Abstract
Among protein families, carbonic anhydrases (CAs, EC 4.2.1.1) are metalloenzymes characterized by a common reaction mechanism in all life domains: the carbon dioxide hydration to bicarbonate and protons (CO2+H2O ⇔ HCO3−+H+). Six genetically distinct CA families are known to date, the α-, β-, γ-, δ-, ζ- and η-CAs. The last CA class was recently discovered analyzing the amino acid sequences of CAs from Plasmodia. Bacteria encode for enzymes belonging to the α-, β-, and γ-CA classes and recently, phylogenetic analysis revealed an interesting relationship regarding the evolution of bacterial CA classes. This result evidenced that the three bacterial CA classes, in spite of the high level of the structural similarity, are evolutionarily distinct, but we noted that the primary structure of some β-CAs identified in the genome of Gram-negative bacteria present a pre-sequence of 18 or more amino acid residues at the N-terminal part. These observations and subsequent phylogenetic data presented here prompted us to propose that the β-CAs found in Gram-negative bacteria with a periplasmic space and characterized by the presence of a signal peptide might have a periplasmic localization and a role similar to that described previously for the α-CAs.
Introduction
Protein evolution
In the course of evolution, protein primary structure was subject to modifications due to a series of events, such as mutations, substitutions, insertions, deletions and other rearrangements of the genomic materialCitation1. Generally, to avoid the loss of the most important protein features, the aforementioned modifications did not change residues located in those positions considered important for the function, stability and folding of the macromolecule. Proteins having similar structure and function but a lower sequence similarity (e.g. <30% sequence identity) are grouped together into broader evolutionary families or superfamiliesCitation1. Sometimes, it is very difficult to classify and characterize the evolutionary relationship of the individual protein family members due to several reasons: (a) two evolutionarily-related proteins appear similar because they descend with divergence from a common ancestor (i.e. they are homologous); (b) the two relatives come from two different ancestral genes but have both converged on the same structural arrangement or fold (i.e. they are analogous)Citation1,Citation2. As a consequence, proteins sharing structural similarity but lacking of distinctive sequence characteristics might be related by a divergent evolution from a common ancestor or by a convergent evolution from two different ancestorsCitation2. As reported in literature, gene duplication represents an important and frequent event during evolution. It may give rise to orthologous genes if they encode for proteins retaining their function (because this is important for the integrity of the organism), but it may lead to paralog genes, if one of the duplicate genes keeps the original function whereas the other one acquires a new protein function, which brings a benefit to the organism. This means that orthologs retain the same function in the course of evolution, whereas paralogs evolve new functions, even if these are related to the original oneCitation2.
Carbonic anhydrase family
At the highest level in a structural classification, proteins are grouped to the same class, if they have similar secondary structure, compositions and packing. Among protein families, carbonic anhydrases (CAs, EC 4.2.1.1) are metalloenzymes catalyzing a common reaction in all life domains: the carbon dioxide hydration to bicarbonate and protons (CO2+H2O ⇔ HCO3−+H+)Citation3. These macromolecules are grouped in different classes mainly on the basis of their structural fold and arrangement of the active site residues. In fact, six genetically distinct CA families are known to date, the α-, β-, γ-, δ-, ζ- and η-CAsCitation3–6. The last CA class was discovered recently analyzing the amino acid sequences of CAs from PlasmodiaCitation5,7–11. The α-, β-, δ- and η-CAs use Zn(II) ions at the active site, the γ-CAs are probably Fe(II) enzymes (but they are active also with bound Zn(II) or Co(II) ions), whereas the ζ-class are cambialistic enzymes, active both with Cd(II) or Zn(II) bound within the active siteCitation3,4,12–19. The metal ion from the enzyme active site is coordinated by three histidine (His) residues (in the α-, γ- δ- class enzymes) or by one His, and two cysteine (Cys) residues (in the β- and ζ-CAs), with the fourth ligand being a water molecule/hydroxide ion acting as nucleophile in the catalytic cycle of the enzyme. Although η-CAs retain many structural features of the α-class, they present a distinctive zinc coordination mode, making their catalytic site unique and different from that found in the other CAs and thus justifying their classification in a new class. Our findings have shown that the metal ion coordination pattern of this η-CAs involved two His and one glutamine (Gln) residues, in addition to the water molecule/hydroxide ion acting as nucleophile in the catalytic cycleCitation20. Some of the catalytically active α-CAs also catalyze the hydrolysis of esters/thioesters (e.g. 4-nitrophenyl acetate (4-NpA) hydrolysis as well as other hydrolytic reactions). However, no esterase activity was detected so far for enzymes belonging to the other five CA genetic families. The 3D fold of the five CA classes is very different: α-CAs are normally monomers and rarely dimersCitation21–23; β-CAs are dimersCitation24, tetramers or octamers; γ-CAs are trimers. The only ζ-CA crystallized so far has three slightly different active sites on the same polypeptide chain, whereas no X-ray crystal structures of δ- and η-and CAs are available so far.
Results and discussion
Bacterial carbonic anhydrase evolution
Bacteria encode for enzymes belonging to the α-, β-, and γ-CA classes and recently, we have conducted a thorough phylogenetic analysis to better understand the evolutionary relationship among the bacterial CA classes ()Citation25–37. reports the list of the bacterial species and the accession numbers of the amino acid sequences of CAs examined for constructing the phylogenetic tree of . The dendrogram was obtained using PhyML 3.0 program and applying the neighbor-joining method (NJ)Citation38, whose topology has been also confirmed by the maximum likelihood (ML) approachCitation39. The results of the phylogenetic analysis showed the presence of well-supported nodes characterizing two distinct clades. Starting from the bottom of the tree, the first clade included bacterial γ-CAs, whereas the second large clade (top of the ) contained β- and α-CAs. This inferred phylogenetic analysis has evidenced that the three bacterial CA classes are evolutionarily distinct, in spite of the rather high level of the structural similarity that characterizes the CA protein family (at least considering the active site architecture). α-CAs, in fact, formed a distinct cluster, which was positioned in the same cluster of the β-CAs. We have hypothesized earlierCitation3 that these two classes have arisen from the γ-CAs cluster (). Probably, the common ancestor was subject to an event of gene duplication, which separated γ-CAs from β-CAs. Subsequently, another duplication event led to the α-CAs starting from β-CAs (). Here, the set of the homologous amino acid sequences of CAs are considered to descend from a common ancestor with divergent sequences, because the primary structure of the three classes shows different hallmarks, but with a convergent function because all the CA classes are mainly involved in the reversible hydration reaction of CO2. This information prompted us to speculate that the most ancestral CA is the γ-class, which was nominated as the “Ur-CA” (Ur in German means the ancestral form)Citation3. In fact, although the γ class is widely distributed among all three domains of life, it is the only CA class mainly identified in Archaea, the most ancient microorganisms that exist on earth. The phylogenetic analysis is corroborated by the promiscuity theory, which is a key factor in the evolution of a new protein functionCitation40,Citation41. Looking at the substrate used by γ-, β- and α-CAs, we noticed that γ-CAs use only CO2 as substrate, β-CAs can hydrolyze CO2, COS and CS2, whereas the α-CAs not only hydrate CO2, CS2, COS, cyanamide and cyanate, but also possess esterase activity, with a range of esters of carboxylic, sulfonic or phosphate estersCitation27,32,42–47. We conclude that α-CA is the class which acquires a high catalytic versatility resulting in the most recent form as suggested by the phylogenetic analysis.
Figure 1. Phylogenetic analysis was obtained using all three classes of CAs identified in the genome of Gram-positive and Gram-negative bacteria reported in . It was carried out using PhyML program. Bootstrap values on 100 replicates are reported at branch points.
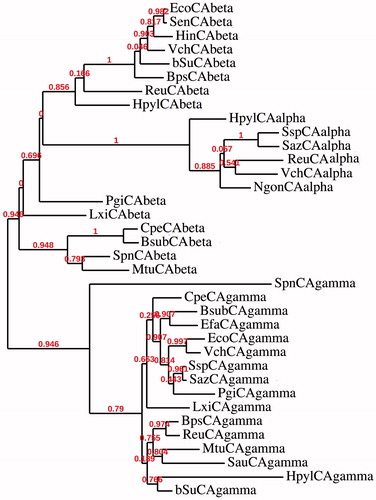
Table 1. List of the proteins, organisms, accession numbers and cryptonyms of the sequences used in the phylogenetic analysis.
New insight in the β-CAs evolution
A common feature of all bacterial α-CAs known to date is the presence of an N-terminal signal peptide, which suggests a periplasmic or extracellular location and a possible physiological role in CO2 uptake processesCitation3. Recently, our groups described that the bacterial α-CAs are mainly localized in the periplasmic space of the Gram-negative bacteria and their function is to convert the diffused CO2 into bicarbonate necessary for the bacterial metabolismCitation3. On the contrary, β- or γ-classes have a cytoplasmic localization and are responsible of CO2 supply for carboxylase enzymes, pH homeostasis and other intracellular functionsCitation3. The most interesting aspect appearing from the phylogenetic analysis reported in was the amino acid sequence of the β-CA identified in the genome of Porphyromonas gingivalis (PgiCAbeta), a Gram-negative oral pathogenic bacteriumCitation29,30,33,48–53. This enzyme can be considered as a “turning sequence”, which led to the bacterial α-CAs, which are present only in Gram-negative bacteria. This observation prompted us to investigate the primary structure of the β-CA encoded by the genome of Porphyromonas gingivalis (). The full nucleotide sequence showed an open reading frame encoding a 242 residues polypeptide chain which contained all the typical features of a β-CA, including the three residues Cys95, Cys151 and His148, which are involved in the catalytic mechanism of the enzyme (as they coordinate the Zn(II) ion), and the residue of the catalytic dyad Asp97 and Arg99 involved in the activation of the metal ion coordinated water molecule (). From the previous alignment of the bacterial β-CAs, we noted that the primary structure of PgiCAbeta had a long stretches of 18 amino acids, as indicated in . Interesting, this pre-sequence was present in other, although not in all β-CAs identified in the genome of Gram-negative bacteria. This observation evoked our curiosity and a strong interest in the automated determination of a possible signal peptide in β-CAs, similar to that identify in the α-CA class enzymes. The secretory signal peptide is a protein signal that targets its passenger protein for translocation across the cytoplasmic membrane in prokaryotesCitation54. Here, we used SignalP version 4.1 (Center for Biological Sequence Analysis, Kongens Lyngby, Denmark), which is a program designed to discriminate between signal peptides and transmembrane regions of proteins. The program is available as a web tool at http://www.cbs.dtu.dk/services/SignalP/Citation55. The obtained result was that PgiCAbeta (encoded by the genome of P. gingivalis) possessed a putative signal peptide of 18 amino acids at its N-terminal amino acid sequence and a predicted cleavage site located between Gly18 and Asn19 (). The two residues involved in the predicted cleavage were indicated as P1′ (the residue just before the cleavage) and P1 (the residue after the cleavage). Proteins have specific amino acids in the cleavage sites. For example, in bacteria P1′ is mostly occupied by these residues: Ala (36–41%), Asp (7–11%), Ser (11%) and Glu (6–10%)Citation56. Following the automated identification of the signal peptide of the α-CAs identified in Gram-negative bacteria, such as HpyCA (from Helicobacter pylori), VchCA (from Vibrio cholerae), SspCA (from Sulfurihydrogenibium yellowstonense), NgonCA (from Neisseria gonorrhoeae), SsalCA (from Streptococcus salivarius), and ReuCA (from Ralstonia eutropha) we noted that the predicted cleavage site showed in P1′ was always an Ala residue (). This observation is in good agreement with the typical periplasmic localization of the α-CAs. In fact, the genome of H. pylori encodes for three CA classes: α-, β- and γ-CACitation57. The α-CA was shown to possess a periplasmic localization, the β-CA has been found in the cytoplasm, whereas no information is available on the expression/localization of the γ-CA from this pathogenic bacteriumCitation3,4,15,58,59. It was suggested that the activity of periplasmic α- and β-CAs could be an additional requirement for the gastric colonization of the bacteriumCitation57,Citation60,Citation61. We want stress the fact that the primary sequence of the β-CA (HpyCAβ) identified in the genome of H. pylori did not show a signal peptide at its N-terminal region when subject to the automated identification of the predicted signal peptide. In fact, HpyCA has a cytoplasmic localization, as described in literatureCitation57.
Figure 2. Nucleotide and amino acid sequence of β-CA from P. gingivalis. Amino acid residues Cys95, His148 and Cys151 are the typical residues of a β-CA participating in the coordination of metal ion, whereas Asp97 and Arg99 form the catalytic dyad involved in the activation of the metal ion coordinated water molecule. In red is reported the signal peptide. The asterisk (*) indicates the stop codon. P1′- P1 indicate the position of the predicted cleavage site.

Table 2. Identification of the predicted signal peptide in β- and α-CAs using the SignalP 4.1 program.
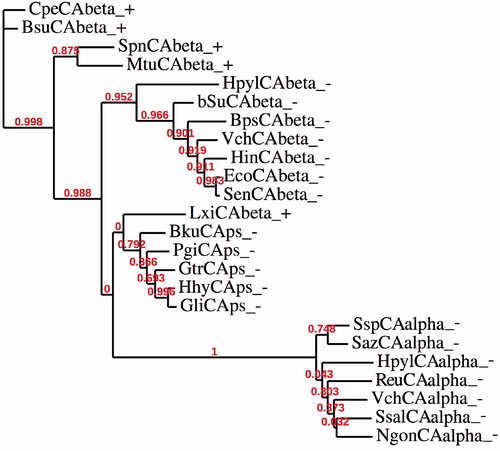
But now a new question arises: what is the cellular localization of the β-CAs characterized by the presence of a signal peptide, which up to date was considered to be a common feature of only α-CAs? In , we present a number of bacterial β-CAs which possess stretches of pre-sequences of 18 or more than 20 amino acid residues before the signal cleavage site. Among the β-CAs considered in this study, only three of them show in position P1′ an Ala residue, which is the most frequent one found in the P1′ cleavage site of bacterial proteins. These two such β-CAs, PgiCA and HhyCA, were identified in the genome of two Gram-negative bacteria Porphyromonas gingivalis and Haliscomenobacter hydrossis, respectively, and show a glycine or an asparagine residue in P1′. These two residues are never found in position P1′ of other bacterial proteins. We have hypothesized that β-CAs with an Ala residue in position P1′ (the same residue found in the P1′ position of the α-CA) could be able, likely the α-CAs, to move toward the periplasmic space, whereas β-CAs having a different residues in P1′ (residue different from Ala, Asp, Ser and Glu; see the text above), probably are not capable to cross the inner membrane of the periplasmic space. We drew this conclusion because Gruber et al. demonstrated that the substitution of the cleavage site motif, in particular when the Ala residues is replaced by a Gly (see cleavage sequence of PgiCAbeta in ), blocks the translocation of proteins to the plastidCitation62. Perhaps, this is the reason why most of the α-CAs show an Ala in the P1′ position ().
Thus, we have constructed a phylogenetic tree in order to better investigate the evolutionary relationship of β-CAs sequences found in Gram-negative bacteria and characterized by the presence of a signal peptide. The dendrogram shown in has been obtained by aligning the amino acid sequences of α-CAs with the amino acid sequences of β-CAs identified in the genome of Gram-positive and Gram-negative bacteria. Bacterial β-CAs having a predicted signal peptide were indicated adding “ps” near the cryptonym ( and ). The complete list of organisms, CA classes, accession numbers and cryptonyms used in the phylogenetic analysis are indicated in . shows that β-CAs from Gram-negative bacteria clustered in a distinct branch: one clade is made of α-CAs and β-CAs with “ps”, while the other clade contained β-CAs without a signal peptide. Interestingly, LxiCAbeta_+ seemed closely associated to the β-CAs “ps” but this CA did not show a predicted cleavage motif, as expected for a β-CAs from a Gram-positive bacteria because in these microorganism the periplasmic space is missing. Probably the β-CAs “ps” have arisen from an ancestor having the amino acid sequence similar to that of a β-CA identified in Gram-positive bacteria. This event might be due to a mutation, which introduced the cleavage site in the β-CAs “ps”. This hypothesis is in agreement with Gupta’s theory sustaining that Gram-positive bacteria occupy an intermediate position between Archaea and Gram-negative bacteria, and this latter group has evolved from themCitation63,Citation64.
Figure 3. Phylogenetic analysis was obtained using the amino acid sequences of α-CAs from Gram-negative bacteria and those of β-CAs identified in the genome of Gram-positive and Gram-negative bacteria. Legend: see and for sequence accession numbers, cryptonyms and microorganisms considered; + indicate a Gram-positive bacterium; − indicate a Gram-negative bacterium. Bootstrap values on 100 replicates are reported at branch points.
Conclusions
We propose here that the β-CAs found in bacteria with a periplasmic space (Gram-negative) and characterized by a signal peptide having an Ala residue (or, with lower frequency, Asp, Ser or Glu) in position P1′ of the cleavage site might have a periplasmic localization. In this case, probably, their physiological function is similar to that suggested for the α-CAs from Gram-negative bacteria, such as involvement in bacterial metabolism or in the acclimatization of the microorganisms to a hostile environment (as it is the case for Helicobacter pylori). Moreover, our non-mainstream analyses are thought in such a way as to give new inputs to the readers, evoking their curiosity in this fascinating world of the carbonic anhydrases.
Declaration of interest
The authors declare no conflict of interest.
References
- Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem 2005;74:867–900
- Makarova KS, Wolf YI, Koonin EV. Archaeal clusters of orthologous genes (arCOGs): an update and application for analysis of shared features between thermococcales, methanococcales, and methanobacteriales. Life (Basel) 2015;5:818–40
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32
- Capasso C, Supuran CT. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–9
- Capasso C, Supuran CT. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Targets 2015;19:1689–704
- De Luca V, Del Prete S, Supuran CT, Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:277–82
- Supuran CT, Capasso C. The η-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63
- Del Prete S, Vullo D, Fisher GM, et al. Discovery of a new family of carbonic anhydrases in the malaria pathogen Plasmodium falciparum – the eta-carbonic anhydrases. Bioorg Med Chem Lett 2014;24:4389–96
- Rodrigues GC, Feijo DF, Bozza MT, et al. Design, synthesis, and evaluation of hydroxamic acid derivatives as promising agents for the management of Chagas disease. J Med Chem 2014;57:298–308
- Syrjanen L, Vermelho AB, Rodrigues Ide A, et al. Cloning, characterization, and inhibition studies of a beta-carbonic anhydrase from Leishmania donovani chagasi, the protozoan parasite responsible for leishmaniasis. J Med Chem 2013;56:7372–81
- Pan P, Vermelho AB, Scozzafava A, et al. Anion inhibition studies of the alpha-carbonic anhydrase from the protozoan pathogen Trypanosoma cruzi, the causative agent of Chagas disease. Bioorg Med Chem 2013;21:4472–6
- Buzas GM, Supuran CT. The history and rationale of using carbonic anhydrase inhibitors in the treatment of peptic ulcers. In memoriam Ioan Puscas (1932–2015). J Enzyme Inhib Med Chem 2016;31:527–33
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs - antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87
- Supuran CT. Carbonic anhydrases-an overview. Curr Pharm Des 2008;14:603–14
- Supuran CT. Carbonic anhydrases: novel therapeutic applications for inhibitors and activators. Nat Rev Drug Discov 2008;7:168–81
- Supuran CT. Carbonic anhydrase inhibitors: an editorial. Expert Opin Ther Pat 2013;23:677–9
- Supuran CT. Acetazolamide for the treatment of idiopathic intracranial hypertension. Expert Rev Neurother 2015;15:851–6
- Supuran CT. How many carbonic anhydrase inhibition mechanisms exist? J Enzyme Inhib Med Chem 2016;31:345–60
- De Simone G, Di Fiore A, Capasso C, Supuran CT. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg Med Chem Lett 2015;25:1385–9
- De Simone G, Monti SM, Alterio V, et al. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2015;25:2002–6
- Di Fiore A, Capasso C, De Luca V, et al. X-ray structure of the first `extremo-alpha-carbonic anhydrase', a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9
- Lomelino CL, Mahon BP, McKenna R, et al. Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111. Bioorg Med Chem 2016;24:976–81
- Ferraroni M, Del Prete S, Vullo D, et al. Crystal structure and kinetic studies of a tetrameric type II β-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Acta Crystallogr D Biol Crystallogr 2015;71:2449–56
- Vullo D, De Luca V, Del Prete S, et al. Sulfonamide inhibition studies of the gamma-carbonic anhydrase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg Med Chem Lett 2015;25:3550–5
- De Luca V, Vullo D, Del Prete S, et al. Cloning, characterization and anion inhibition studies of a new gamma-carbonic anhydrase from the Antarctic bacterium Pseudoalteromonas haloplanktis. Bioorg Med Chem 2015;23:4405–9
- Dedeoglu N, De Luca V, Isik S, et al. Cloning, characterization and anion inhibition study of a beta-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorg Med Chem 2015;23:2995–3001
- Dedeoglu N, DeLuca V, Isik S, et al. Sulfonamide inhibition study of the beta-class carbonic anhydrase from the caries producing pathogen Streptococcus mutans. Bioorg Med Chem Lett 2015;25:2291–7
- Alafeefy AM, Ceruso M, Al-Tamimi AM, et al. Inhibition studies of quinazoline-sulfonamide derivatives against the gamma-CA (PgiCA) from the pathogenic bacterium, Porphyromonas gingivalis. J Enzyme Inhib Med Chem 2015;30:592–6
- Prete SD, Vullo D, Osman SM, et al. Sulfonamide inhibition study of the carbonic anhydrases from the bacterial pathogen Porphyromonas gingivalis: the beta-class (PgiCAb) versus the gamma-class (PgiCA) enzymes. Bioorg Med Chem 2014;22:4537–43
- Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of the delta-carbonic anhydrase from the marine diatom Thalassiosira weissflogii, TweCA. J Enzyme Inhib Med Chem 2014;29:906–11
- Vullo D, Sai Kumar RS, Scozzafava A, et al. Anion inhibition studies of a β-carbonic anhydrase from Clostridium perfringens. Bioorg Med Chem Lett 2013;23:6706–10
- Del Prete S, Vullo D, De Luca V, et al. A highly catalytically active gamma-carbonic anhydrase from the pathogenic anaerobe Porphyromonas gingivalis and its inhibition profile with anions and small molecules. Bioorg Med Chem Lett 2013;23:4067–71
- Del Prete S, Isik S, Vullo D, et al. DNA cloning, characterization, and inhibition studies of an alpha-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. J Med Chem 2012;55:10742–8
- Luca VD, Vullo D, Scozzafava A, et al. An alpha-carbonic anhydrase from the thermophilic bacterium Sulphurihydrogenibium azorense is the fastest enzyme known for the CO2 hydration reaction. Bioorg Med Chem 2013;21:1465–9
- Vullo D, De Luca V, Scozzafava A, et al. Anion inhibition studies of the fastest carbonic anhydrase (CA) known, the extremo-CA from the bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2012;22:7142–5
- Vullo D, Luca VD, Scozzafava A, et al. The alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1 is highly susceptible to inhibition by sulfonamides. Bioorg Med Chem 2013;21:1534–8
- Guindon S, Dufayard JF, Lefort V, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 2010;59:307–21
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 2003;52:696–704
- Sanchez-Tarin M, Swiderek K, Roca M, Tunon I. Enzyme promiscuity in enolase superfamily. Theoretical study of o-succinylbenzoate synthase using QM/MM methods. J Phys Chem B 2015;119:1899–911
- Alderson RG, De Ferrari L, Mavridis L, et al. Enzyme informatics. Curr Top Med Chem 2012;12:1911–23
- Tanc M, Carta F, Scozzafava A, Supuran CT. α-Carbonic anhydrases possess thioesterase activity. ACS Med Chem Lett 2015;6:292–5
- Scozzafava A, Supuran CT. Hydroxyurea is a carbonic anhydrase inhibitor. Bioorg Med Chem 2003;11:2241–6
- Guerri A, Briganti F, Scozzafava A, et al. Mechanism of cyanamide hydration catalyzed by carbonic anhydrase II suggested by cryogenic X-ray diffraction. Biochemistry 2000;39:12391–7
- Vullo D, Del Prete S, De Luca V, et al. Anion inhibition studies of the beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2016;26:1406–10
- De Luca V, Vullo D, Del Prete S, et al. Cloning, characterization and anion inhibition studies of a gamma-carbonic anhydrase from the Antarctic bacterium Colwellia psychrerythraea. Bioorg Med Chem 2016;24:835–40
- Del Prete S, Vullo D, Osman SM, et al. Anion inhibition studies of the dandruff-producing fungus Malassezia globosa β-carbonic anhydrase MgCA. Bioorg Med Chem Lett 2015;25:5194–8
- Ceruso M, Del Prete S, AlOthman Z, et al. Synthesis of sulfonamides with effective inhibitory action against Porphyromonas gingivalis gamma-carbonic anhydrase. Bioorg Med Chem Lett 2014;24:4006–10
- Del Prete S, De Luca V, Vullo D, et al. Biochemical characterization of the gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis, PgiCA. J Enzyme Inhib Med Chem 2014;29:532–7
- Vullo D, Del Prete S, Osman SM, et al. Sulfonamide inhibition studies of the gamma-carbonic anhydrase from the oral pathogen Porphyromonas gingivalis. Bioorg Med Chem Lett 2014;24:240–4
- Vullo D, Del Prete S, Osman SM, et al. Anion inhibition study of the beta-class carbonic anhydrase (PgiCAb) from the oral pathogen Porphyromonas gingivalis. Bioorg Med Chem Lett 2014;24:4402–6
- Del Prete S, De Luca V, Iandolo E, et al. Protonography, a powerful tool for analyzing the activity and the oligomeric state of the gamma-carbonic anhydrase identified in the genome of Porphyromonas gingivalis. Bioorg Med Chem 2015;23:3747–50
- Del Prete S, Vullo D, De Luca V, et al. Biochemical characterization of recombinant beta-carbonic anhydrase (PgiCAb) identified in the genome of the oral pathogenic bacterium Porphyromonas gingivalis. J Enzyme Inhib Med Chem 2015;30:366–70
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng 1997;10:1–6
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011;8:785–6
- Dhanaraj P, James JV, Michael PG, Muthiah I. SIGLOCPRED: an algorithm to predict bacterial signal peptides and OMPS. Bioinformation 2012;8:970–3
- Bury-Mone S, Mendz GL, Ball GE, et al. Roles of alpha and beta carbonic anhydrases of Helicobacter pylori in the urease-dependent response to acidity and in colonization of the murine gastric mucosa. Infect Immun 2008;76:497–509
- Capasso C, Supuran CT. An overview of the carbonic anhydrases from two pathogens of the oral cavity: Streptococcus mutans and Porphyromonas gingivalis. Curr Top Med Chem 2016. 16. [Epub ahead of print]. doi: 10.2174/1568026616666160413135522
- Supuran CT. Carbonic anhydrases as drug targets-an overview. Curr Top Med Chem 2007;7:825–33
- Compostella ME, Berto P, Vallese F, Zanotti G. Structure of alpha-carbonic anhydrase from the human pathogen Helicobacter pylori. Acta Crystallogr F Struct Biol Commun 2015;71:1005–11
- Marcus EA, Moshfegh AP, Sachs G, Scott DR. The periplasmic alpha-carbonic anhydrase activity of Helicobacter pylori is essential for acid acclimation. J Bacteriol 2005;187:729–38
- Gruber A, Vugrinec S, Hempel F, et al. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol 2007;64:519–30
- Gupta RS. Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among archaebacteria, eubacteria, and eukaryotes. Microbiol Mol Biol Rev 1998;62:1435–91
- Gupta RS. Protein phylogenies and signature sequences: evolutionary relationships within prokaryotes and between prokaryotes and eukaryotes. Antonie Van Leeuwenhoek 1997;72:49–61
