Abstract
The inhibition of xanthine oxidase (XO) activity by the purine analogue 6-(N-benzoylamino)purine was evaluated and compared with the standard inhibitor, allopurinol and the parent compound adenine. 6-(N-benzoylamino)purine is a highly potent inhibitor of XO and comparable to allopurinol
. Furthermore, 6-(N-benzoylamino)purine neither produced any enzymatic superoxide nor reduced XO by an electron transfer reaction unlike allopurinol. 6-(N-benzoylamino)purine
is about 10000-fold more potent as a XO inhibitor compared to the only known purine analogue 8-bromoxanthine
. 6-(N-Benzoylamino)purine is a competitive inhibitor of XO and the inhibition was not completely reversed even at 100 μM xanthine concentration. The calculated interaction energy [Ecomplex − (Eligand+Eprotein)] of − 30.5, − 22.6, and − 17.2 kcal/mol, respectively, of 6-(N-benzoylamino)purine, 8-bromoxanthine and the parent compound adenine provided the rationale for the better enzyme inhibitory activity of 6-(N-benzoylamino)purine. To understand the role of the benzamido group in the inhibition process, molecular docking studies were carried out and it was revealed that the hydrogen bonding interactions involving N-7 of the purine ring and the N–H of Arg880, N–H of the purine ring and OH of Thr1010, as well as non-bonded interactions of the benzamido group of 6-(N-benzoylamino)purine with amino acid residues Gly799, Glu802, Phe914, Ala1078, Ala1079 and Glu1261 in the active site of XO play an important role in the stabilization of the E–I complex.
1 Introduction
XO is a complex molybdoflavoprotein that catalyzes the hydroxylation of xanthine to uric acid [Citation1]. In humans the enzyme catalyzes the last two steps of purine catabolism, the oxidative hydroxylation of hypoxanthine to xanthine and xanthine to uric acid [Citation2]. The redox reaction occurs at the molybdopterin center and from there the electrons are passed via two Fe2S2 centre to the isoalloxazine ring of FAD and finally transferred to the acceptor substrate, oxygen [Citation3]. The enzyme has been recognized as a drug target for gout [Citation4] and oxygen radical-induced tissue damage [Citation5]. Allopurinol, a pyrazolopyrimidine derivative, was launched in 1969 and has been widely prescribed for the treatment of hyperuricemia, gout and post-ischemic tissue damage. Allopurinol is a suicide inhibitor of XO but, oxypurinol, the oxidation product of allopurinol, is the actual inhibiting species [Citation6] and is under phase II trial for the treatment of gout [Citation7]. However both allopurinol and oxypurinol exhibit some toxicity. This has led to the need for the structural modification of the pyrazolopyrimidine moiety to develop new inhibitors such as BOF-4272 [Citation8]. Further search for novel inhibitors of XO led to the discovery of pteridines [Citation9], TMX-67 [Citation7] or TEI-6720 [Citation10] (thiazole derivatives), 1-phenylpyrazoles [Citation11], flavanoids Citation12-14 and 1,3-diaryltriazoles [Citation15]. Since the purine nucleus is structurally similar to the pyrazolopyrimidine moiety, purine derivatives are expected to exhibit XO inhibitory activity. However, there has only been one example of a purine derivative e.g., 8-bromoxanthine as XO inhibitor and this was found to exhibit uncompetitive inhibition with respect to xanthine and non-competitive inhibition with respect to molecular oxygen [Citation16].
The potential of purine analogues as XO inhibitors was supported by Rastelli et al. [Citation17] and Hernandez et al. [Citation18] who studied the interactions of substrates and inhibitors with the motifs and active site residues of XO by molecular modeling and predicted that the functional group present at the C-6 position of the purines undergoes hydrogen bond formation with the enzyme. This recognition is expected to be stronger when this functional group is capable of exhibiting additional interactions with the enzyme. Therefore, we hypothesized that purine derivative having a C-6 substituent possessing the capability of hydrogen bond formation, a π–π stacking interaction and hydrophobic interactions, may serve as a potent inhibitor of XO. In the present work we describe 6-(N-benzoylamino)purine as a novel and potent inhibitor of XO.
2 Material and methods
Xanthine oxidase, xanthine and allopurinol were purchased from Sigma Chemical Co., USA. 6-(N-Benzoylamino)purine and adenine were obtained from Lancaster.
2.1 XO activity assay
Enzyme activity was measured spectrophotometrically by measuring uric acid formation at 293 nm with xanthine as the substrate [Citation19] using a Perkin Elmer Lambda 25 UV-visible spectrophotometer at 25°C.
2.2 Super oxide assay
Super oxide formation was measured by monitoring the reduction of cytochrome c as described by Hodges et al. [Citation20]. This reaction was inhibitable by superoxide dismutase. The extinction coefficient of reduced cytochrome c at 550 nm was taken as 21,000 M− 1cm− 1.
2.3 Hydrogen peroxide assay
Hydrogen peroxide formation was measured using a coupled assay system containing o-dianisidine and peroxidase following the protocol of Liochev et al. [Citation21].
2.4 Electron transfer assay
The standard reaction mixture contained 1 ml of a solution containing 980 μl of 100 mM sodium phosphate buffer (pH 7.5), 15 μM of dichlorophenol-indophenol (DCPIP) and 10 μM of xanthine. The reaction was initiated by the addition of 2.25 units xanthine oxidase to the reaction mixture and the time-dependent absorbance change was monitored at 605 nm.
2.5 XO inhibition
Inhibition of the xanthine oxidase reaction by various purine based inhibitors was measured in terms of the decrease in uric acid formation at 293 nm. The nature of the inhibition was determined using a Lineweaver-Burk plot and the Ki of the inhibitors was calculated using the standard equation for competitive inhibition.
2.6 Unit definition
One unit of activity of xanthine oxidase is defined as the amount of enzyme that produces 1.0 μM of uric acid min− 1 at 25°C. The concentration of the enzyme was calculated using the molar extinction coefficient of XO [Citation22].
2.7 Software and dtandard deviation
All the kinetic and IC50 data were fitted using Grafit (Erithacus software limited, UK. Version 4.0 written by Dr. Robin J. Leatherbarrow). Values are expressed as means ± SEM. Statistical significance was set as P < 0.05.
2.8 Molecular docking
All the molecular modeling studies were performed using SYBYL 6.9 [Citation23] installed on an Octane 2 workstation. The coordinates of bovine milk xanthine oxidase complexed with salicylate (Protein Data Bank entry 1fiq) were retrieved from the PDB [Citation24]. Chain A and B were deleted. Using chain C, amino acid residues within 8 Å of the inhibitor (salicylate) and water molecules within the active site region were selected for docking. The ligands were modeled and optimized using the Tripos forcefield method [Citation25]. The partial charges were calculated using the Gasteiger-Huckel method. FlexX [Citation26] method was used for the docking. After docking, the small molecules were removed and protein-ligand complexes were subjected to optimization using the MMFF94 method [Citation27]. Refinement of the enzyme-inhibitor complex was performed in a stepwise manner. In the initial step the hydrogen atoms of the complex were refined which was then followed by refinement of the active site and finally of the whole complex. The distance-dependent dielectric function was used.
3 Results and discussion
3.1 Steady state analysis of the inhibition of XO with the purine based inhibitor 6-(N-benzoylamino)purine
Since XO has a broad substrate specificity, initially the substrate specificity of 6-(N-Benzoylamino) purine was verified by employing time-dependent scans between 200 to 400 nm and any enzymatic hydroxylation was assessed from the time-dependent decrease in the absorbance at the corresponding λmax (285 nm) value. This compound was not found be substrate of XO. However, unlike many purine based compounds that we have tested (data not shown-available on request), 6-(N-Benzoylamino) purine displayed a very strong inhibitory effect on XO with an IC50 value of 0.45 μM. 6-(N-Benzoylamino) purine was found to be a competitive inhibitor of XO with a Ki value of 0.0475 μM as shown in .
Figure 1 (a). (A) Lineweaver-Burk plot for the inhibition of xanthine oxidase by 6-(N-benzoylamino)purine. Enzyme concentration used was 2.25 units/ml. Xanthine concentration was varied (0.30 μM, 0.50 μM, 0.80 μM, 1.00 μM, 2.00 μM) at a series of fixed concentrations of 6-(N-benzoylamino)purine. (B) Inhibition type and Kii value of 6-(N-benzoylamino)purine. (b). Time course of xanthine oxidase-catalyzed reaction in the absence and presence of allopurinol and 6-(N-benzoylamino)purine. 1) 0 μM inhibitor, 2) 2.5 μM allopurinol, 3) 5 μM allopurinol, 4) 2.5 μM 6-(N-benzoylamino)purine, 5) 5 μM 6-(N-benzoylamino)purine, 6) 7.5 μM allopurinol, 7) 7.5 μM 6-(N-benzoylamino)purine. Enzyme concentration used was 2.25 units/ml. Level of enzymatically generated uric acid formation was measured at 293 nm.
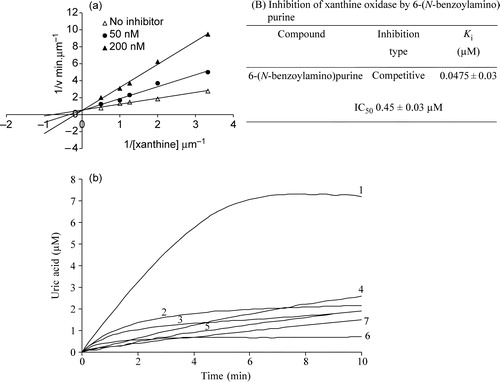
In comparison, the parent compound adenine exhibited an IC50 value of 75.61 μM (Table II) suggesting thereby that the benzamido group present in this inhibitor has a potential role in the inhibition mechanism. shows the percentage inhibition of the XO reaction (xanthine to uric acid) in the presence of 10 μM of 6-(N-benzoylamino) purine using variable xanthine concentrations of 10, 20, 50 and 100 μM. The experiments were performed following two different protocols. In the set 1 protocol the enzyme was added to a mixture of buffer, substrate and inhibitor and the inhibition was monitored. In the set 2 protocol the enzyme was preincubated with the inhibitor followed by addition of this enzyme-inhibitor mixture to the buffer containing the substrate. The inhibition was a little more pronounced in the set 2 experiments (). It was found that in both protocols the inhibition was not reversed completely even with a very high concentration of xanthine. Therefore it seems that this competitive inhibitor binds strongly with the enzyme and that a very high concentration of substrate is required to displace the inhibitor from the active site. In order to assess the XO inhibitory activity of 6-(N-benzoylamino)purine in comparison to that of the standard XO inhibitor allopurinol, inhibition was carried out under similar conditions with allopurinol. It was found that 6-(N-benzoylamino)purine possesses comparable XO inhibitory activity to that of allopurinol
. The inhibitory effect of allopurinol on XO activity is known to be time-dependent. As is commonly observed for tight binding inhibitors, both allopurinol and 6-(N-benzoylamino)purine show time-dependent inhibition (). However, the underlying mechanisms for the time-dependent inhibition by these two inhibitors are fundamentally different. Allopurinol is a suicide substrate of XO and the intermediate enzymatic product oxypurinol, binds to the reduced form of the enzyme and prevents the reaction proceeding further. In contrast, being a competitive inhibitor of XO, 6-(N-benzoylamino)purine caused a time-and concentration-dependent decline in the catalytic rate.
Table I. Percentage inhibition of XO in the presence of increasing concentrations of xanthine and 10 μM of 6-(N-benzoylamino)purine. In Set1 experiments, 2.25 units of XO was added to 1 ml of 100 mM Sodium phosphate buffer, pH 7.5 containing different concentrations of xanthine (10–100 μM) in presence and absence of inhibitor. In set 2 experiments, 2.25 units of XO and 10 μM of 6-(N-benzoylamino)purine was incubated and added to 1 ml sodium phosphate buffer containing different concentrations of xanthine as indicated.
3.2 Xanthine, allopurinol and 6-(N-benzoylamino)purine induced inhibition of ROS formation and electron transfer by XO
In XO reaction mechanism, oxidative hydroxylation of xanthine takes place at the molybdopterin centre of the enzyme and the released electrons from the substrate are introduced into the enzyme. These electrons are finally taken up by the oxygen at the enzyme's flavin motif and generate superoxide (one electron species) and hydrogen peroxide (two electron species) as common products for all purine substrates. On the other hand, the electron acceptor dye 2,6-dichlorophenol-indophenol scavenges electrons from the molybdopterin centre and is reduced from a ‘chromo’ to its ‘leuko’ form. It was found () that both allopurinol and xanthine reduced 2,6-dicholorophenol-indophenol in a time-dependent manner but 6-(N-benzoylamino)purine did not show any appreciable DCPIP reduction suggesting that 6-(N-benzoylamino)purine did not transfer two electron to xanthine oxidase.
Figure 2 (a). Xanthine, allopurinol and 6-(N-benzoylamino)purine induced enzymatic electron transfer reaction. The reaction mixture contained 100 mM sodium phosphate, pH 7.4, 10 μM xanthine or 10 μM allopurinol or 10 μM 6-(N-benzoylamino)purine and 15 μM DCPIP. The reaction was initiated with the addition of 2.25 units of xanthine oxidase and the decrease in absorbance with time was measured at 605 nm. (b). Xanthine, allopurinol and 6-(N-benzoylamino)purine induced enzymatic superoxide formation. The reaction mixture contained 100 mM sodium phosphate, pH 7.4, 10 μM xanthine or 10 μM allopurinol or 10 μM 6-(N-benzoylamino)purine and 15 μM Cyt c. The reaction was initiated with the addition of 2.25 units of xanthine oxidase and the increase in absorbance with time was measured at 550 nm.
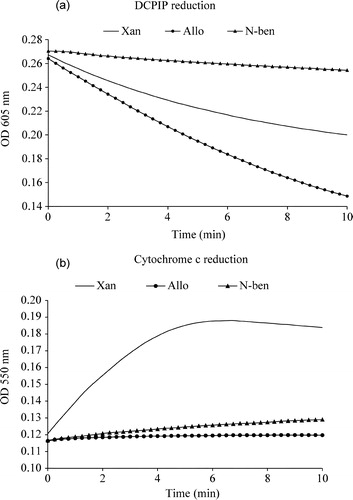
Enzymatic production of superoxide with time using saturated concentrations of xanthine, allopurinol and 6-(N-benzoylamino)purine is shown in . It is seen that out of these three compounds, only xanthine produces superoxide, while allopurinol and 6-(N-benzoylamino)purine do not. Therefore it is concluded that 6-(N-benzoylamino)purine neither generated any superoxide nor reduced the enzyme by electron transfer. Both xanthine and allopurinol produced hydrogen peroxide, but, as expected, the inhibitor 6-(N-benzoylamino)purine did not (data not shown). The above results therefore suggest that this compound is a potent competitive inhibitor of XO and binds very strongly to the enzyme without producing any enzyme-mediated electrons or ROS.
3.3 Molecular basis for XO inhibition by 6-(N-benzoylamino)purine-Modeling and docking studies
The binding orientation of 6-(N-benzoylamino)purine at the active site of XO is shown in . The interaction energy [Ecomplex − (Eligand+Eprotein)] was calculated to be − 30.5 kcal/mol (). The purine ring was oriented parallel to Phe914 and was involved in a π-π stacking interaction. Two hydrogen bonding interactions were observed involving the N-7 of the purine ring and the N–H of the Arg880 [(N…H–N) = 2.741 Å], N–H of the purine ring and OH of the Thr1010 [(N–H…O) = 2.128 Å]. The benzamido group of 6-(N-benzoylamino)purine is involved in a number of favourable interaction with various amino acid residues. The –NH group was placed within interacting distance with the phenyl ring of Phe914 and the –CO group of the inhibitor was positioned so as to interact with Ala1079 and the aromatic residue of Phe914. The phenyl ring of the benzamido group was surrounded by various amino acid residues such as Gly799, Glu802, Ala1078 and Glu1261 in the active site residue. Enroth et al. [Citation28] suggested that Phe1009 interacts with the six-membered ring of xanthine and Phe914 interacts with the five membered ring of xanthine. Here, however, Phe1009 was present beyond the interacting distance from the benzamido group of the inhibitor. On the other hand both Phe1009 and Phe914 were making interactions with adenine and 8-bromoxanthine as discussed below.
Figure 3 Binding mode of 6-(N-benzoylamino)purine in the active site of bovine milk XO. The hydrogen bonding interactions are shown as broken lines. Purine nucleus is coloured in blue and green. The nitrogen atoms of the purine are labeled in blue colour. Benzoyl group of 6-(N-Benzoylamino)purine is labelled in green and oxygen moiety in general (both for the inhibitor and the enzyme) is shown in red colour.
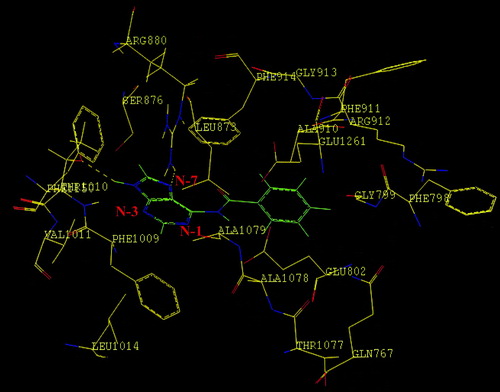
Table II. The IC50, Ki and binding energy values of adenine, 8-bromoxanthine and 6-(N-benzoylamino)purine with XO.
In order to understand the importance of the benzamido group of 6-(N-benzoylamino)purine in imparting specific binding interactions with the active site of the enzyme, we delineated the various interactions of the parent compound, adenine, with the enzyme. The calculated interaction energy [Ecomplex − (Eligand+Eprotein)] of − 17.2 kcal/mol for adenine compared with a value of − 30.5 kcal/mol observed for 6-(N-benzoylamino)purine provides the rationale for the superior inhibitory effect of 6-(N-benzoylamino)purine (). Much similarity between adenine and 6-(N-benzoylamino)purine was observed for the binding interaction of the purine moiety of these ligands with the enzyme. Aromatic-aromatic interaction was observed involving the 5- and 6-membered rings of adenine with Phe914 and Phe1009, respectively. Three hydrogen bonds were observed involving the N-3 of the ligand and NH of Ser876 (N…H–O = 3.487 Å), N–H of the purine ring of the ligand and the OH of Thr1010 (N–H…O = 2.972 Å) and N–H of the amino group at C-6 of the ligand and Glu802 (N–H…O = 3.016). Thus, the superior inhibitory activity of 6-(N-benzoylamino)purine is due to additional favorable non-bonded interactions involving the benzamido group and the amino acid residues.
Interestingly we observed that 6-(N-benzoylamino)purine was approximately 104-fold more potent as an XO inhibitor than 8-bromoxanthine as revealed by the Ki value of 400 μM for 8-bromoxanthine[Citation16] compared to the Ki value of 0.0475 μM of 6-(N-benzoylamino)purine. The mechanisms of inhibition of XO by the two compounds are also fundamentally different [Citation16]. 8-Bromoxanthine was found to be a non-competitive inhibitor with respect to molecular oxygen but was uncompetitive with respect to Xanthine [Citation16]. Whereas, 6-(N-benzoylamino)purine was purely a competitive inhibitor of xanthine oxidase. Moreover, 6-(N-benzoylamino)purine did not generate any reactive oxygen species or electrons to reduce the enzyme. This encouraged us to compare the various binding interactions of these two inhibitors with XO so as to understand the crucial binding interactions for designing more potent inhibitors. The calculated interaction energy [Ecomplex − (Eligand+Eprotein)] of − 22.604 kcal/mol of 8-bromoxanthine compared to that of 6-(N-benzoylamino)purine ( − 30.5 kcal/mol) provides the rationale for superiority of the latter as an XO inhibitor (). 8-Bromoxanthine was nestled between two aromatic residues Phe914 and Phe1009. Hydrogen bonding interactions were observed involving the C-2 carbonyl oxygen (of the tautomeric form of 8-bromoxanthine) and NH of Arg880 (O…H–N = 2.910 Å), H of N-3 of the ligand and OH of Thr1010 (N–H…O = 3.124 Å), and H of N-7 and Glu802 (N–H…O = 3.160 Å). The bromine atom was oriented towards Leu873, a hydrophobic residue in the active site. The distance between the bromine atom and one of the methyl groups of Leu873 was found to be 3.745 Å. van der Waals contact was observed between the 5-membered ring of the purine and Phe914. The comparison of various interactions of 8-bromoxanthine and 6-(N-benzoylamino)purine with the active site residues of XO clearly reveals that the benzamido group at C-6 of the purine ring provides crucial anchoring effects for stronger inhibitory activity.
In conclusion, the present study describes 6-(N-benzoylamino)purine as a highly potent competitive inhibitor of XO being approximately 104-fold more active than the only reported purine analogue inhibitor 8-bromoxanthine and has comparable XO inhibitory activity as compared to allopurinol. The calculation of interaction energies [Ecomplex − (Eligand+Eprotein)] involving 6-(N-benzoylamino) purine, adenine and 8-bromoxanthine provides a rationale for the superiority of 6-(N-benzoylamino)purine as an inhibitor of XO (). The determination of various molecular level interactions between the ligand and the enzyme provided crucial information for the future design of more potent inhibitors. Among the two inhibitors described, 8-bromoxanthine and 6-(N-Benzoylamino)puine are positioned differently within the Xanthine oxidase active site region. In case of 8-bromoxanthine the carbonyl group at the C-2 position of this inhibitor makes a H-bonding interaction with the –NH of guanido group of Arg880. On the other hand, in 6-(N-Benzoylamino)purine the N7 of the purine ring and –NH of Arg 880 make a H-bonding interaction. The N7 of the purine ring of 8-bromoxanthine was however making a H bonding interaction to the –NH of Arg880.
Therefore the five-membered ring of 6-(N-benzoylamino)purine and the 6-membered ring of 8-bromoxanthine are positioned in a way that makes hydrogen bonding interactions with the –NH of Arg880 and –OH of Thr1010 respectively. The different positioning of the 6-membered ring in 8-bromoxanthine may also additionally be due the presence of the keto group at C-2.
We further found that orientation of 8-bromoxanthine within the active site moiety of Xanthine oxidase was also different to that of 6-(N-Benzoylamino)purine because of the presence of the benzamido group in the latter inhibitor.
The molecular docking studies predicted that the hydrogen bonding interactions involving N-7 of the purine ring and the N–H of the Arg880, N–H(N-9) of the purine ring and –OH of the Thr1010, and non-bonded interactions of the benzamido group of 6-(N-benzoylamino) purine with amino acid residues Gly799, Glu802, Phe914, Ala1078, Ala1079 and Glu1261 in the active site of XO play important roles in the stabilization of the E–I complex.
| Abbreviations | ||
| DCPIP: | = | Dichlorophenol-indophenol |
| ROS: | = | Reactive oxygen species |
| XO: | = | Xanthine oxidase |
Acknowledgements
We thank Council of Scientific and Industrial Research (CSIR, New Delhi, Govt. of India) Grant # 01(1823)/02/EMR-II) for funding of the project to Dr. A.K.M.
Notes
‡ When we submitted our manuscript a review article entitled “ Progress towards the discovery of Xanthine oxidase inhibitors” by F. Borges, E. Fernandes and F. Roleira appeared in the Current Medicinal Chemistry:-2002, 9, 195–217. ([email protected]). There was no mention of any purine inhibitor which was even structurally similar to 6-(N-Benzoylamino)purine in this review either.
References
- Hille R, Nishino T. Flavoprotein structure and mechanism. 4. Xanthine oxidase and xanthine dehydrogenase. FASEB J 1995; 9: 995–1003
- Krenitsky TA, Spector T, Hall WW. Xanthine oxidase from human liver: purification and characterization. Arch Biochem Biophys 1986; 247: 108–119
- McWhirter RB, Hille R. The reductive half-reaction of xanthine oxidase. Identification of spectral intermediates in the hydroxylation of 2-hydroxy-6-methylpurine. J Biol Chem 1991; 266: 23724–23731
- Elion GB. The purine path to chemotherapy. Science 1989; 244: 41–47
- McCord JM. Oxygen-derived free radicals in postischemic tissue injury. New Engl J Med 1985; 312: 159–163
- Nakamura M. Allopurinol-insensitive oxygen radical formation by milk xanthine oxidase systems. J Biochem (Tokyo) 1991; 110: 450–456
- Sorbera LA, Revel L, Rabasseda X, Castaner J. TMX-67 Treatment of gout and hyperuricemia—Xanthine oxidase inhibitor. Drug Future 2001; 26: 32–38
- Okamoto K, Nishino T. Mechanism of inhibition of xanthine oxidase with a new tight binding inhibitor. J Biol Chem 1995; 270: 7816–7821
- Oettl K, Reibnegger G. Pteridines as inhibitors of xanthine oxidase: structural requirements Biochim. Biophys Acta 1999; 1430: 387–395
- Okamoto K, Eger BT, Nishino T, Kondo S, Pai EF. An extremely potent inhibitor of xanthine oxidoreductase. Crystal structure of the enzyme-inhibitor complex and mechanism of inhibition. J Biol Chem 2003; 278: 1848–1855
- Ishibuchi S, Morimoto H, Oe T, Ikebe T, Inoue H, Fukunari A, Kamezawa M, Yamada I, Naka Y. Synthesis and structure-activity relationships of 1-phenylpyrazoles as xanthine oxidase inhibitors. Bioorg Med Chem Lett 2001; 11: 879–882
- Lin CM, Chen CS, Chen CT, Liang YC, Lin JK. Molecular modeling of flavonoids that inhibits xanthine oxidase. Biochem Biophys Res Commun 2002; 294: 167–172
- Van Hoorn DE, Nijveldt RJ, Van Leeuwen PA, Hofman Z, M'Rabet L, De Bont DB, Van Norren K. Accurate prediction of xanthine oxidase inhibition based on the structure of flavonoids. Eur J Pharmacol 2002; 451: 111–118
- Cos P, Ying L, Calomme M, Hu JP, Cimanga K, Poel BV, Pieters L, Vlietinck AJ, Berghe DV. Structure-activity relationship and classification of flavonoids as inhibitors of xanthine oxidase and superoxide scavengers. J Nat Prod 1998; 61: 71–76
- Okamoto K, Matsumoto K, Hille R, Eger BT, Pai EF, Nishino T. The crystal structure of xanthine oxidoreductase during catalysis: implications for reaction mechanism and enzyme inhibition. Proc Natl Acad Sci USA 2004; 101: 7931–7936
- Hille R, Stewart RC. The inhibition of xanthine oxidase by 8-bromoxanthine. J Biol Chem 1984; 259: 1570–1576
- Rastelli G, Costantino L, Albasini A. A model of the interaction of substrates and inhibitors with xanthine oxidase. J Am Chem Soc 1997; 119: 3007–3016
- Hernandez B, Orozco M, Luque FJ. Tautomerism of xanthine and alloxanthine: a model for substrate recognition by xanthine oxidase. J Comput Aided Mol Des 1996; 10: 535–544
- Escribano J, Gracia-Canovas F, Gracia-Carmona F. A kinetic study of hypoxanthine oxidation by milk xanthine oxidase. Biochem J 1988; 254: 829–833
- Hodges GR, Young MJ, Paul T, Ingold KU. How should xanthine oxidase-generated superoxide yields be measured?. Free Radic Biol Med 2000; 29: 434–441
- Liochev SI, Fridovich I. Copper, zinc superoxide dismutase and H2O2. Effects of bicarbonate on inactivation and oxidations of NADPH and urate, and on consumption of H2O2. J Biol Chem 2002; 277: 34674–34678
- Godner BLJ, Doel JJ, Goult TA, Eisenthal R, Harrison R. Suicide inactivation of xanthine oxidoreductase during reduction of inorganic nitrite to nitrite oxide. Biochem J 2001; 358: 325–333
- SYBYL 6.9 Molecular Modelling Software; Tripos Associates Inc.: 1699 S. Hanley, St. Louis MI 63144, USA.
- Abola EE, Berstein FC, Bryant SH, Koetzle TF, Weng J. Protein Data Bank. Crystallographic Databases-Information Content, Software Systems, Scientific Applications, FH Allen, G Berjerhoff, RR Sievers. Data Commission of the International Union of Crystallography, Bonn 1987; 107–132
- Clark M, Cramer RD, Van opdenbosch N. Validation of the general purpose Tripose 5.2 Force Field. J Comp Chem 1989; 10: 982–1012
- Rarey M, Kramer B, Lengauer T, Kleb GA. A fast flexible docking method using an incremental construction algorithm. J Mol Biol 1996; 261: 470–489
- Halgren T. Merck Molecular Force Field. II. MMFF94 van der Waals and electrostatic parameters for intermolecular interactions. J Comp Chem 1996; 17: 520–552
- Enroth C, Eger BY, Okamoto K, Nishino T, Pai EF. Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion. Proc Natl Acad Sci USA 2000; 97: 10723–10728