, TMPRSS2-ERG was not predictive for response. Conversely, the TMPRSS2-ERG and KLK3 levels in chemo-naïve patients decreased along with docetaxel cycles [Citation50]. In an independent study, docetaxel treated mCRPC patients that were negative for TMPRSS2-ERG at baseline switched to a positive status at time of progression (41%) [Citation51]. These findings hint at a possible role of TMPRSS2-ERG in chemotherapy resistance. Large prospective studies are needed to validate these results prior to clinical implementation.
3.4.1.2. MicroRNAs
MicroRNAs (miRNAs) are short (~21 nucleotide) single-stranded RNAs that can control gene expression at the posttranscriptional level, by binding to corresponding binding sites in their mRNA targets [Citation52]. MiRNAs may be able to drive tumor initiation and progression by controlling expression of tumor suppressor genes and oncogenes. MiRNAs are found in blood or urine. In body fluids miRNAs are surprisingly stable, as they are protected from RNase degradation via incorporation in ribonucleoprotein complexes or by their presence in extracellular vesicles (e.g. exosomes) [Citation53,Citation54]. It has been shown that exosome isolation from liquid biopsies improves the sensitivity of miRNA amplification and detection [Citation53]. Cumulating data suggest that miRNAs have potential as diagnostic and/or prognostic biomarkers in PCa [Citation55–Citation59]. Several miRNAs have been studied in plasma and serum of mCRPC patients. Unfortunately, the available data is conflicting and there is limited consensus on the most valuable miRNA. So far, the mir-200-family (including miRNA-141-3p) and miR-375 seem to be most promising. In two clinical studies, levels of miR-21, miR-200a, miR-200c, and miR-375 were related to OS of CRPC patients [Citation60,Citation61]. In combination with clinical markers, these miRNAs have demonstrated diagnostic and prognostic potential for discrimination between high risk PCa, low risk PCa and healthy controls [Citation62,Citation63]. Despite the numerous miRNA studies that have been performed, at present no miRNA-based test is clinically available for PCa.
3.4.1.2.1. MiRNA-141-3p
Expression of miR-141-3p has studied extensively in PCa liquid biomarker research. The biological effects of miR-141-3p are context specific and may be oncogenic or tumor suppressive [Citation64]. Increased miR-141-3p levels in urine can distinguish benign from malignant PCa, while elevated levels in blood has been associated with disease progression and metastasis [Citation65–Citation68]. Exosomal miR-141-3p likely primes bone-tissue to produce a tumor-favorable microenvironment while its expression in CTCs induces MET and promotes PCa cells to settle new metastatic sites [Citation66,Citation67]. Two independent studies have demonstrated that increased miR-141-3p levels are predictive for metastatic PCa [Citation63,Citation69]. Monitoring plasma miR-141-3p levels in PCa patients could be predictive for metastatic risk.
3.4.1.2.2. MiRNA-375
MiRNA-375 (miR-375) was first described as a tumor suppressor, but recent evidence suggests its biological effects are context and tissue specific [Citation69]. When measured in serum, miR-375 levels were found to be significantly increased in PCa when compared to healthy controls [Citation68,Citation70]. Moreover, expression levels of miR-375 have been correlated with PCa progression, metastasis, and docetaxel resistance in CRPC [Citation62,Citation65,Citation71]. Like miR-141-3p, miR-375 also has been shown to induce MET in CTCs to promote formation of distant metastasis [Citation72]. These findings are in line with the findings of Bryant et al., who showed that serum-derived exosomes from metastatic PCa patients (n = 47) contained significantly higher miR-375 copy numbers than exosomes from a control cohort (n = 72) [Citation73]. In summary, increased plasma/serum miR-375 levels could be predictive for the presence of metastasis and chemotherapy resistance in PCa.
3.4.1.3. Long non-coding RNAs
Long non-coding RNAs (lncRNAs) are RNA transcripts that do not code for proteins and are longer than 200 nucleotides. Current estimates suggest there to be more than 60,000 lncRNA genes in humans. Aberrant expression of many lncRNAs has been associated with human disease including PCa. However, the only lncRNA-based diagnostic test that is clinically available for PCa is the Progensa PCA3 urine test (see ). Nonetheless, more than a dozen other PCa-related lncRNAs have been detected in tissue, blood or urine [Citation74,Citation75].
3.4.1.3.1. SCHLAP1
In a multicenter study, SWI/SNF Complex Antagonist Associated With Prostate Cancer 1 (SCHLAP1) lncRNA expression levels in primary PCa tissues was highly correlated with metastasis and biochemical recurrence [Citation76]. Upon measuring expression in post-DRE urine sediments, SCHLAP1 could only be detected in a subset of patients [Citation76]. This is likely due to the patient cohort in which it was investigated. High stage (T2 and higher) patients are more likely to be detected using SCHLAP1 as a biomarker, and hence SCHLAP1 testing could be promising for monitoring metastatic risk. The presence of SCHLAP1 in blood samples has not yet been confirmed.
3.4.1.3.2. MALAT1
Metastasis Associated Lung Adenocarcinoma Transcript 1 (MALAT1) is a lncRNA that is overexpressed in 82.5% of PCa tissue specimens compared with normal adjacent tissue [Citation77]. MALAT1 was found to be stable in plasma after 24h storage at room temperature, repeated freeze-thaw cycles, and acid-base treatment. Plasma MALAT1 levels were measured in 87 PCa, 82 benign prostatic hyperplasia (BPH), and 23 healthy controls. Plasma MALAT1 levels were a better predictor of disease than sPSA. Diagnostic accuracy improved when both MALAT1 and sPSA were combined [Citation78]. Development of novel liquid biopsy-based tests should focus on treatment monitoring and patient stratification. In this light, MALAT1 has been shown to induce enzalutamide resistance by promoting AR-v7 production [Citation79]. Preclinical evidence suggests that MALAT1 may also contribute to docetaxel resistance via indirect upregulation of A-kinase anchor protein 12 (AKAP12) expression [Citation80]. MALAT1 in plasma is stable, measurable, outperforms sPSA, and may induce first-line mCRPC therapy resistance. Taken together, measuring MALAT1 in plasma could be a promising platform for treatment monitoring in mCRPC patients.
3.4.2. Cell-free DNA
Circulating cell-free tumor DNA (ctDNA) is emerging as a biomarker in PCa. Cell-free DNA (cfDNA) is shed into the bloodstream by cancer cells and nonmalignant cells trough apoptosis and necrosis of (tumor) cells and possibly also by active secretion [Citation81]. CfDNA is found as double-stranded fragments of approximately 150 to 200 base pairs in length [Citation82]. Molecules of cfDNA are rapidly cleared from the circulation, with a half-life ranging from minutes to several hours. Healthy individuals have cfDNA levels of 1–10 ng/ml which can increase by intense exercise, trauma, infections and inflammatory conditions [Citation83]. Due to the small size it is expected that cfDNA can also be analyzed in urine, but concentrations will be influenced by the glomerular filtration rate [Citation84]. One of the main challenges is the contamination of ctDNA with nonmalignant cfDNA. Furthermore, the yield of ctDNA is often low, especially in early disease. Therefore, ultrasensitive methods to detect aberrations in ctDNA are necessary. Another challenge is the low stability of cfDNA, which necessitates its collection in specialized collection tubes.
3.4.2.1. Quantification of cfDNA
Quantitative assessment of cfDNA can be used as a diagnostic biomarker and can assess tumor burden and response to therapy. High cfDNA concentrations prior to therapy may indicate aggressive disease. In two phase III clinical trials, a decline in cfDNA levels was positively correlated with both radiographic PFS and OS for mCRPC patients treated with taxane-based chemotherapy [Citation85]. Furthermore, a decline in total cfDNA was associated with treatment response [Citation85]. In a separate study, high cfDNA levels prior to chemotherapy was associated with poor sPSA response and could act as an independent predictor of shorter OS [Citation86]. Thus, quantification of cfDNA reflects disease burden, prognosis and treatment response.
3.4.2.2. DNA mutations
A number of studies have shown that sequencing ctDNA for genomic alterations provides important biomarkers for predicting therapy response and resistance. The genetic profile of ctDNA has shown a high degree of concordance with metastatic tissue biopsies in men with mCRPC [Citation87]. Due to the short half-life of ctDNA, accurate real-time tumor profiling is possible. Several pathways are involved in the development and progression of mCRPC, including androgen receptor aberrations (3.4.2.2.1), DNA repair gene aberrations (3.4.2.2.2), TP53 aberrations (3.4.2.2.3), and PTEN (3.4.2.2.4) loss. The genomic landscape of mCRPC evolves after treatment with systemic therapies [Citation88]. Therefore, detection of specific gene alterations in ctDNA prior to therapy could steer treatment strategies to target the underlying molecular pathology and improve treatment response.
3.4.2.2.1. AR gene aberrations
AR gene alterations are rarely detected before hormone therapy, but are present in up to 60% of patients with mCRPC [Citation89]. AR gene aberrations include AR gain, AR mutations and AR genomic structural variations. AR genomic structural variations (GSV) have not been studied widely, but are associated with expression of splice variants (see above). A correlation of AR GSV in ctDNA and splice variants in CTCs has been found [Citation61]. The presence of two AR point mutations (L702H and T878A) in ctDNA were identified as predictive for resistance to abiraterone treatment [Citation90–Citation94]. Resistance to enzalutamide has been associated with another AR point mutation: c.2629T>C (F877L), although this mutation is very rare [Citation95,Citation96]. Multiple studies have revealed that detection of AR gain in ctDNA is related to poor outcomes in CRPC patients treated with abiraterone or enzalutamide [Citation90,Citation91,Citation96–Citation99]. However, Annala et al. demonstrated that the detection of AR gain lost its prognostic effect after adjustment for routine prognostic clinical markers and ctDNA levels [Citation100]. Only high AR copy number gain (≥ 8 copies) was associated with poor responses in first-line mCRPC patients [Citation100]. It is reasonable to suggest that AR copy number gain evolves during treatment leading to cross-resistance between next generation ARTAs. Sumiyoshi et al. reported that AR aberrations were only associated with poor response to abiraterone, but not to enzalutamide [Citation101]. Recent data suggest that patients with AR gain should be treated with docetaxel [Citation102]. However, the number of patients in these studies is very limited and methods of detecting AR copy number gains differ. In conclusion, evaluation of AR status prior to treatment might be useful in predicting response to therapy and therefore guiding treatment choice. Analytical standardization and validation of methods is required before such tests can be used in clinical practice.
3.4.2.2.2. DNA damage repair gene aberrations
Almost a quarter of mCRPC tumors have mutations in the DNA repair genes including BRCA1 or BRCA2 and ATM [Citation103]. Defects in those genes are strongly associated with poor clinical response to next generation ARTAs. Alterations in homologous recombination repair genes BRCA2 or ATM were independently and significantly associated with shorter time to progression [Citation100]. Loss of function of homologous recombination repair genes (HRR) predicts response to poly ADP ribose polymerase (PARP) inhibitors and platinum-based chemotherapies [Citation9]. PARP inhibitors target mutated BRCA or DNA repair deficient cancers. The TOPARP trial showed a good response rate for the PARP inhibitor olaparib in mCRPC patients with DNA repair mutations BRCA1/2 or ATM [Citation9]. Goodall et al. demonstrated that all somatic (tissue) DNA repair mutations were detectable in ctDNA and even new mutations were detected at time of disease progression. The allele frequency of somatic mutations decreased selectively in responding patients [Citation104]. Moreover, another study identified BRCA2 reversion mutations in ctDNA at the time of tumor progression on olaparib or talazoparib [Citation105]. Thus, detection of DNA repair mutations in ctDNA can guide treatment choice and could monitor treatment response and resistance.
3.4.2.2.3. TP53 aberrations
Aberrations in tumor protein 53 (TP53) are associated with poor prognosis [Citation100,Citation106,Citation107]. TP53 is a tumor suppressor gene of which alterations are frequently observed, up to 50%, in mCRPC patients [Citation103]. Loss of TP53 can also be detected in ctDNA, and it has shown to be a predictor for worse response to next generation ARTAs [Citation100]. Interestingly, De Laere et al. recently showed that TP53 inactivation outperforms any AR-derived biomarker as negative prognosticator for next generation ARTAs [Citation108]. They collected CTCs and ctDNA of 168 mCRPC patients prior to next generation ARTAs for the detection of AR, AR splice variant and TP53 alterations. Stratification of mCRPC patients starting next generation ARTAs based on TP53 alterations revealed two groups: a poor prognosis (PFS median ≤ 2.5 months) and good prognosis (PFS median ≥ 14.0 months) group [Citation108]. These data suggest that TP53 inactivation is an important biomarker for predicting treatment response. However, at this stage patients harboring TP53 inactivation should not be withheld next generation ARTAs. Instead, these patients should be monitored closely and clinicians should be aware of possible treatment resistance and rapid disease progression.
3.4.2.2.4. Pi3k pathway defects
The phosphatidylinositol-3-kinase (PI3K) pathway is one of the most commonly activated signaling pathways in PCa. Approximately half of all mCRPC patients have aberrations in PIK3CA/B, PTEN or AKT [Citation103]. These aberrations can be well detected in ctDNA, and PI3K pathway defects have been associated with a poor response to ARTAs [Citation100]. Carver et al. showed that inhibition of the PI3K pathway can activate the AR pathway and vice-versa by reciprocal negative feedback [Citation109]. This suggests that tumor cells are able to adapt and survive when either single pathway is inhibited pharmacologically. De Bono et al. studied the effect of an AKT inhibitor ipatasertib combined with abiraterone in mCRPC patients with and without PTEN loss [Citation10,Citation110]. The combination therapy showed superior antitumor activity compared to abiraterone alone, especially in patients with PTEN loss [Citation10,Citation110]. In summary, detection of PIK3CA/B, PTEN or AKT alterations in ctDNA can be used as a predictive biomarker for response to anti-androgens and can guide treatment choice (e.g. AKT inhibitors in combination with next generation ARTAs).
4. Conclusions
Liquid biopsies can be obtained using minimally invasive techniques and have shown their potency as diagnostic, prognostic and predictive biomarker. Several urine biomarker tests are commercially and clinically available and are currently being used in many clinics worldwide. Urine biomarkers have shown their great potential for diagnostic, and more importantly prognostic purposes. For blood-based liquid biopsies only a few biomarker tests are commercially available (see ), and hence their clinical use is currently limited. However, the potential of blood-based liquid biomarkers is very high. Particularly CTCs and ctDNA have gained remarkable attention as predictive biomarkers for therapy response and treatment choice. A central problem for blood based biomarkers is the low abundance of cancer-derived material, especially in the early stage disease setting. Therefore, more sensitive measuring techniques or enrichment techniques are required. Most importantly, the clinical validation of liquid biomarkers is needed to assure their value in clinical practice.
Table 2. Clinically available blood based liquid biomarker tests for PCa.
5. Expert opinion
The area of biomarker research in cancer is developing quickly. As new biomarkers are being discovered, innovative platforms are being developed to detect them. Unfortunately, there is no consensus on the use of specific technologies, methods of normalization, sample collection and processing. This hampers comparison of different pre-clinical and clinical studies resulting in the failure of most novel biomarkers to be implemented clinically. To ensure promising biomarkers are tested in clinical trials and reach clinical practice, these obstacles need to be tackled. Aside from sPSA measurements, commercially-available liquid biopsy tests are not common in PCa clinical practice. What underlies this problem? At first, there are questions surrounding the validity, reproducibility and utility of many liquid biomarkers in clinical practice. Only a small number of liquid biopsy-based assays have been validated. Furthermore, the clinical trials used to test new biomarkers have known limitations due to selection bias and limited external validation. Even if a biomarker has shown its value in a large clinical trial, will it be implemented in routine practice? For example: can a clinician withhold a patient with a AR-v7 positive status next generation ARTA therapy? There will always be a significant chance that this specific patient does respond, albeit much less likely than AR-v7 negative patients. The impact on clinical decision making remains to be established, which brings up an ethical discussion. Translating results based on large clinical trials to a single patient is difficult. Secondly, only a small number of clinicians are involved in the field of liquid biopsy development. For the use of these biomarkers in routine patient care, knowledge is needed for deciding when to use and how to interpret the results. Finally, who pays for the implementation of novel liquid biomarker assays in routine patient care? Besides, the procedures for blood sampling, processing and storage can be complicated even when the analysis itself is outsourced.
In our opinion the most critical step before implementing novel liquid biomarkers in routine patient care is the validation of the assays. Quality management and reproducibility must be determined prior to clinical use. Next to small number of commercially available PCa diagnostic tests, there are only two FDA approved liquid biopsy tests, one in blood and one in urine (see and ). At present, there is a large gap in the PCa biomarker discovery, and prospective validation of these biomarkers in phase II and III clinical trials. Typically the aim of these phase II and III trials is to identify the largest possible patient cohort suited for newly discovered drugs. Conversely, the use of biomarkers aims to categorize patients, steer clinical decision making, and prevent overtreatment. This brings forth the question: will the infrastructure used for drug marketing be accessible for liquid biomarker validation?
Another gap we see in the biomarker research field is the lack of studies investigating the relation between the pharmacokinetics of a drug and the predictive biomarker for treatment response. This is called the pharmacokinetic/pharmacodynamic relationship. Based on the pharmacological mechanism of a drug, a certain drug exposure is needed for clinical effect. For many drugs, the maximum exposure-response is related to a trough level (Ctrough). Upregulation of specific targets of drugs, which can be detected as a biomarker, might influence the defined minimal plasma level. In almost all biomarker studies this has not been investigated.
Finally, much of the focus in biomarker research has been on the discovery of the next ‘golden egg’ biomarker. However, to address heterogeneous nature of metastatic PCa, biomarker panels in combination with clinical parameters is preferred. This will likely enhance the successful use of novel biomarkers in clinical practice. Nonetheless, we must emphasize the opportunities and promising outlook for clinical use of liquid biomarkers in PCa. The PCA3, SelectMDx, and CellSearch® platforms have demonstrated the potency to outperform sPSA in diagnostic and prognostic settings. Hence it is time to implement the next generation of liquid biomarkers in clinical practice.
In the past decade, many new treatment options have become available for advanced PCa. The heterogeneous nature of this disease has driven a push to develop personalized treatment strategies. In this light, the validation of actionable and predictive biomarkers for clinical utility is the key. Due to the overlap in actionable targets in many diseases, therapies used in the treatment of other cancers will become increasingly available to PCa patients (e.g. PARP inhibitors, immunotherapy/checkpoint inhibitors). Therefore, profiling ctDNA will become extremely important to guide personalized medicine. To make personalized PCa therapy a reality, new assays need to be validated, convenient to perform, and easy to be analyzed and interpreted. In this regard, the measurement of lncRNAs and miRNAs in blood offers promising prospects, since both are often tumor-specific and relatively easy to detect, but these are still far from clinical implementation. Molecular characterization of CTCs at the single cell level can provide deeper insight into tumor biology. However, it is less likely to do this routinely in clinical practice in the near future due to complicated sampling and pre-treatment procedures.
In the next five years, treatment of PCa will not only be based on histopathological determined tumor stage, grade and on sPSA response, but will also include biomarker profiles. Furthermore, the integration of biomarker profiling and pharmacokinetic sampling will help fine tuning therapy by monitoring key parameters such as drug distribution and metabolism in relation to the tumor profile. In this light, we are at the beginning of the biomarker profiling landscape. We expect the integration of multiple markers to improve patient stratification, reconsider therapeutic strategies and improve clinical outcomes.
Article Highlights
Liquid biopsies are minimally invasive tools for detecting tumor-derived material, which can serve as a diagnostic, prognostic and/or predictive biomarker.
Four urine-based molecular biomarker tests for PCa diagnostics and prognostics are available clinically.
Blood-based biomarkers for PCa include enumeration and characterization of circulating tumor cells (CTCs), tumor-educated platelets, exosomes and cell-free nucleic acids (incl. ctDNA, lncRNAs, mRNAs and microRNAs).
Blood-based liquid biopsies have a high potential to be used as a prognostic and predictive biomarker for treatment response and guidance of treatment choice.
There is a large gap between biomarker discovery and clinical validation. Only a few blood-based biomarkers are clinically available for PCa.
Declaration of interest
J.A.S. and G.W.V. are inventors on PCA3-related IP. The IP is owned by their employer, Radboud university medical center, which has licensed the technology and receives royalty payments. J.A.S. is advisor to the board of MDxHealth. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Note from the Authors
Since the landscape of liquid biomarker development is rapidly moving, we did not perform a systematic review. This review is based on published articles indexed in the National Library of Medicine (NLM) PubMed database and Embase database. We searched for articles using the following search terms and their synonyms: prostate cancer, biomarker and CTC/ctDNA/mRNA/miRNA/lncRNA/exosomes/platelets. The most relevant articles published before April 2019 were selected by the authors’ assessment. We did not include the articles representing biomarkers for research use only, but selected the biomarkers that are clinically used or in advanced stages of clinical validation.
Reviewers Disclosure
Peer reviewers on this manuscript have no relevant financial relationships or otherwise to disclose.
Additional information
Funding
References
- de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011 May 26;364(21):1995–2005. PubMed PMID: 21612468; PubMed Central PMCID: PMCPMC3471149. eng.
- Ryan CJ, Smith MR, de Bono JS, et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N Engl J Med. 2013 Jan 10;368(2):138–148. PubMed PMID: 23228172; PubMed Central PMCID: PMCPMC3683570. eng.
- Scher HI, Fizazi K, Saad F, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012 Sep 27;367(13):1187–1197. PubMed PMID: 22894553; eng.
- Beer TM, Armstrong AJ, Rathkopf DE, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014 Jul 31;371(5):424–433. PubMed PMID: 24881730; PubMed Central PMCID: PMCPMC4418931. eng.
- Tannock IF, de Wit R, Berry WR, et al. Docetaxel plus prednisone or mitoxantrone plus prednisone for advanced prostate cancer. N Engl J Med. 2004 Oct 7;351(15):1502–1512. PubMed PMID: 15470213; eng.
- de Bono JS, Oudard S, Ozguroglu M, et al. Prednisone plus cabazitaxel or mitoxantrone for metastatic castration-resistant prostate cancer progressing after docetaxel treatment: a randomised open-label trial. Lancet. 2010 Oct 2;376(9747):1147–1154. PubMed PMID: 20888992; eng.
- Parker C, Nilsson S, Heinrich D, et al. Alpha emitter radium-223 and survival in metastatic prostate cancer. N Engl J Med. 2013 Jul 18;369(3):213–223. PubMed PMID: 23863050; eng.
- Kantoff PW, Higano CS, Shore ND, et al. Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med. 2010 Jul 29;363(5):411–422. PubMed PMID: 20818862; eng.
- Mateo J, Carreira S, Sandhu S, et al. DNA-repair defects and olaparib in metastatic prostate cancer. N Engl J Med. 2015 Oct 29;373(18):1697–1708. PubMed PMID: 26510020; PubMed Central PMCID: PMCPMC5228595. eng.
- de Bono JS, De Giorgi U, Rodrigues DN, et al. Randomized phase II study evaluating Akt blockade with ipatasertib, in combination with abiraterone, in patients with metastatic prostate cancer with and without PTEN loss. Clin Cancer Res off J Am Assoc Cancer Res. 2019 Feb 1;25(3):928–936. PubMed PMID: 30037818; eng.
- Hendriks RJ, van Oort IM, Schalken JA. Blood-based and urinary prostate cancer biomarkers: a review and comparison of novel biomarkers for detection and treatment decisions. Prostate Cancer Prostatic Dis. 2017 Mar;20(1):12–19. . PubMed PMID: 27922627; eng.
- Marks LS, Fradet Y, Deras IL, et al. PCA3 molecular urine assay for prostate cancer in men undergoing repeat biopsy. Urology. 2007 Mar;69(3):532–535. PubMed PMID: 17382159; eng.
- Groskopf J, Aubin SM, Deras IL, et al. APTIMA PCA3 molecular urine test: development of a method to aid in the diagnosis of prostate cancer. Clin Chem. 2006 Jun;52(6):1089–1095. PubMed PMID: 16627561; eng.
- Gittelman MC, Hertzman B, Bailen J, et al. PCA3 molecular urine test as a predictor of repeat prostate biopsy outcome in men with previous negative biopsies: a prospective multicenter clinical study. J Urol. 2013 Jul;190(1):64–69. PubMed PMID: 23416644; eng.
- Van Neste L, Hendriks RJ, Dijkstra S, et al. Detection of high-grade prostate cancer using a urinary molecular biomarker-based risk score. Eur Urol. 2016 Nov;70(5):740–748. PubMed PMID: 27108162; eng.
- Haese A, Trooskens G, Steyaert S, et al. Multicenter optimization and validation of a 2-gene mRNA urine test for detection of clinically significant prostate cancer prior to initial prostate biopsy. J Urol. 2019 Apr;26:101097ju0000000000000293. PubMed PMID: 31026217; eng.
- Leyten GH, Hessels D, Smit FP, et al. Identification of a candidate gene panel for the early diagnosis of prostate cancer. Clin Cancer Res off J Am Assoc Cancer Res. 2015 Jul 1;21(13):3061–3070. PubMed PMID: 25788493; eng.
- Tomlins SA, Day JR, Lonigro RJ, et al. Urine TMPRSS2: eRGPlus PCA3 for individualized prostate cancer risk assessment. Eur Urol. 2016 Jul;70(1):45–53. PubMed PMID: 25985884; PubMed Central PMCID: PMCPMC4644724. eng.
- Salami SS, Schmidt F, Laxman B, et al. Combining urinary detection of TMPRSS2: eRGand PCA3 with serum PSA to predict diagnosis of prostate cancer. Urol Oncol. 2013 Jul;31(5):566–571. PubMed PMID: 21600800; PubMed Central PMCID: PMCPMC3210917. eng.
- McKiernan J, Donovan MJ, O’Neill V, et al. A novel urine exosome gene expression assay to predict high-grade prostate cancer at initial biopsy. JAMA Oncol. 2016 Jul 1;2(7):882–889. PubMed PMID: 27032035; eng.
- Latosinska A, Frantzi M, Merseburger AS, et al. Promise and implementation of proteomic prostate cancer biomarkers. Diagnostics (Basel). 2018 Aug 29;8(3). PubMed PMID: 30158500; PubMed Central PMCID: PMCPMC6174350. eng. doi: 10.3390/diagnostics8030057
- Scher HI, Morris MJ, Stadler WM, et al. The prostate cancer working group 3 (PCWG3) consensus for trials in castrationresistant prostate cancer (CRPC) [Conference Abstract]. J. Clin Oncol Conf. 2015;33(15 SUPPL. 1)
- Heller G, McCormack R, Kheoh T, et al. Circulating tumor cell number as a response measure of prolonged survival for metastatic castration-resistant prostate cancer: a comparison with prostate-specific antigen across five randomized phase III clinical trials. J Clin Oncol. 2018 Feb 20;36(6):572–580. PubMed PMID: 29272162; PubMed Central PMCID: PMCPMC5815402. eng.
- Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol. 2015 Apr 20;33(12):1348–1355. PubMed PMID: 25800753; PubMed Central PMCID: PMCPMC4397279. eng.
- Scher HI, Jia X, de Bono JS, et al. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009 Mar;10(3):233–239. PubMed PMID: 19213602; PubMed Central PMCID: PMCPMC2774131. eng.
- Okegawa T, Itaya N, Hara H, et al. Circulating tumor cells as a biomarker predictive of sensitivity to docetaxel chemotherapy in patients with castration-resistant prostate cancer. Anticancer Res. 2014 Nov;34(11):6705–6710. PubMed PMID: 25368278; eng.
- Goldkorn A, Ely B, Quinn DI, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol. 2014 Apr 10;32(11):1136–1142. PubMed PMID: 24616308; PubMed Central PMCID: PMCPMC3970171. eng.
- Amato RJ, Melnikova V, Zhang Y, et al. Epithelial cell adhesion molecule-positive circulating tumor cells as predictive biomarker in patients with prostate cancer. Urology. 2013 Jun;81(6):1303–1307. PubMed PMID: 23622774; eng.
- Resel Folkersma L, San Jose Manso L, Galante Romo I, et al. Prognostic significance of circulating tumor cell count in patients with metastatic hormone-sensitive prostate cancer. Urology. 2012 Dec;80(6):1328–1332. PubMed PMID: 23063057; eng.
- Goodman OB Jr., Symanowski JT, Loudyi A, et al. Circulating tumor cells as a predictive biomarker in patients with hormone-sensitive prostate cancer. Clin Genitourin Cancer. 2011 Sep;9(1):31–38. PubMed PMID: 21705286; eng.
- Okegawa T, Nutahara K, Higashihara E. Immunomagnetic quantification of circulating tumor cells as a prognostic factor of androgen deprivation responsiveness in patients with hormone naive metastatic prostate cancer. J Urol. 2008 Oct;180(4):1342–1347. PubMed PMID: 18707699; eng.
- Nakazawa M, Antonarakis ES, Luo J. Androgen receptor splice variants in the era of enzalutamide and abiraterone. Horm Cancer. 2014 Oct;5(5):265–273. . PubMed PMID: 25048254; PubMed Central PMCID: PMCPMC4167475. eng.
- Antonarakis ES, Lu C, Wang H, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014 Sep 11;371(11):1028–1038. PubMed PMID: 25184630; PubMed Central PMCID: PMCPMC4201502. eng.
- Antonarakis ES, Lu C, Luber B, et al. Androgen receptor splice variant 7 and efficacy of taxane chemotherapy in patients with metastatic castration-resistant prostate cancer. JAMA Oncol. 2015 Aug;1(5):582–591. PubMed PMID: 26181238; PubMed Central PMCID: PMCPMC4537351. eng.
- Onstenk W, Sieuwerts AM, Kraan J, et al. Efficacy of cabazitaxel in castration-resistant prostate cancer is independent of the presence of AR-V7 in circulating tumor cells. Eur Urol. 2015 Dec;68(6):939–945. PubMed PMID: 26188394; eng.
- Scher HI, Lu D, Schreiber NA, et al. Association of AR-V7 on circulating tumor cells as a treatment-specific biomarker with outcomes and survival in castration-resistant prostate cancer. JAMA Oncol. 2016 Nov 1;2(11):1441–1449. PubMed PMID: 27262168; PubMed Central PMCID: PMCPMC5206761. eng.
- Antonarakis ES, Lu C, Luber B, et al. Clinical significance of androgen receptor splice variant-7 mRNA detection in circulating tumor cells of men with metastatic castration-resistant prostate cancer treated with first- and second-line abiraterone and enzalutamide. J Clin Oncol. 2017 Jul 1;35(19):2149–2156. PubMed PMID: 28384066; PubMed Central PMCID: PMCPMC5493048. eng.
- Bernemann C, Schnoeller TJ, Luedeke M, et al. Expression of AR-V7 in circulating tumour cells does not preclude response to next generation androgen deprivation therapy in patients with castration resistant prostate cancer. Eur Urol. 2017 Jan;71(1):1–3. PubMed PMID: 27471164; eng.
- Hu R, Denmeade SR, Luo J. Molecular processes leading to aberrant androgen receptor signaling and castration resistance in prostate cancer. Expert Rev Endocrinol Metab. 2010 Sep;5(5):753–764. PubMed PMID: 21318111; PubMed Central PMCID: PMCPMC3035007. eng.
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr Rev. 2004 Apr;25(2):276–308. . PubMed PMID: 15082523; eng.
- Del Re M, Biasco E, Crucitta S, et al. The detection of androgen receptor splice variant 7 in plasma-derived exosomal RNA strongly predicts resistance to hormonal therapy in metastatic prostate cancer patients. Eur Urol. 2017 Apr;71(4):680–687. PubMed PMID: 27733296; eng.
- To SQ, Kwan EM, Fettke HC, et al. Expression of androgen receptor splice variant 7 or 9 in whole blood does not predict response to androgen-axis-targeting agents in metastatic castration-resistant prostate cancer. Eur Urol. 2018 Jun;73(6):818–821. PubMed PMID: 29398263; eng.
- Seitz AK, Thoene S, Bietenbeck A, et al. AR-V7 in peripheral whole blood of patients with castration-resistant prostate cancer: association with treatment-specific outcome under abiraterone and enzalutamide. Eur Urol. 2017 Nov;72(5):828–834. PubMed PMID: 28818355; eng.
- Todenhofer T, Azad A, Stewart C, et al. AR-V7 transcripts in whole blood RNA of patients with metastatic castration resistant prostate cancer correlate with response to abiraterone acetate. J Urol. 2017 Jan 01;197(1):135–142.
- Markowski MC, Silberstein JL, Eshleman JR, et al. Clinical utility of CLIA-grade AR-V7 testing in patients with metastatic castration-resistant prostate cancer. JCO Precis Oncol. 2017;2017. PubMed PMID: 29170762; PubMed Central PMCID: PMCPMC5697780. eng. DOI:10.1200/po.17.00127
- Armstrong AJ, Halabi S, Luo J, et al. Prospective multicenter validation of androgen receptor splice variant 7 and hormone therapy resistance in high-risk castration-resistant prostate cancer: the PROPHECY study. J Clin Oncol. 2019 Mar;(13):Jco1801731. PubMed PMID: 30865549; eng. DOI:10.1200/jco.18.01731.
- Tjon-Kon-Fat LA, Lundholm M, Schroder M, et al. Platelets harbor prostate cancer biomarkers and the ability to predict therapeutic response to abiraterone in castration resistant patients. Prostate. 2018 Jan;78(1):48–53. PubMed PMID: 29094381; eng.
- Gurunathan S, Kang MH, Jeyaraj M, et al. Review of the isolation, characterization, biological function, and multifarious therapeutic approaches of exosomes. Cells. 2019 Apr 3;8(4). PubMed PMID: 30987213. doi: 10.3390/cells8040307.
- Danila DC, Anand A, Sung CC, et al. TMPRSS2-ERG status in circulating tumor cells as a predictive biomarker of sensitivity in castration-resistant prostate cancer patients treated with abiraterone acetate. Eur Urol. 2011 Nov;60(5):897–904. PubMed PMID: 21802835; PubMed Central PMCID: PMCPMC3185163. eng.
- Dijkstra S, Leyten GH, Jannink SA, et al. KLK3, PCA3, and TMPRSS2-ERG expression in the peripheral blood mononuclear cell fraction from castration-resistant prostate cancer patients and response to docetaxel treatment. Prostate. 2014 Sep;74(12):1222–1230. PubMed PMID: 25043536; eng.
- Reig O, Marin-Aguilera M, Carrera G, et al. TMPRSS2-ERG in blood and docetaxel resistance in metastatic castration-resistant prostate cancer. Eur Urol. 2016 Nov;70(5):709–713. PubMed PMID: 26948395; eng.
- Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012 Apr 18;13(5):358–369. PubMed PMID: 22510765; PubMed Central PMCID: PMCPMC4517822. eng.
- Gallo A, Tandon M, Alevizos I, et al. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7(3):e30679. PubMed PMID: 22427800; PubMed Central PMCID: PMCPMC3302865. eng.
- Zhang Z, Qin YW, Brewer G, et al. MicroRNA degradation and turnover: regulating the regulators. Wiley Interdiscip Rev RNA. 2012 Jul–Aug;3(4):593–600. PubMed PMID: 22461385; PubMed Central PMCID: PMCPMC3635675.
- Lin HM, Castillo L, Mahon KL, et al. Circulating microRNAs are associated with docetaxel chemotherapy outcome in castration-resistant prostate cancer. Br J Cancer. 2014 May 13;110(10):2462–2471. PubMed PMID: 24714754; PubMed Central PMCID: PMCPMC4021524. eng.
- Lin HM, Mahon KL, Spielman C, et al. Phase 2 study of circulating microRNA biomarkers in castration-resistant prostate cancer. Br J Cancer. 2017 Apr 11;116(8):1002–1011.
- Corcoran C, Rani S, O’Driscoll L. miR-34a is an intracellular and exosomal predictive biomarker for response to docetaxel with clinical relevance to prostate cancer progression. Prostate. 2014 Sep;74(13):1320–1334. . PubMed PMID: 25053345; eng.
- Cheng HH, Plets M, Li H, et al. Circulating microRNAs and treatment response in the Phase II SWOG S0925 study for patients with new metastatic hormone-sensitive prostate cancer. Prostate. 2018 Feb;78(2):121–127. PubMed PMID: 29105802; PubMed Central PMCID: PMCPMC5728359. eng.
- Urabe F, Matsuzaki J, Yamamoto Y, et al. Large-scale circulating microRNA profiling for the liquid biopsy of prostate cancer. Clin Cancer Res off J Am Assoc Cancer Res. 2019 Feb 26;25:3016–3025. PubMed PMID: 30808771; eng.
- Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol. 2015 Jan;67(1):33–41. PubMed PMID: 25129854; PubMed Central PMCID: PMCPMC4252606.
- De Laere B, van Dam PJ, Whitington T, et al. Comprehensive profiling of the androgen receptor in liquid biopsies from castration-resistant prostate cancer reveals novel intra-ar structural variation and splice variant expression patterns. Eur Urol. 2017 Aug;72(2):192–200. PubMed PMID: 28104311; eng.
- Bijnsdorp IV, van Royen ME, Verhaegh GW, et al. The Non-Coding Transcriptome of Prostate Cancer: implications for Clinical Practice. Mol Diagn Ther. 2017 Aug;21(4):385–400. PubMed PMID: 28299719; PubMed Central PMCID: PMCPMC5511609. eng.
- Kelly BD, Miller N, Sweeney KJ, et al. A circulating microRNA signature as a biomarker for prostate cancer in a high risk group. J Clin Med. 2015 Jul 7;4(7):1369–1379. PubMed PMID: 26239681; PubMed Central PMCID: PMCPMC4519795.
- Gao Y, Feng B, Han S, et al. The roles of microRNA-141 in human cancers: from diagnosis to treatment. Cell Physiol Biochem. 2016;38(2):427–448. PubMed PMID: 26828359.
- Brase JC, Johannes M, Schlomm T, et al. Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer. 2011;128(3):608–616.
- Zheng Q, Peskoe SB, Ribas J, et al. Investigation of miR-21, miR-141, and miR-221 expression levels in prostate adenocarcinoma for associated risk of recurrence after radical prostatectomy. Prostate. 2014 Dec;74(16):1655–1662. PubMed PMID: 25252191; PubMed Central PMCID: PMCPMC4205269.
- Ghorbanmehr N, Gharbi S, Korsching E, et al. miR-21-5p, miR-141-3p, and miR-205-5p levels in urine-promising biomarkers for the identification of prostate and bladder cancer. Prostate. 2019 Jan;79(1):88–95. PubMed PMID: 30194772.
- Porzycki P, Ciszkowicz E, Semik M, et al. Combination of three miRNA (miR-141, miR-21, and miR-375) as potential diagnostic tool for prostate cancer recognition. Int Urol Nephrol. 2018 Sep;50(9):1619–1626. PubMed PMID: 30014459; PubMed Central PMCID: PMCPMC6133127.
- Mitchell PS, RKP E, Kroh M, et al. Circulating microRNAs as stable blood-based markers for cancer detection. PNAS. 2008;105:30.
- He S, Shi J, Mao J, et al. The expression of miR-375 in prostate cancer: A study based on GEO, TCGA data and bioinformatics analysis. Pathol Res Pract. 2019 Jun;215(6):152375. PubMed PMID: 30879885.
- Wang Y, Lieberman R, Pan J, et al. miR-375 induces docetaxel resistance in prostate cancer by targeting SEC23A and YAP1. Mol Cancer. 2016 Nov 10;15(1):70. PubMed PMID: 27832783; PubMed Central PMCID: PMCPMC5105253.
- Selth LA, Das R, Townley SL, et al. A ZEB1-miR-375-YAP1 pathway regulates epithelial plasticity in prostate cancer. Oncogene. 2017 Jan 5;36(1):24–34. PubMed PMID: 27270433.
- Bryant RJ, Pawlowski T, Catto JW, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012 Feb 14;106(4):768–774. PubMed PMID: 22240788; PubMed Central PMCID: PMCPMC3322952.
- Helsmoortel H, Everaert C, Lumen N, et al. Detecting long non-coding RNA biomarkers in prostate cancer liquid biopsies: hype or hope? Noncoding RNA Res. 2018 Jun;3(2):64–74. PubMed PMID: 30159441; PubMed Central PMCID: PMCPMC6096408. eng.
- Mohammed AGWV, Jiménez, Erho N, et al. Low PCA3 expression is a marker of poor differentiation in localized prostate tumors: exploratory analysis from 12,076 patients. Oncotarget. 2017;8(31):50804–50813.
- Prensner JR, Zhao S, Erho N, et al. RNA biomarkers associated with metastatic progression in prostate cancer: a multi-institutional high-throughput analysis of SChLAP1. Lancet Oncol. 2014;15(13):1469–1480.
- Ren S, Peng Z, Mao JH, et al. RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 2012 May;22(5):806–821. PubMed PMID: 22349460; PubMed Central PMCID: PMCPMC3343650. eng.
- Ren S, Wang F, Shen J, et al. Long non-coding RNA metastasis associated in lung adenocarcinoma transcript 1 derived miniRNA as a novel plasma-based biomarker for diagnosing prostate cancer. Eur J Cancer. 2013;49(13):2949–2959.
- Wang R, Sun Y, Li L, et al. Preclinical study using malat1 small interfering RNA or androgen receptor splicing variant 7 degradation enhancer ASC-J9((R)) to suppress enzalutamide-resistant prostate cancer progression. Eur Urol. 2017 Nov;72(5):835–844. PubMed PMID: 28528814; PubMed Central PMCID: PMCPMC5802348. eng.
- Xue D, Lu H, Xu HY, et al. Long noncoding RNA MALAT1 enhances the docetaxel resistance of prostate cancer cells via miR-145-5p-mediated regulation of AKAP12. J Cell Mol Med. 2018 Jun;22(6):3223–3237. PubMed PMID: 29633510; PubMed Central PMCID: PMCPMC5980122. eng.
- Stroun M, Lederrey JL,C, Olson-Sand A, et al. About the possible origin and mechanism of circulating DNA Apoptosis and active DNA release. Clin Chim Acta. 2001;313:139–142.
- Huang CC, Du M, Wang L. Bioinformatics analysis for circulating cell-free DNA in cancer. Cancers (Basel). 2019 Jun 11;11(6):805. . PubMed PMID: 31212602; PubMed Central PMCID: PMCPMC6627444.
- Barbany G, Arthur C, Lieden A, et al. Cell-free tumour DNA testing for early detection of cancer - a potential future tool. J Intern Med. 2019 Aug;286(2):118–136. PubMed PMID: 30861222.
- Siravegna G, Marsoni S, Siena S, et al. Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol. 2017 Sep;14(9):531–548. PubMed PMID: 28252003.
- Mehra N, Dolling D, Sumanasuriya S, et al. Plasma cell-free DNA concentration and outcomes from taxane therapy in metastatic castration-resistant prostate cancer from two phase III trials (FIRSTANA and PROSELICA). Eur Urol. 2018 September;74(3):283–291.
- Kienel A, Porres D, Heidenreich A, et al. cfDNA as a prognostic marker of response to taxane based chemotherapy in patients with prostate cancer. J Urol. 2015 Oct;194(4):966–971. PubMed PMID: 25896555; eng.
- Wyatt AW, Annala M, Aggarwal R, et al. Concordance of circulating tumor DNA and matched metastatic tissue biopsy in prostate cancer. J Natl Cancer Inst. 2017 Dec 1;109(12). PubMed PMID: 29206995; eng. doi: 10.1093/jnci/djx118.
- Hahn AW, Stenehjem D, Nussenzveig R, et al. Evolution of the genomic landscape of circulating tumor DNA (ctDNA) in metastatic prostate cancer over treatment and time. Cancer Treat Res Commun. 2019 Feb 6;19:100120. PubMed PMID: 30743187; eng.
- Kumar A, Coleman I, Morrissey C, et al. Substantial interindividual and limited intraindividual genomic diversity among tumors from men with metastatic prostate cancer. Nat Med. 2016 Apr;22(4):369–378. PubMed PMID: 26928463; PubMed Central PMCID: PMCPMC5045679. eng.
- Conteduca V, Wetterskog D, Sharabiani MTA, et al. Androgen receptor gene status in plasma DNA associates with worse outcome on enzalutamide or abiraterone for castration-resistant prostate cancer: a multi-institution correlative biomarker study. Ann Oncol. 2017 Jul 1;28(7):1508–1516. PubMed PMID: 28472366; PubMed Central PMCID: PMCPMC5834043. eng.
- Romanel A, Gasi Tandefelt D, Conteduca V, et al. Plasma AR and abiraterone-resistant prostate cancer. Sci Transl Med. 2015 Nov 4;7(312):312re10. PubMed PMID: 26537258; PubMed Central PMCID: PMCPMC6112410. eng.
- Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med. 2014 Sep 17;6(254):254ra125. PubMed PMID: 25232177; PubMed Central PMCID: PMCPMC4422178. eng.
- Taplin ME, Balk SP. Has the time arrived for biomarker-directed therapy in castration-resistant prostate cancer? JAMA Oncol. 2015 Aug;1(5):577–579. PubMed PMID: 26181796; eng.
- Chen EJ, Sowalsky AG, Gao S, et al. Abiraterone treatment in castration-resistant prostate cancer selects for progesterone responsive mutant androgen receptors. Clin Cancer Res off J Am Assoc Cancer Res. 2015 Mar 15;21(6):1273–1280. PubMed PMID: 25320358; PubMed Central PMCID: PMCPMC4359958. eng.
- Joseph JD, Lu N, Qian J, et al. A clinically relevant androgen receptor mutation confers resistance to second-generation antiandrogens enzalutamide and ARN-509. Cancer Discov. 2013 Sep;3(9):1020–1029. PubMed PMID: 23779130; eng.
- Wyatt AW, Azad AA, Volik SV, et al. Genomic alterations in cell-free DNA and enzalutamide resistance in castration-resistant prostate cancer. JAMA Oncol. 2016 Dec 1;2(12):1598–1606. PubMed PMID: 27148695; PubMed Central PMCID: PMCPMC5097690. eng.
- Salvi S, Casadio V, Conteduca V, et al. Circulating cell-free AR and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br J Cancer. 2015 May 12;112(10):1717–1724. PubMed PMID: 25897673; PubMed Central PMCID: PMCPMC4430717. eng.
- Azad AA, Volik SV, Wyatt AW, et al. Androgen receptor gene aberrations in circulating cell-free DNA: biomarkers of therapeutic resistance in castration-resistant prostate cancer. Clin Cancer Res off J Am Assoc Cancer Res. 2015 May 15;21(10):2315–2324. PubMed PMID: 25712683; eng.
- Salvi S, Casadio V, Conteduca, et al. Copy number variations of AR gene in serum circulating cell-free DNA as biomarker predicting outcome in patients with metastatic castration-resistant prostate cancer treated with enzalutamide [Conference Abstract]. Tumor Biol. 2015;36(Supplement 1):S46–S47.
- Annala M, Vandekerkhove G, Khalaf D, et al. Circulating tumor DNA genomics correlate with resistance to abiraterone and enzalutamide in prostate cancer. Cancer Discov. 2018 Apr;8(4):444–457. PubMed PMID: 29367197; eng.
- Sumiyoshi T, Mizuno K, Yamasaki T, et al. Clinical utility of androgen receptor gene aberrations in circulating cell-free DNA as a biomarker for treatment of castration-resistant prostate cancer. Sci Rep. 2019;9:1.
- Conteduca V, Jayaram A, Romero-Laorden N, et al. Plasma androgen receptor and docetaxel for metastatic castration-resistant prostate cancer. Eur Urol. 2019 Mar;75(3):368–373. PubMed PMID: 30773204; PubMed Central PMCID: PMCPMC6377278. eng.
- Robinson D, Van Allen EM, Wu YM, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015 Jul 16;162(2):454. PubMed PMID: 28843286; eng.
- Goodall J, Mateo J, Yuan W, et al. Circulating Cell-Free DNA to Guide Prostate Cancer Treatment with PARP Inhibition. Cancer Discov. 2017 Sep;7(9):1006–1017. PubMed PMID: 28450425; PubMed Central PMCID: PMCPMC6143169. eng.
- Quigley D, Alumkal JJ, Wyatt AW, et al. Analysis of circulating cell-free DNA identifies multiclonal heterogeneity of BRCA2 reversion mutations associated with resistance to PARP inhibitors. Cancer Discov. 2017 Sep;7(9):999–1005. PubMed PMID: 28450426; PubMed Central PMCID: PMCPMC5581695. eng.
- Maughan BL, Guedes LB, Boucher K, et al. p53 status in the primary tumor predicts efficacy of subsequent abiraterone and enzalutamide in castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2018 Jun;21(2):260–268. PubMed PMID: 29302046; eng.
- Hussain M, Daignault-Newton S, Twardowski PW, et al. Targeting androgen receptor and DNA repair in metastatic castration-resistant prostate cancer: results from NCI 9012. J Clin Oncol. 2018 Apr 1;36(10):991–999. PubMed PMID: 29261439; PubMed Central PMCID: PMCPMC6075827. eng.
- De Laere B, Oeyen S, Mayrhofer M, et al. TP53 outperforms other androgen receptor biomarkers to predict abiraterone or enzalutamide outcome in metastatic castration-resistant prostate cancer. Clin Cancer Res off J Am Assoc Cancer Res. 2019 Mar 15;25(6):1766–1773. PubMed PMID: 30209161; PubMed Central PMCID: PMCPMC6330086. eng.
- Carver BS, Chapinski C, Wongvipat J, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011 May 17;19(5):575–586. PubMed PMID: 21575859; PubMed Central PMCID: PMCPMC3142785. eng.
- de Bono JS, De Giorgi U, Massard C, et al. PTEN loss as a predictive biomarker for the Akt inhibitor ipatasertib combined with abiraterone acetate in patients with metastatic castration-resistant prostate cancer (mCRPC). Ann Oncol. 2016;27(suppl_6). DOI:10.1093/annonc/mdw372.02
- Schweizer MT, Gulati R, Beightol M, et al. Clinical determinants for successful circulating tumor DNA analysis in prostate cancer. Prostate. 2019 Mar 13 PubMed PMID: 30865311; eng. DOI:10.1002/pros.23778.
- Vandekerkhove G, Struss WJ, Annala M, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur Urol. 2019 Apr;75(4):667–675. PubMed PMID: 30638634; eng.
