Abstract
The cytosolic human carbonic anhydrase (hCA, EC 4.2.1.1) isozyme III (hCA III) has been cloned and purified by the GST-fusion protein method. Recombinant pure hCA III had the following kinetic parameters for the CO2 hydration reaction at 20°C and pH 7.5: kcat of 1.3 × 104 s− 1 and kcat/KM of 2.5.105 M− 1 s− 1. The first detailed inhibition study of this enzyme with anions is reported. Inhibition data of the cytosolic isozymes hCA I - hCA III with a large number of anions (halides, pseudohalides, bicarbonate, carbonate, nitrate, nitrite, hydrosulfide, sulfate, sulfamic acid, sulfamide, etc.), were determined and these values are comparatively discussed for these three cytosolic isoforms. Fluoride, nitrate, nitrite, phenylboronic acid and phenylarsonic acid (as anions) were weak hCA III inhibitors (KIs of 21–78.5 mM), whereas bicarbonate, chloride, bromide, sulfate and several other simple anions showed KIs around 1 mM. The best hCA III inhibitors were carbonate, cyanide, thiocyanate, azide and hydrogensulfide, which showed KIs in the range of 10–90 μM. It is difficult to explain the inhibitory activity of carbonate (KI of 10 μM) against hCA III, also considering the fact that this ion has an affinity of 15–73 mM for hCA I and II and is in equilibrium with one of the substrates of this enzyme, i.e., bicarbonate, which is a much weaker inhibitor (KI of 0.74 mM against hCA III, of 12 mM against hCA I and of 85 mM against hCA II).
Introduction
Among the 16 carbonic anhydrase (CA, EC 4.2.1.1) isoforms described so far in mammals Citation1–4, CA III is the least understood and investigated one, and the worst catalyst for CO2 hydration as compared to other cytosolic, mitochondrial or membrane-associated isozymes [Citation5,Citation6]. In analogy with the highly abundant CA I and II, CA III is a cytosolic isoform Citation1–5, but it has a catalytic activity of around 1% that of CA II for the physiologic reaction catalyzed by these enzymes, i.e., hydration of carbon dioxide to bicarbonate and a proton [Citation7]. However, unlike the ubiquitous isozymes I and II, CA III is mainly present in slow skeletal muscles (24% of the cytosolic protein content) and liver, where its primary functions remain largely unknown [Citation7,Citation8]. Recent studies with CA III knockout mice showed CA III to be involved in mitochondrial ATP synthesis [Citation8], whereas its levels were found to be significantly decreased in mutant mice lacking the gene SULT1E1, indicating a role of CA III in cystic fibrosis liver disease [Citation9]. CA III is also considered as one of the proteins involved in oxidative stress response both in liver [Citation10] and skeletal muscle [Citation11], probably by scavenging reactive oxygen species (ROS) and thus protecting cells from oxidative damage [Citation12]. CA III seems to play an important role (together with E-cadherin) also in disruption of the intercellular barrier associated with the down-regulation of E-cadherin in the laryngopharyngeal reflux disease [Citation13].
Biochemically, these physiologic/pathophysiologic functions of CA III are poorly understood, except for the antioxidant role of this enzyme, which has been shown to be modulated by the S-glutathionylation of two cysteine residues (Cys181 and Cys186) present on the surface of the protein (but not within its active site) [Citation14,Citation15]. Indeed, oxidants such as hydrogen peroxide, peroxy radicals or hypochlorous acid oxidize these two cysteine residues to sulfinic/sulfenic acids (in the absence of glutathione), but when this tripeptide was present in the medium, the S-glutathionylation of the two Cys residues occurred, without damage to the protein [Citation14,Citation15]. It is thus probable that one of the main in vivo functions of CA III, is that of protecting proteins from irreversible oxidation processes with subsequent cellular damage [Citation4,Citation14,Citation15].
Another research line showed some interesting connections between obesity and CA III. Thus, Lynch et al [Citation16] reported a decrease in CA III expression in obese Zucker rats, possibly related to hyperinsulinemia, whereas Keha's group [Citation17,Citation18] showed that leptin, another protein involved in the genesis of obesity, decreased CA III expression (whereas insulin increased it). Since some CA inhibitors are known to act as effective agents for the management of obesity [Citation19,Citation20], mainly targeting the mitochondrial isoforms CA VA and CA VB, it might be of great interest to better understand the biochemical/physiologic processes connecting obesity and various CAs, including the less investigated CA III.
Up to now CA III from various organisms (e.g., bovines [Citation5,Citation6], rodents [Citation7], or humans [Citation21]) was obtained by extracting/purifying the enzyme from muscles or liver. Here we report the first cDNA cloning, purification and characterization of recombinant human CA III (hCA III), as well as a detailed inhibition study of the enzyme with anions, as very few such data [Citation5] are available in the literature.
Materials and methods
Chemistry
Buffers and metal salts (sodium or potassium fluoride, chloride, bromide, iodide, cyanide, cyanate, thiocyanate, azide, bicarbonate, carbonate, bisulfite, nitrate, nitrite, hydrosulfide and sulfate) were from Sigma-Aldrich (Milan, Italy) of highest purity available, and were used without further purification. Sulfamide, sulfamic acid, phenylboronic acid and phenylarsonic acid were also commercially available reagents from Sigma-Aldrich (Milan, Italy) being used with no supplementary purification.
CA III cloning
The cDNA fragment encoding the open reading frame of hCA III was amplified from polyA(+) RNA obtained from human pancreas (Clontech, Palo Alto, CA, USA) by using a commertial RT-PCR kit (Takara, Kyoto, Japan) with adopter primers including EcoRI and SalI recognition sequences (underlined in the following sequences, respectively): 5′-CGGAATTCCCATGGCCAAGGAGTGGGGC-3′ and 5′-GCAGTCGACCCTCATTTGAAGGAAGCTCT-3′. The PCR reaction was hot-started with incubation for 5 min at 94°C and consisted of 35 cycles of 30 s at 94°C, 30 sec at 57°C and 90 sec at 72°C. The PCR products were cleaved with EcoRI and SalI, purified and cloned in-frame into the pGEX-4T2 vector (Amersham). The cDNA sequence of the hCA III insert included in the vector was reconfirmed by DNA sequencing. The constructs were then transfected into E. coli strain BL21 for production of the GST-hCA III fusion protein, similarly to the procedure already described for hCA VB, IX and XII Citation22–24. Following induction of the protein expression by addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG), the bacteria were harvested and sonicated in PBS. The cell homogenate was incubated at room temperature for 15 min and homogenized twice with a Polytron (Brinkmann) for 30 s each at 4°C. Centrifugation at 30,000 g for 30 min afforded the supernatant containing the soluble proteins. The obtained supernatant was then applied to a prepacked Glutathione Sepharose 4B column (Amersham). The column was extensively washed with buffer and then the GST-hCA III fusion protein was eluted with a buffer consisting of 5 mM reduced glutathione in 50 mM Tris-HCl pH 8.0. Finally the GST part of the fusion protein was cleaved with thrombin Citation22–24. The advantage of this method is that hCA III is purified easily and the procedure is quite simple. The obtained hCA III was further purified by prontosil affinity column chromatography [Citation25], the elution being achieved with sodium azide 5 mM in 50 mM Hepes-HCL pH 7.5 buffer. The amount of enzyme was determined by spectrophotometric measurements and its activity by stopped-flow experiments, with CO2 as substrate [Citation26].
Human CA I and CA II cDNAs were expressed in E.coli strain BL21 (DE3) from the plasmids pACA/hCA I and pACA/hCA II as described earlier [Citation22].
CA catalytic/inhibition assay
An SX.18MV-R Applied Photophysics stopped-flow instrument has been used for assaying the CA I, II and III CO2 hydration activity [Citation26]. Phenol red (at a concentration of 0.2 mM) has been used as indicator, working at the absorbance maximum of 557 nm, with 10 mM Hepes (pH 7.5) as buffer, 0.1 M NaClO4 (for maintaining constant the ionic strength–this anion is not inhibitory anyhow) [Citation26], following the CA-catalyzed CO2 hydration reaction for a period of 10–100 s. Saturated CO2 solutions in water at 20°C were used as substrate. Stock solutions of inhibitors were prepared at a concentration of 10–50 mM (in the assay buffer) and dilutions up to 0.1 μM done with the assay buffer mentioned above. Enzyme concentrations were 0.09 μM for CA I, 0.06 μM for CA II and 0.10 μM for CA III. Kinetic parameters and inhibition constants were calculated as described in refs. Citation22–24.
Results and discussion
hCA III cloning, sequence and purification
To date, the complete amino acid sequences in the open reading frame of three hCA III clones have been deposited in GenBank (accession numbers BC004897, NM(005181 and AK096880). The amino acid sequence of our clone was identical to the other three clones mentioned above, except for one amino acid substitution at position 70, i.e., Phe(TTT):Ser(TCT). Another amino acid substitution was found in the NM(005181 clone at position 31, i.e., Ile(ATT):Val(GTT). Considering the fact that these amino acids are not within the active site cavity of this enzyme, they are probably normal (neutral) polymorphic substitutions.
The amino acid sequence deduced from the cDNA sequence of our hCA III clone, was aligned with that of other cytoplasmic CA isozymes, i.e., hCA I and hCA II (). hCA III shows a sequence similarity of 55% with hCA I and of 58% with hCA II. In , the previously defined thirty-six CA active site amino acid residues [Citation27] are indicated by a mixture of asterisk, “plus” and “z” signs above the hCA I sequence. Among these residues, 24 amino acids are conserved between hCA III and hCA II. These two isozymes are those with the lowest and highest activity for the CO2 hydration Citation1–5, respectively, among the mammalian CAs (). Among these active site residues seventeen are known to form a network of hydrogen bonds (they are indicated by “plus” and “z”; the latter indicating the three zinc-liganded His residues, i.e., His94, 96 and 119) important for the binding of the substrates, inhibitors and activators [Citation27,Citation28]. 15 of these amino acids are conserved between hCA III and hCA II. However, two of the remaining three amino acids (i.e., the residue 64, which is His in CA II and I, and Lys in CA III, and 198, which is Leu in CA II and I, and Phe in CA III, respectively), play a very important role in catalysis/binding of inhibitors, and they may explain the tremendous differences between these proteins Citation1,Citation2,Citation4,Citation5–7,Citation28. Thus, His64 acts as a proton shuttle residue in the catalytically active CA isoforms (such as among others CA I, II, IV, VI, VII, IX, XII, XIII and XIV) Citation1–4,Citation24,Citation28, favoring the transfer of a proton from the zinc-bound water molecule to the reaction medium, with formation of the nucleophilic, zinc-hydroxide species of the enzyme (this is the rate-determining step of the entire catalytic cycle for the CO2 hydration reaction catalyzed by these enzymes) [Citation4]. Lys64 present in CA III is less efficient as a proton shuttling residue as compared to His, due to the inappropriate pKa of the ϵ-NH2 moiety of this residue (pKa around 9) as compared to the imidazole of a histidine (pKa around 7) [Citation4]. On the other hand, the residue in position 198 is situated just in the middle of the active site cavity Citation28–31. When this residue is a relatively compact Leu (such as in CA I and II), there is enough space for the binding of inhibitors (and substrates), as shown by detailed X-ray crystallographic studies from this and other laboratories Citation28–31. However, the bulky Phe198 present in CA III, unlike Leu198, interferes with the binding of most inhibitors/substrates, due to the steric impairment engendered by the phenyl moiety of the Phe residue. As a consequence of these two factors, CA III presents a quite low catalytic activity as compared to CA II (and also CA I), and is difficultly inhibited by most sulfonamide CA inhibitors Citation1,Citation2,Citation4–7.
Figure 1. Alignment of the amino acid sequence of isoform CA III with that of isozymes CA I and II (CA I numbering system used). Thirty-six active site residues previously defined as forming the active site [Citation27] are indicated by a mixture of asterisk, “plus” and “z” signs above the CA I sequence. Seventeen residues known to participate in a network of hydrogen bonds and being involved in the binding of inhibitors/activators [Citation28] are indicated by “plus” and “z” above the sequence; the latter sign indicates the three zinc-liganded histidine residues (His94, 96 and 119). Conserved amino acids in the three isoforms are indicated by a closed box.
![Figure 1. Alignment of the amino acid sequence of isoform CA III with that of isozymes CA I and II (CA I numbering system used). Thirty-six active site residues previously defined as forming the active site [Citation27] are indicated by a mixture of asterisk, “plus” and “z” signs above the CA I sequence. Seventeen residues known to participate in a network of hydrogen bonds and being involved in the binding of inhibitors/activators [Citation28] are indicated by “plus” and “z” above the sequence; the latter sign indicates the three zinc-liganded histidine residues (His94, 96 and 119). Conserved amino acids in the three isoforms are indicated by a closed box.](/cms/asset/03b7a2a9-37ac-43f3-9a97-b76334008966/ienz_a_290880_f0001_b.gif)
Table I. Kinetic parameters for the CO2 hydration reaction catalysed by the recombinant cytosolic hCA isozymes I-III, at 20°C and pH 7.5, and their inhibition data with acetazolamide AAZ(5-acetamido-1,3,4-thiadiazole-2-sulfonamide), a clinically used drug [Citation1].
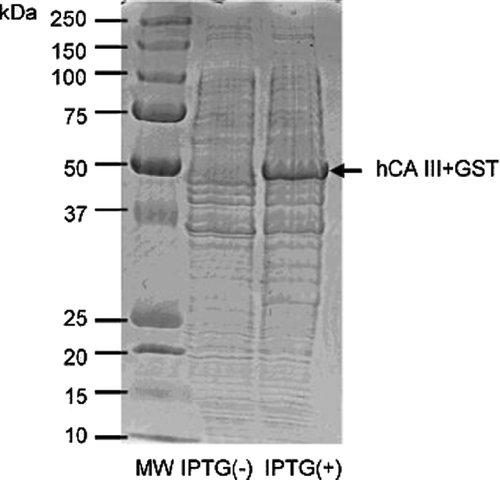
A GST-hCA III fusion protein construct has been then obtained by the procedure already described by us for the production of other isoforms such as hCA VB, VI; IX and XII among others Citation22–24. This fusion protein with the molecular weight of 50 kDa () has been thereafter purified in two steps by affinity chromatography: the first one involved a Glutathione Sepharose 4B column (which binds the GST part of the fusion protein with high affinity), followed by cleavage of the GST part by thrombin Citation22–24. The second step consisted in sulfonamide affinity chromatography, which was actually complicated by the low affinity of hCA III for the normally used columns for purification of other CA isoforms, based on p-aminomethyl-benzenesulfonamide derivatized columns Citation22–24. However, hCA III has high enough affinity for prontosil-based affinity columns [Citation25], and we used such a column for purification of our protein. Elution of hCA III from the column was then achieved with sodium azide (which is a rather strong CA III inhibitor, see later in the text), and extensive dialysis in Hepes buffer afforded the pure protein in rather good yield (4.5 mg protein/L of culture), as shown by the SDS PAGE of , in which the molecular weight of CA III is of 29 kDa, as reported in the literature [Citation7].
hCA III catalytic activity
Since all CA III preparations reported so far in the literature were isolating the enzyme from various organs, such as muscle or liver Citation5–7, sometimes involving rather harsh treatments which potentially lead to protein denaturation/unfolding, we were interested to measure the kinetic parameters for the physiologic reaction (CO2 hydration to bicarbonate) catalyzed by our enzyme, which has been produced in much milder, non-denaturating conditions. These parameters are shown in , where data for the other cytosolic, recombinant isozymes hCA I and II are also included for comparison.
Data of show that the recombinant hCA III produced by us is indeed a very poor catalyst for CO2 hydration (kcat of 1.3 × 104 s− 1) as compared to the highly active hCA II (kcat of 1.4 × 106 s− 1) or the slower hCA I (kcat of 2.0 × 105 s− 1). Indeed, the Km for CO2 of this isoform (hCA III) is higher as compared to those of the other two related isozymes, which is clearly reflected in the kcat/Km values presented in . Considering these values, hCA III shows 0.16% of the catalytic activity of hCA II (the best catalyst among all known CAs Citation1–4) and 0.50% of the catalytic activity of hCA I. It is difficult to explain why Nature preserved during evolution such a “bad” catalyst or CO2 hydration, when the much more efficient and highly abundant hCA I and II were clearly available (in addition to the remaining 9 other catalytically active human CAs). Thus, our data reinforce the idea that probably CA III has different physiological functions which are not connected to its catalytic function for CO2 hydration Citation1–4. It may be also seen that whereas hCA I and II are inhibited by the clinically used sulfonamide CA inhibitor acetazolamide (5-acetamido-1,3,4-thiadiazole-2-sulfonamide), with inhibition constants in the range of 12–250 nM, hCA III has a much weaker affinity for this compound, with an inhibition constant of 200 μM. As outlined above, this is probably due to the presence of the bulky Phe198 in the middle of the hCA III active site, which interferes with the binding of compounds possessing an organic scaffold attached to the sulfonamide zinc-binding moiety.
hCA III inhibition by anions
Data of show that similarly to isozymes hCA I and II previously investigated [Citation32,Citation33], hCA III is also susceptible to inhibition by metal-complexing anions, which being less bulky than the organic sulfonamides, may have an easier access to the catalytically vital Zn(II) ion where most CA inhibitors bind Citation1–4,Citation28–31. The following three anion categories are established from the data of : (i) Fluoride, nitrate, nitrite, sulfamic acid (as sulfamate anion), phenylboronic- and phenylarsonic acid act as very weak hCA III inhibitors, with KIs in the range of 21.3–117 mM. Whereas fluoride is also a very weak hCA I and II inhibitor, the other anions act as more efficient inhibitors against these two cytosolic isozymes as compared to hCA III (e.g., sulfamic acid, whose X-ray crystal structure in adduct with hCA II has been reported [Citation34] is a 1766-times more effective hCA I and an 80-times better hCA II than hCA III inhibitor). (ii) Another group of anions, including the remaining heavier halides, cyanate, bicarbonate, bisulfite, sulfate and sulfamide, showed a better inhibitory activity against hCA III, with inhibition constants in the range of 0.57–1.09 mM. It may be observed that the three halides (chloride, bromide and iodide) showed a very similar inhibitory activity against hCA III, whereas their affinities for hCA I and II vary considerably with the atomic weight of the halogen. Bisulfite, sulfate and sulfamide also showed quite comparable inhibitory power against hCA III, all of them with KIs of around 1 mM. Particularly interesting is the sulfate activity, which acts as a relatively potent hCA III inhibitor but it is a much weaker hCA I and II inhibitor (KIs of 63 - >200 mM). It is also interesting to note that our data for hCA III inhibition with sulfate are in very good agreement with the bCA III (b = bovine enzyme) inhibition data reported by Rowlett et al [Citation5] who found basically the same KI as the one reported by us. However our data greatly disagree with the proposal of these scientists [Citation5] regarding the effect of the pKa of the anion (actually the conjugated acid of these bases) on its behaviour either as CA III inhibitor or activator. In fact Rowlett et al. claimed that dianions with a pKa (of the conjugated acid) around 7 act as CA III activators, whereas those derived from stronger acids (for example sulfate, oxalate, etc) act as CA III inhibitors, and that their binding site within the cavity is situated somewhere near Lys64, not interacting thus with the zinc ion. We wish to stress that this is not true for any cytosolic CAs examined so far. For example, we demonstrated by means of X-ray crystallography [Citation34] that the very strong acid sulfamic acid (pKa 1.2) as well as the very weak one sulfamide (pKa of 12) bind very similarly to each other (as anions) to the CA II active site, coordinating to the Zn(II) ion by means of their deprotonated NH2 moiety, and that both of them behave as relatively weak hCA II (and also hCA III) inhibitors (see ). We also did not observe any hCA III activating properties for the few dianions we investigated here such as carbonate or sulfate, or such as silicate, molybdate or wolframate investigated earlier against isoforms hCA I, II, IV, VA and IX [Citation35]. In fact such dianions always acted as CA inhibitors and not activators. (iii) The third group of anions, including cyanide, thiocyanate, azide, carbonate and hydrosulfide, behave as potent hCA III inhibitors, with inhibition constants in the range of 10–90 μM. It is quite unexpected that the best anion inhibitor that we detected is carbonate, which is a 74-times better hCA III inhibitor than bicarbonate. It is difficult to find an interpretation to these data both from the biochemical and physiological viewpoints, also considering the fact that this ion has an affinity of 15–73 mM for hCA I and II and is in equilibrium with one of the substrates of this enzyme, i.e., bicarbonate (which shows a KI of 0.74 mM against hCA III, of 12 mM against hCA I and of 85 mM against hCA II). The remaining potent hCA III anion inhibitors belong to the well known type of “metal poisons”, i.e., anions possessing a high affinity for complexing heavy metal ions in solution or in metalloenzyme active sites, such as cyanide, azide, thiocyanate or hydrosulfide [Citation36].
Table II. Inhibition of recombinant isozymes hCA I, II and III with anions by a stopped-flow kinetic assay monitoring the CO2 hydration reaction, at 20°C and pH 7.5 [Citation26].
In conclusion, we report here the cloning and purification of hCA III. The enzyme shows low catalytic activity as compared to other cytosolic isoforms (such as hCA I and II), and a very characteristic inhibition profile with physiologic and non-physiologic anions. Fluoride, nitrate, nitrite, phenylboronic acid and phenylarsonic acid (as anions) were weak hCA III inhibitors (KIs of 21–78.5 mM), whereas bicarbonate, chloride, bromide, sulfate and several other simple anions showed KIs around 1 mM. The best hCA III inhibitors were carbonate, cyanide, thiocyanate, azide and hydrosulfide, which showed KIs in the range of 10–90 μM. It is difficult to explain the stronger inhibitory activity of carbonate (KI of 10 μM) against hCA III, also considering the fact that this ion has an affinity of 15–73 mM for hCA I and II and is in equilibrium with one of the substrates of this enzyme, i.e., bicarbonate, which is a much weaker inhibitor (KI of 0.74 mM against hCA III, of 12 mM against hCA I and of 85 mM against hCA II).
Acknowledgements
This work was financed in part by a EU project of the 6th framework programme (DeZnIT project, contract No. LSHB-CT-2007-037303).
References
- CT Supuran, A Scozzafava, and A Casini. (2003). Carbonic anhydrase inhibitors. Med Res Rev 23:146–189.
- S Pastorekova, S Parkkila, J Pastorek, and CT Supuran. (2004). Carbonic anhydrases: Current state of the art, therapeutic applications and future prospects. J Enz Inhib Med Chem 19:199–229.
- M Hilvo, CT Supuran, and S Parkkila. (2007). Characterization and inhibition of the recently discovered carbonic anhydrase isoforms CA XIII, XIV and XV. Curr Top Med Chem 7:893–899.
- CT Supuran, and A Scozzafava. (2007). Carbonic anhydrases as targets for medicinal chemistry. Bioorg Med Chem 15:4336–4350.
- RS Rowlett, NJ GargiuloIII, FA Santoli, JM Jackson, and AH Corbett. (1991). Activation and inhibition of bovine carbonic anhydrasde III by dianions. J Biol Chem 266:933–941.
- AE Eriksson, and A Liljas. (1993). Refined structure of bovine carbonic anhydrase III at 2.0 A resolution. Proteins 16:29–42.
- PJ Wistrand. (2002). Carbonic anhydrase III in liver and muscle of male rats purification and properties. Ups J Med Sci 107:77–88.
- M Liu, GA Walter, NC Pathare, RE Forster, and K Vandenborne. (2007). A quantitative study of bioenergetics in skeletal muscle lacking carbonic anhydrase III using 31P magnetic resonance spectroscopy. Proc Natl Acad Sci USA 104:371–376.
- L Li, and CN Falany. (2007). Elevated hepatic SULT1E1 activity in mouse models of cystic fibrosis alters the regulation of estrogen responsive proteins. J Cyst Fibros 6:23–30.
- T Yamamoto, R Kikkawa, H Yamada, and I Horii. (2006). Investigation of proteomic biomarkers in in vivo hepatotoxicity study of rat liver: Toxicity differentiation in hepatotoxicants. J ToxicolSci 31:49–60.
- UJ Zimmerman, P Wang, X Zhang, S Bogdanovich, and R Forster. (2004). Anti-oxidative response of carbonic anhydrase III in skeletal muscle. IUBMB Life 56:343–347.
- SR Räisänen, P Lehenkari, M Tasanen, P Rahkila, PL Härkönen, and HK Väänänen. (1999). Carbonic anhydrase III protects cells from hydrogen peroxide-induced apoptosis. FASEB J 13:513–522.
- GA Gill, N Johnston, A Buda, M Pignatelli, J Pearson, PW Dettmar, and J Koufman. (2005). Laryngeal epithelial defenses against laryngopharyngeal reflux: Investigations of E-cadherin, carbonic anhydrase isoenzyme III, and pepsin. Ann Otol Rhinol Laryngol 114:913–921.
- G Kim, and RL Levine. (2005). Molecular determinants of S-glutathionylation of carbonic anhydrase III. Antioxid Redox Signal 7:849–854.
- RJ Mallis, MJ Hamann, W Zhao, T Zhang, S Hendrich, and JA Thomas. (2002). Irreversible thiol oxidation in carbonic anhydrase III: Protection by S-glutathiolation and detection in aging rats. Biol Chem 383:649–662.
- CJ Lynch, WA Brennan, TC Vary, N Carter, and SJ Dodgson. (1993). Carbonic anhydrase III in obese Zucker rats. Am J Physiol 264:E621–E630.
- A Alver, F Uçar, EE Keha, E Kalay, and E Ovali. (2004). Effects of leptin and insulin on CA III expression in rat adipose tissue. J Enz Inhib Med Chem 19:279–281.
- A Alver, EE Keha, F Uçar, and E Ovali. (2004). The effect of carbonic anhydrase inhibition on leptin secretion by rat adipose tissue. J Enz Inhib Med Chem 19:181–184.
- CT Supuran. (2003). Carbonic anhydrase inhibitors in the treatment and prophylaxis of obesity. Expert Opin Ther Pat 13:1545–1550.
- G De Simone, and CT Supuran. (2007). Antiobesity carbonic anhydrase inhibitors. Curr Top Med Chem 7:879–884.
- N Carter, A Shiels, and R Tashian. (1978). Carbonic anhydrase III isoenzyme from human and bovine muscle. Biochem Soc Trans 6:552–553.
- I Nishimori, D Vullo, A Innocenti, A Scozzafava, A Mastrolorenzo, and CT Supuran. (2005). Carbonic anhydrase inhibitors. The mitochondrial isozyme VB as a new target for sulfonamide and sulfamate inhibitors. J Med Chem 48:7860–7866.
- S Pastorekova, D Vullo, A Casini, A Scozzafava, J Pastorek, I Nishimori, and CT Supuran. (2005). Carbonic anhydrase inhibitors: Inhibition of the tumor-associated isozymes IX and XII with polyfluorinated aromatic/heterocyclic sulfonamides. J Enz Inhib Med Chem 20:211–217.
- D Vullo, A Innocenti, I Nishimori, J Pastorek, A Scozzafava, S Pastorekova, and CT Supuran. (2005). Carbonic anhydrase inhibitors. Inhibition of the transmembrane isozyme XII with sulfonamides-a new target for the design of antitumor and antiglaucoma drugs?. Bioorg Med Chem Lett 15:963–969.
- I Nishimori, T Minakuchi, S Onishi, D Vullo, A Cecchi, A Scozzafava, and CT Supuran. (2007). Carbonic anhydrase inhibitors: Cloning, characterization, and inhibition studies of the cytosolic isozyme III with sulfonamides. Bioorg Med Chem 15:7229–7236.
- RG Khalifah. (1971). The carbon dioxide hydration activity of carbonic anhydrase. I. Stop-flow kinetic studies on the native human isoenzymes B and C. J Biol Chem 246:2561–2573.
- RE Tashian. (1992). Genetics of the mammalian carbonic anhydrases. Adv. Genet 30:321–356.
- CT Supuran, A Scozzafava, and J Conway. Carbonic Anhydrase – Its Inhibitors and Activators. Boca Raton, New York, London: CRC Press; (2004). p 1–363.
- PA Boriack-Sjodin, S Zeitlin, HH Chen, L Crenshaw, S Gross, A Dantanarayana, P Delgado, JA May, T Dean, and DW Christianson. (1998). Structural analysis of inhibitor binding to human carbonic anhydrase II. Protein Sci 7:2483–2489.
- G De Simone, A Di Fiore, V Menchise, C Pedone, J Antel, A Casini, A Scozzafava, M Wurl, and CT Supuran. (2005). Carbonic anhydrase inhibitors. Zonisamide is an effective inhibitor of the cytosolic isozyme II and mitochondrial isozyme V: Solution and X-ray crystallographic studies. Bioorg Med Chem Lett 15:2315–2320.
- C Temperini, A Scozzafava, and CT Supuran. (2006). Carbonic anhydrase activators: The first X-ray crystallographic study of an adduct of isoform I. Bioorg Med Chem Lett 16:5152–5156.
- D Vullo, M Franchi, E Gallori, J Pastorek, A Scozzafava, S Pastorekova, and CT Supuran. (2003). Carbonic anhydrase inhibitors. Inhibition of cytosolic isozymes I and II and transmembrane, cancer-associated isozyme IX with anions. J Enz Inhib Med Chem 18:403–406.
- A Innocenti, JM Lehtonen, S Parkkila, A Scozzafava, and CT Supuran. (2004). Carbonic anhydrase inhibitors. Inhibition of the newly isolated murine isozyme XIII with anions. Bioorg Med Chem Lett 14:5435–5439.
- F Abbate, CT Supuran, A Scozzafava, P Orioli, MT Stubbs, and G Klebe. (2002). Nonaromatic sulfonamide group as an ideal anchor for potent human carbonic anhydrase inhibitors: Role of hydrogen-bonding networks in ligand binding and drug design. J Med Chem 45:3583–3587.
- A Innocenti, D Vullo, A Scozzafava, and CT Supuran. (2005). Carbonic anhydrase inhibitors. Inhibition of isozymes I, II, IV, V and IX with anions isosteric and isoelectronic with sulfate, nitrate and carbonate. Bioorg Med Chem Lett 15:567–571.
- MA Ilies, and MD Banciu. Nonsulfonamide carbonic anhydrase inhibitors In: CT Supuran, A Scozzafava, and J Conway. editors. Carbonic anhydrase - Its inhibitors and activators. Boca Raton (FL), USA: CRC Press; (2004). p 209–242.