Abstract
Carbonic anhydrases (CAs, EC 4.2.1.1) are a superfamily of ubiquitous metalloenzymes present in all living organisms on the planet. They are classified into seven genetically distinct families and catalyse the hydration reaction of carbon dioxide to bicarbonate and protons, as well as the opposite reaction. CAs were proposed to be used for biotechnological applications, such as the post-combustion carbon capture processes. In this context, there is a great interest in searching CAs with robust chemical and physical properties. Here, we describe the enhancement of thermostability of the α-CA from Sulfurihydrogenibium yellowstonense (SspCA) by using the anchoring-and-self-labelling-protein-tag system (ASLtag). The anchored chimeric H5-SspCA was active for the CO2 hydration reaction and its thermostability increased when the cells were heated for a prolonged period at high temperatures (e.g. 70 °C). The ASLtag can be considered as a useful method for enhancing the thermostability of a protein useful for biotechnological applications, which often need harsh operating conditions.
Graphical Abstract
1. Introduction
The hydration/dehydration reaction involving carbon dioxide, water, bicarbonate, and protons (CO2 + H2O ⇄ HCO3− + H+) is a fundamentally important process for the planet and all its associated forms of lifeCitation1–9. The dissolution of CO2 in the aqueous phase develops carbonic acid (H2CO2), which is subject to an ionisation reaction producing bicarbonate (HCO3−), whereas this last species then generates carbonate (CO2) through a second dissociation reaction. These species are disseminated in the fluids of the all living organisms and are involved in a large number of physiological processes, such as some biosynthetic pathways, photosynthesis, respiration, pH homeostasis, secretion of electrolytes, etc.Citation9–11. At physiological pH, the naturally uncatalysed CO2 hydration reaction has a catalytic constant (kcat) of 0.15 s−1, whereas the uncatalysed dehydration shows a kcat of 50.0 s−1 Citation12,Citation13. These values are typical of slow reactions and are not sufficient for accomplishing fast cellular physiological processes which support metabolic activities dependent on the dissolved inorganic carbon species (CO2, HCO3−, CO32−)Citation9. Probably this is the reason why living organisms evolved a superfamily of ubiquitous metalloenzymes, the carbonic anhydrases (CAs, EC 4.2.1.1), which catalyse, and highly accelerate, the above-mentioned reactions, at a very high rate with respect to the non-catalysed reaction. CAs show kinetic constants kcat varying from 104 to 106 s−1 for the hydration reactionCitation12,Citation13.
Up to date, CA superfamily contains seven genetically distinct families (or classes), named α-, β-, γ-, δ-, ζ-, η-, and ɵ-CAsCitation9,Citation14,Citation15, characterised by multiple transcript variants and protein isoforms, with different biochemical properties and specific tissue/organ and sub-cellular localisationsCitation7,Citation9,Citation12,Citation16–19. Generally, only α-class enzymes are present in the animalsCitation20,Citation21, whereas α-, β-, γ-, δ- and θ-classes are found in plants and algae; α- and β-CAs in fungi; α-, β-, and/or η-CAs in protozoans; α-, β-, and γ-CA classes in bacteriaCitation7,Citation19,Citation15,Citation22–25.
Studies carried out on the bacterial CAs concern two main aspects. They are considered an attractive and rather new drug target, because their inhibition affects the growth or virulence of many pathogensCitation4,Citation7,Citation26–28. Furthermore, they are biocatalysts often used in biotechnological applicationsCitation29,Citation30, such as the post-combustion carbon capture process, artificial lungs, and biosensorsCitation31,Citation32. Many such processes are characterised by conditions, which may be deleterious to an enzyme belonging to the mesophilic organismsCitation25,Citation33–45. In the field of biotechnology, there is a great interest in searching proteins with robust chemical and physical properties, which resist the hard conditions of industrial processes. In this context, our groups identified in the genome of the extreme thermophiles Sulfurihydrogenibium yellowstonense and Sulfurihydrogenibium azorense two CAs, indicated with the acronyms SspCA and SazCA, respectively. It has been demonstrated that these two CAs belong to the α-CA class and showed an excellent activity as catalysts for the CO2 hydratase reaction (kcat =105–106 s−1)Citation30,Citation46–52. Interestingly, the two extreme enzymes resulted to be highly thermostable, retaining an excellent catalytic activity when heated for a prolonged period at a temperature higher than 80 °CCitation30,Citation46–52. The X-ray tridimensional structures of the two proteins demonstrated that the high compactness of the dimeric structure, the higher content of secondary-structural elements, the increased number of charged residues on the protein surface, and the vast number of ionic networks with respect to the mesophilic counterparts, are the main structural elements responsible for the protein thermostabilityCitation29,Citation30. Moreover, Russo et al. reported the use of free SspCA in experiment of CO2 absorptionCitation53 demonstrating that it is an excellent candidate for the biomimetic capture of CO2. Subsequently, the necessity to use this biocatalyst repeatedly and continuously, led to the immobilisation of the recombinant SspCA on polyurethane foam (PU), a pre-polymer of polyethylene glycolCitation54; onto supported ionic liquid membranes (SMLs), in order to realise a system able to selectively separate and transform CO2Citation55. Furthermore, the immobilisation onto magnetic support for recovering the biocatalyst from the bioreactor effortlessly and practically, for example through the use of a magnet, was also proposed for these thermostable CAsCitation56. Unfortunately, these strategies may discourage the wide utilisation of enzymes in industrial applications because of the high costs connected to the biocatalyst production and purification, and the expenses for the preparation of the immobilisation support. Thus, to overcome this limitation, a one-step immobilisation procedure has been proposed, which consists in the overexpression of SspCA directly onto the surface of bacterial hosts, by using the ice nucleation protein (INP) from the Gram-negative bacterium Pseudomonas syringaeCitation57.
In this article, we describe the improvement of the thermostability of SspCA by using a novel protein-tag system, the ASLtagCitation58. The anchored SspCA was fused to the thermostable variant of the alkylguanine-DNA-alkyl-transferase (H5) from the hyperthermophilic archaeon Sulfolobus solfataricus. The chimeric H5-SspCA was efficiently overexpressed on the bacterial surface of Escherichia coli. The protonography technique showed that the neosynthetised H5-SspCA was active for the CO2 hydration reaction. Even more intriguing, the chimeric H5-SspCA expressed onto the bacterial surface resulted to be more stable with respect to the non-chimeric SspCA, when treated at high temperatures (50.0 and 70.0 °C) for a prolonged time. The ASLtag system may thus be considered as a brilliant strategy to further increase the thermostability of proteins to be used in biotechnological applications, in which a highly effective and thermostable catalyst is needed.
2. Materials and methods
2.1 Construction of vectors for surface fusion and H5-SspCA overexpression
The vector pET-22b/INPN-SspCA was used to produce the pET-ASLtag-SspCA vector, which overexpressed onto the bacterial surface the chimeric H5-SspCA. The pET-22b/INPN-SspCA and pET-ASLtag-SspCA vectors were prepared as described previouslyCitation57,Citation58. For overexpressing the chimeric H5-SspCA or SspCA on the bacterial cell surface, competent E. coli BL21 (DE3) cells were transformed with the above-mentioned constructs. They were grown at 37.0 °C and induced with 1.0 mM isopropyl-thio-β-D-galactoside (IPTG) and 0.5 mM ZnSO4 at an OD600 of 0.6–0.7. After additional growth for 6 h, the cells were harvested by centrifugation and washed three times with PBS 1×. Aliquots of cells were resuspended in 25 mM Tris/HCl and used to determine the enzyme activity and for the preparation of the outer membrane fraction.
2.2 Carbonic anhydrase assay and SDS-PAGE
CA activity assay was a modification of the procedure described by Capasso et al.Citation59. Briefly, the assay was performed at 0 °C using CO2 as substrate and following the pH variation due to the catalysed conversion of CO2 to bicarbonate. Bromothymol blue was used as the indicator of pH variation. The production of hydrogen ions during the CO2 hydration reaction lowers the pH of the solution until the colour transition point of the dye is reached. The time required for the colour change is inversely related to the quantity of CA present in the sample. Wilbur-Anderson units (WAU) were calculated according to the following definition: one WAU of activity is defined as (T0−T)/T, where T0 (uncatalysed reaction) and T (catalysed reaction) are recorded as the time (in seconds) required for the pH to drop from 8.3 to the transition point of the dye in a control buffer and in the presence of enzyme, respectively. Assay of the membrane-bound enzyme (H5-SspCA or SspCA) was carried out using an amount of whole cells or outer membranes ranging from 1.0 to 5.0 mg. Sodium dodecyl sulphate (SDS)-polyacrylamide gel electrophoresis (PAGE) was performed as described by Laemmli using 12% gels.Citation60 Samples were dissolved in buffer with 5% β-mercaptoethanol. The gel was stained with Coomassie blue and protein concentration was determined by Bio-Rad assay kit (Bio-Rad, Hercules, CA).
2.3 Protonography and his-tag Western blotting
To perform the protonography, wells of 12% SDS-gel were loaded with solubilised outer membranes having on their surface H5-SspCA or SspCA, and a solution of free SspCA (enzyme overexpressed as cytoplasmic protein and purified as described previouslyCitation59). Samples were mixed with loading buffer without 2-mercaptoethanol and without boiling the samples, to solubilise cells and avoid protein denaturation. The gel was run at 180 V until the dye front moved off the gel. Following the electrophoresis, the 12% SDS-gel was subject to protonography to detect the cytoplasmic SspCA, the surface-SspCA, and surface-H5-SspCA hydratase activity on the gel as described by Del Prete et al.Citation61,Citation62 and De Luca et al.Citation63. To perform the Western-Blot, after a 12% (w/v) SDS-PAGE, the overexpressed cytoplasmic SspCA and the membrane-bound enzymes (SspCA and H5-SspCA) were also electrophoretic transferred to a PVDF membrane with transfer buffer (25 mM Tris, solubilised whole cells 192 mM glycine, 20% methanol) by using Trans-Plot SD Cell (Bio-Rad, Hercules, CA). His-tag Western blot was carried out using the Pierce Fast Western Blot Kit (Thermo Scientific, Waltham, MA). The blotted membrane has been placed in the wash blot solution Fast Western 1× Wash Buffer to remove transfer buffer. Primary Antibody Working Dilution was added to the blot and incubated for 30.0 min at room temperature (RT) with shaking. After, the blot was removed from the primary antibody solution and incubated for 10.0 min with the Fast Western Optimized HRP Reagent Working Dilution. Subsequently, the membrane was washed two times in about 20 ml of Fast Western 1× Wash Buffer. Finally, the membrane was incubated with the Detection Reagent Working Solution and incubated for 3.0 min at RT and then developed with X-ray film.
2.4 Determination of the H5 activity by an in vitro and in vivo fluorescent assay
Whole overnight inducted E. coli BL21(DE3) cells were collected and the expression of the H5-derived fusion proteins was analysed by an in vitro assay with the fluorescent SNAP-Vista Green™ substrate (New England Biolabs, Ipswich, MA; hereinafter BG-FL), as previously describedCitation58,Citation64. The in vivo imaging was carried out as described by Merlo et al.Citation58. Briefly, bacterial cells expressing the H5-SspCA onto cell surface were washed twice in PBS 1× and resuspended in 50.0 μl of the same buffer supplemented with 5.0 μM of the BG-FL. After incubation at 37.0 °C for 30.0 min, cells were washed twice, resuspended, and again incubated for 30.0 min at 37.0 °C, to allow the external diffusion of the unreacted substrate. Images were collected using a DM6 fluorescence microscope and Hamamatsu camera under the control of Leica LAS AF 6000 software; excitation and emission wavelengths used suitably for AlexaFluor488 dye were λex = 490 nm; λem = 525 nm, respectively.
2.5 Outer membrane preparation
The bacterial outer membranes were fractioned by inner membranes as described previously by Del Prete et al.Citation57. Briefly, 2.0 g of harvested bacterial cells were resuspended and disrupted by sonication on the ice. Cell extract was ultracentrifuged to recover the total membrane fraction. The outer membrane fraction was purified resuspending the pellet in phosphate-buffered saline (PBS 1×) containing 0.01 mM MgCl2 and 2% Triton X-100 and incubated at RT for 30.0 min to solubilise the inner membrane. The outer membrane fraction was then pelleted by ultracentrifugation at 120,000×g and used for further experiments.
2.6 Temperature stability studies
2.6.1 Thermostability
Bacterial cells (2.0 g/20 ml) were incubated at 25.0, 50.0, and 70.0 °C for different time up to 24 h to compare the stability of the membrane-bound enzymes (SspCA and H5-SspCA) at the above-indicated temperatures. Cell membrane-bound enzymes aliquots were withdrawn at appropriate times and the residual activity was measured using CO2 as the substrate. All data have been analysed using GraphPad Prism version 5.0 software (GraphPad Software, San Diego, CA). Curves were obtained by the mean of three independent determinations.
2.6.2 Long-term stability
Membrane-bound SspCA or H5-SspCA were investigated for their long-term stability at different temperatures (25.0, 50.0, and 70.0 °C) by assaying their hydratase residual activities using CO2 as substrate and withdrawing aliquots of cell surface SspCA or H5-SspCA at appropriate times. All the buffers were sterilised by using a sterile 0.22 μm filter, while samples containing the membrane-bound enzymes were treated with a diluted solution of NaN3 to avoid contamination. All data were obtained by the mean of 3 independent determinations.
3. Results and discussion
3.1 Expression and surface localisation of SspCA and H5-SspCA
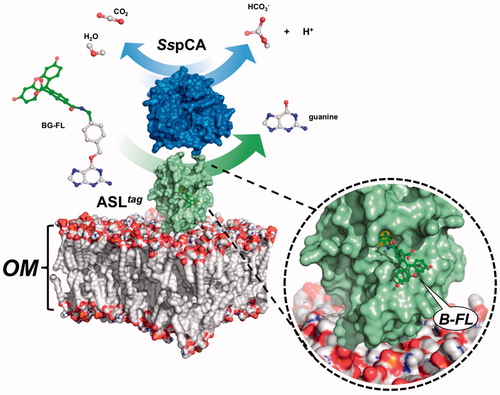
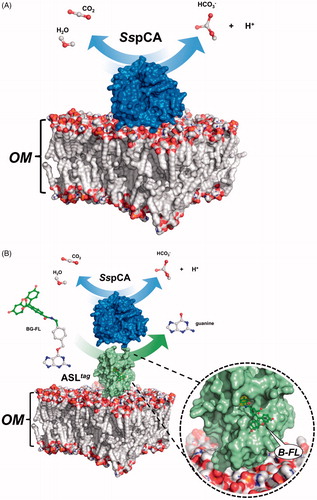
Expression of the SspCA and H5-SspCA was realised through the one-step procedure, by transforming the E. coli cells with the construct expressing a gene composed of a signal peptide (necessary for the periplasmic translocation of the protein), the P. syringae INPN domain (fundamental for displaying the overexpressed protein onto the bacterial surface), and the protein of interest (SspCA or H5-SspCA). This strategy has the advantage to overexpress and directly immobilise in vivo the α-CA or other proteins on the bacterial cell surface. Besides, the system expressing the H5-SspCA, named anchoring-and-self-labelling-protein-tag (ASLtag), allowed the labelling of the neosynthesised protein fused to H5 through the use of the fluorescein derivative of the O6-BG (BG-FL), which is the substrate of H5. As reported in , the expression of the chimeric H5-SspCA on the bacterial surface has been confirmed using the H5 substrate and analysing the whole bacterial cells with fluorescent microscopy. The irreversible reaction of the ASLtag system with a fluorescent substrate allowed the quantitative determination of the immobilised bacterial α-CA or of other proteins fused to H5, by in vitro gel-imaging techniques as described by Del Prete et al.Citation57 and Merlo et al.Citation58. Diversely from H5-SspCA, the expression of the anchored His-tagged SspCA (without the H5) has been confirmed only by the Western Blot analysis using an anti-His-tag antibody (), indicating an expected molecular weight of 50.0 kDa (the sum of the INPN and SspCA polypeptide chains produced with the construct pET-22b/INPN-SspCA; see the experimental section). Anchored His-tagged H5-SspCA showed a higher molecular weight (70.0 kDa) with respect to the non-chimeric protein because of the presence of the H5 protein (158 amino acid residues). The H5-SspCA Western-Blot fully supports the fluorescence microscopy results. Thus, using this one-step procedure, the thermostable proteins α-CA (SspCA) and the chimeric ASLtag -SspCACitation56 were efficiently expressed on the external side of the bacterial outer membrane. reports a model representing the in vivo immobilisation of SspCA (Panel A) and chimeric H5-SspCA (Panel B) on the bacterial external cell surface. Moreover, shows the reactions catalysed by both the biocatalysts (the hydration/dehydration of CO2 and the conversion of BG-FL in the free guanine and the fluorescent benzyl-guanine derivative covalently linked to the active site of H5).
Figure 1. Fluorescence microscopy of E. coli BL21(DE3) cells transformed with pET-22b/INPN-SspCA (left) or with pET-ASLtag-SspCA (right). The cells were incubated with BG-FL and then analysed by fluorescence microscopy. Images show bright field (BHF) and AlexaFluor488 (green). As expected, the fluorescence is only evidenced for the bacterial cell transformed with the ASLtag system.
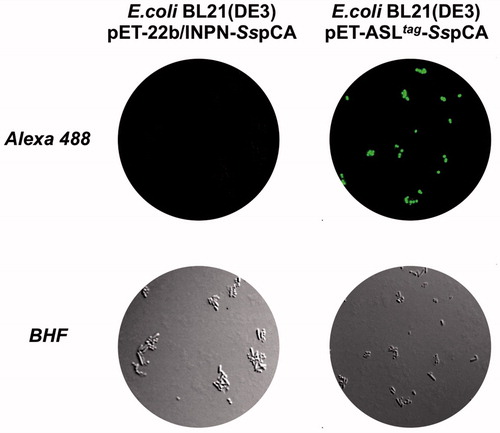
Figure 2. Western Blot performed on the outer membrane purified from the whole bacterial cells. The anti His-tag antibody was raised against the C-terminus of His-tagged SspCA. Legend: Lane Std, molecular markers, M.W. starting from the top: 75.0, 50.0, and 37.0 kDa; Lane 1, anchored SspCA; Lane 2, anchored H5-SspCA.
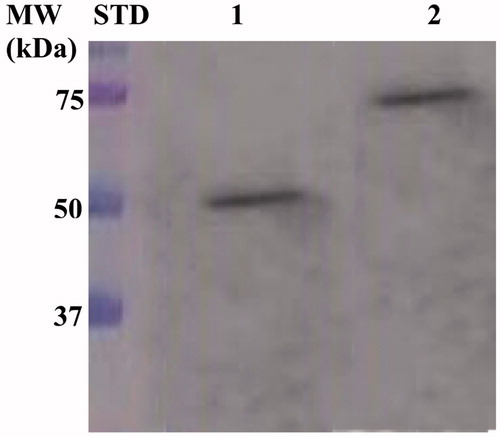
Figure 3. Model representation of an outer membrane fraction (OM; pdb from Tieleman and Berendsen65) describing the in vivo immobilisation of SspCA (in blue; PDB ID: 4G7A; panel A) and in fusion with H5 (in green; PDB ID: 6GA0; panel B). The INPN domain is omitted because inserted in the OM. The catalytic reaction of SspCA (the hydration/dehydration of CO2) and H5 (the conversion of BG-FL in the free guanine and the fluorescent benzyl-guanine derivative, B-FL, covalently linked to the active site of H5) are also shown.
3.2 Hydratase activity of the membrane anchored SspCA and H5-SspCA
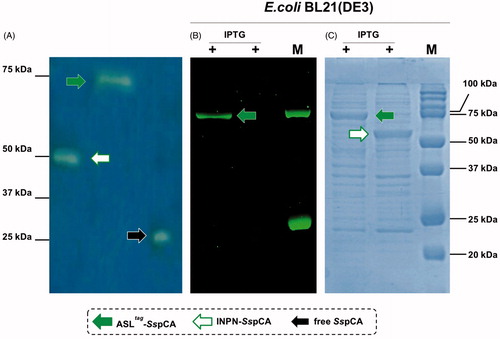
Using CO2 as the substrate, the hydratase activity of all the forms of SspCA has been investigated in solutionCitation57. The results showed that the membrane-bound SspCA with and without H5 was an active enzyme, when immobilised on the bacterial surface. The CO2 hydratase activity of SspCA and H5-SspCA did not show any differences. The results also evidenced that 1.0 µg of bacterial cells had a CO2 hydratase activity corresponding to that of 10.0 ng of free SspCA. Probably, anchored SspCA or H5-SspCA is subjected to various phenomena, which influence the enzymatic reaction, e.g. a reduction of the structural conformational changes (this is typical of an immobilised enzyme); a different substrate access to the active site with respect to the free biocatalyst due to the bacterial cell surface microenvironment, and, finally, an aggregation of the cells or outer membranes used in the assay. Otherwise, the activity of SspCAs was compared by using the protonography, which is a technique able to reveal the hydrogen ions produced by the hydratase activity reaction as a yellow band on the SDS-PAGE. The protonography results showed that the all the forms of SspCA (the two membrane-bound ones and the free enzyme) had a comparable enzyme activity and a different molecular weight on SDS-PAGE (, panel A and C). Protonography corroborated the results obtained with the fluorescent microscopy () and Western Blot (). Interestingly, H5-SspCA fluorescent band at a molecular weight of about 70.0 kDa (, panel B) indicated that the presence of SspCA does not affect the activity of the thermostable H5 enzyme on the BG-FL substrate.
Figure 4. Protonography (Panel A), fluorescence gel-imaging (Panel B) and Coomassie staining (Panel C) of SspCA and H5-SspCA carried out with different amounts of the whole E. coli cells (see Materials and Methods). Filled green, white and black arrows represent the ASLtag-SspCA, INPN-SspCA and the free SspCA, respectively.
3.3 Stability of SspCA and H5-SspCA linked to the bacterial cell surface
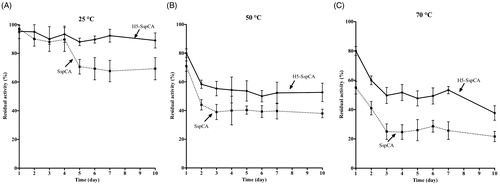
Using the whole bacterial cells expressing on the external surface SspCA or H5-SspCA, the effect of the CO2 hydratase reaction as a function of temperature has been investigated. In , the residual activity of the SspCA and H5-SspCA remained almost constant at 25.0 and 50.0 °C, retaining their residual activity at 100% up to 24 h (panel A) and at 70% up to 6 h of incubation (panel B), respectively. In contrast, it is readily apparent that at higher temperatures (70.0 °C) SspCA and H5-SspCA behave differently (, panel C): the residual activity of SspCA started to decline rapidly after 2 h, getting a value of about 60% after 14 h of incubation; whereas the stabilising effect of H5 on the SspCA showed a residual activity of about 85% and remained almost constant for the rest of the time indicated in the figure (panel C). These results demonstrated that the anchoring ASLtag system, enhanced the SspCA stability of about 20%. On the other hand, it is important to highlight that both anchored enzymes continued to work for several hours at temperatures considered prohibitive for the free enzymes, as SspCA, which Russo et al. demonstrated to show a residual activity of 20% when heated at 70.0 °C for 15 minCitation57. This aspect is crucial in the context of the post-combustion carbon capture process, which requires temperatures ranging from 40.0 and 60.0 °CCitation53. shows the residual activity for the CO2 hydration reaction for SspCA and H5-SspCA when the whole cells were treated at different temperatures for a very long period (up to 10 d). At 25.0 °C, the SspCA residual activity started to decrease after 4 d and reached a value of about 70% after 10 d, while H5-SspCA remained almost constant (panel A). At 50.0 and 70.0 °C, the residual activity of SspCA decreased up to 40 and 20%, respectively (panel B and C), whereas H5-SspCA showed a residual activity of about 60 and 40%, respectively (panel B and C). All these data confirmed that the presence of a thermostable protein-tag between the INPN anchoring domain and the SspCA significantly improved the long-term stability and the storage of this CA.
Figure 5. The thermostability of immobilised SspCA and H5-SspCA on the bacterial surface. Measures were carried out at indicated temperatures, by using aliquots of the whole cells incubated up to 24 h. Legend: continuous line, membrane-bound H5-SspCA; dashed line, membrane-bound SspCA. Each point is the mean of three independent determinations.
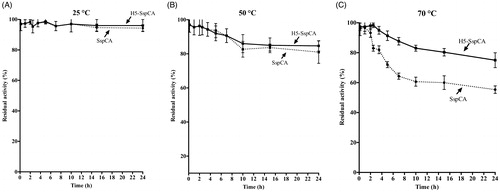
Figure 6. The long-term stability of immobilised SspCA and H5-SspCA on the bacterial surface. Measures were carried out at indicated temperatures up to 10 d, using aliquots of whole bacterial cells. Continuous line: free SspCA; Dashed line: membrane-bound SspCA. Each point is the mean of three independent determinations.
4. Conclusions
The ASLtag system efficiently overexpressed the chimeric H5-SspCA onto to the bacterial cell surface, as demonstrated by fluorescence microscopy and Western-Blot. As expected, the CO2 hydratase assay and the protonography showed that SspCA was still very active, even linked on the bacterial surface and the H5 moiety, showing a CO2 hydratase activity similar to that of its anchored counterpart without H5. Furthermore, by investigating the behaviour of the whole bacterial cells expressing on the external surface SspCA or H5-SspCA at different temperatures, we demonstrated an enhancement in terms of thermal stability of the chimeric protein. In conclusion, the H5-SspCA obtained by the ASLtag system constitutes a valid strategy for further increasing the thermostability of proteins, for processes in which a highly effective, thermostable catalyst is needed.
Acknowledgements
We are grateful to Giovanni Del Monaco for technical assistance.
Disclosure statement
The authors state no conflict of interests.
Additional information
Funding
References
- Capasso C, Supuran CT. Inhibition of bacterial carbonic anhydrases as a novel approach to escape drug resistance. Curr Top Med Chem 2017;17:1237–48.
- Del Prete S, Vullo D, De Luca V, et al. Cloning, expression, purification and sulfonamide inhibition profile of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg Med Chem Lett 2016;26:4184–90.
- Del Prete S, Vullo D, De Luca V, et al. Anion inhibition profiles of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. Bioorg Med Chem 2016;24:4410–4.
- Annunziato G, Angeli A, D’Alba F, et al. Discovery of new potential anti-infective compounds based on carbonic anhydrase inhibitors by rational target-focused repurposing approaches. Chem Med Chem 2016;11:1904–14.
- Del Prete S, Vullo D, De Luca V, et al. Anion inhibition profiles of alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:3413–7.
- Abdel Gawad NM, Amin NH, Elsaadi MT, et al. Synthesis of 4-(thiazol-2-ylamino)-benzenesulfonamides with carbonic anhydrase I, II and IX inhibitory activity and cytotoxic effects against breast cancer cell lines. Bioorg Med Chem 2016;24:3043–51.
- Capasso C, Supuran CT. An overview of the carbonic anhydrases from two pathogens of the oral cavity: streptococcus mutans and porphyromonas gingivalis. Curr Top Med Chem 2016;16:2359–68.
- Del Prete S, Vullo D, De Luca V, et al. Comparison of the sulfonamide inhibition profiles of the alpha-, beta- and gamma-carbonic anhydrases from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem Lett 2016;26:1941–6.
- Supuran CT, Capasso C. Biomedical applications of prokaryotic carbonic anhydrases. Expert Opin Ther Pat 2018;28:745–54.
- Nishimori I, Onishi S, Takeuchi H, Supuran CT. The alpha and beta classes carbonic anhydrases from Helicobacter pylori as novel drug targets. Curr Pharm Des 2008;14:622–30.
- Morishita S, Nishimori I, Minakuchi T, et al. Cloning, polymorphism, and inhibition of beta-carbonic anhydrase of Helicobacter pylori. J Gastroenterol 2008;43:849–57.
- Supuran CT. Advances in structure-based drug discovery of carbonic anhydrase inhibitors. Expert Opin Drug Discov 2017;12:61–88.
- Supuran CT. Structure and function of carbonic anhydrases. Biochem J 2016;473:2023–32.
- Supuran CT, Capasso C. An overview of the bacterial carbonic anhydrases. Metabolites 2017;7:pii: E56.
- Capasso C, Supuran CT. An overview of the alpha-, beta- and gamma-carbonic anhydrases from bacteria: can bacterial carbonic anhydrases shed new light on evolution of bacteria? J Enzyme Inhib Med Chem 2015;30:325–32.
- Supuran CT. Carbonic anhydrase activators. Future Med Chem 2018;10:561–73.
- Supuran CT, Capasso C. An overview of the bacterial carbonic anhydrases. Metabolites 2017;7:56–74.
- Supuran CT, Capasso C. Carbonic anhydrase from Porphyromonas gingivalis as a drug target. Pathogens 2017;6:30–42.
- Capasso C, Supuran CT. Bacterial, fungal and protozoan carbonic anhydrases as drug targets. Expert Opin Ther Targets 2015;19:1689–704.
- Aspatwar A, Tolvanen ME, Ortutay C, Parkkila S. Carbonic anhydrase related proteins: molecular biology and evolution. Subcell Biochem 2014;75:135–56.
- Supuran CT . Carbonic anhydrases as drug targets-an overview. Curr Top Med Chem 2007;7:825–33.
- Supuran CT, Capasso C . The η-class carbonic anhydrases as drug targets for antimalarial agents. Expert Opin Ther Targets 2015;19:551–63.
- Capasso C, Supuran CT. An overview of the selectivity and efficiency of the bacterial carbonic anhydrase inhibitors. Curr Med Chem 2015;22:2130–9.
- Capasso C, Supuran CT. Sulfa and trimethoprim-like drugs - antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors. J Enzyme Inhib Med Chem 2014;29:379–87.
- Capasso C, Supuran CT. Anti-infective carbonic anhydrase inhibitors: a patent and literature review. Expert Opin Ther Pat 2013;23:693–704.
- Ozensoy Guler O, Capasso C, Supuran CT. A magnificent enzyme superfamily: carbonic anhydrases, their purification and characterization. J Enzyme Inhib Med Chem 2016;31:689–94.
- Del Prete S, Vullo D, De Luca V, et al. Sulfonamide inhibition studies of the beta-carbonic anhydrase from the pathogenic bacterium Vibrio cholerae. Bioorg Med Chem 2016;24:1115–20.
- Del Prete S, De Luca V, De Simone G, et al. Cloning, expression and purification of the complete domain of the eta-carbonic anhydrase from Plasmodium falciparum. J Enzyme Inhib Med Chem 2016;31:54–59.
- De Simone G, Monti SM, Alterio V, et al. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2015;25:2002–6.
- Di Fiore A, Capasso C, De Luca V, et al. X-ray structure of the first `extremo-alpha-carbonic anhydrase', a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr D Biol Crystallogr 2013;69:1150–9.
- Alterio V, Langella E, De Simone G, Monti SM. Cadmium-containing carbonic anhydrase CDCA1 in marine diatom Thalassiosira weissflogii. Mar Drugs 2015;13:1688–97.
- Di Fiore A, Alterio V, Monti SM, et al. Thermostable carbonic anhydrases in biotechnological applications. Int J Mol Sci 2015;16:15456–80.
- Supuran CT. CA IX stratification based on cancer treatment: a patent evaluation of US2016/0002350. Expert Opin Ther Pat 2016;26:1105–9.
- Lomelino C, McKenna R. Carbonic anhydrase inhibitors: a review on the progress of patent literature (2011–2016). Expert Opin Ther Pat 2016;26:947–56.
- Monti SM, Supuran CT, De Simone G. Anticancer carbonic anhydrase inhibitors: a patent review (2008–2013). Expert Opin Ther Pat 2013;23:737–49.
- Masini E, Carta F, Scozzafava A, Supuran CT. Antiglaucoma carbonic anhydrase inhibitors: a patent review. Expert Opin Ther Pat 2013;23:705–16.
- Scozzafava A, Supuran CT, Carta F. Antiobesity carbonic anhydrase inhibitors: a literature and patent review. Expert Opin Ther Pat 2013;23:725–35.
- Aggarwal M, Kondeti B, McKenna R. Anticonvulsant/antiepileptic carbonic anhydrase inhibitors: a patent review. Expert Opin Ther Pat 2013;23:717–24.
- Carta F, Supuran CT. Diuretics with carbonic anhydrase inhibitory action: a patent and literature review (2005–2013). Expert Opin Ther Pat 2013;23:681–91.
- Winum JY, Capasso C. Novel antibody to a carbonic anhydrase: patent evaluation of WO2011138279A1. Expert Opin Ther Pat 2013;23:757–60.
- Aggarwal M, McKenna R. Update on carbonic anhydrase inhibitors: a patent review (2008–2011). Expert Opin Ther Pat 2012;22:903–15.
- Carta F, Scozzafava A, Supuran CT. Sulfonamides: a patent review (2008–2012). Expert Opin Ther Pat 2012;22:747–58.
- Carta F, Supuran CT, Scozzafava A. Novel therapies for glaucoma: a patent review 2007–2011. Expert Opin Ther Pat 2012;22:79–88.
- Poulsen SA. Carbonic anhydrase inhibition as a cancer therapy: a review of patent literature, 2007–2009. Expert Opin Ther Pat 2010;20:795–806.
- Boone CD, Habibzadegan A, Gill S, McKenna R. Carbonic anhydrases and their biotechnological applications. Biomolecules 2013;3:553–62.
- Alafeefy AM, Abdel-Aziz HA, Vullo D, et al. Inhibition of carbonic anhydrases from the extremophilic bacteria Sulfurihydrogenibium yellostonense (SspCA) and Sulfurihydrogenibium azorense (SazCA) with a new series of sulfonamides incorporating aroylhydrazone-, [1,2,4]triazolo[3,4-b][1,3,4]thiadiazinyl- or 2-(cyanophenylmethylene)-1,3,4-thiadiazol-3(2H)-yl moieties. Bioorg Med Chem 2014;22:141–7.
- Vullo D, De Luca V, Scozzafava A, et al. The extremo-alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium azorense is highly inhibited by sulfonamides. Bioorg Med Chem 2013;21:4521–5.
- Vullo D, Luca VD, Scozzafava A, et al. The alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1 is highly susceptible to inhibition by sulfonamides. Bioorg Med Chem 2013;21:1534–8.
- Akdemir A, Vullo D, De Luca V, et al. The extremo-alpha-carbonic anhydrase (CA) from Sulfurihydrogenibium azorense, the fastest CA known, is highly activated by amino acids and amines. Bioorg Med Chem Lett 2013;23:1087–90.
- De Luca V, Vullo D, Scozzafava A, et al. Anion inhibition studies of an alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Bioorg Med Chem Lett 2012;22:5630–4.
- Vullo D, De Luca V, Scozzafava A, et al. Anion inhibition studies of the fastest carbonic anhydrase (CA) known, the extremo-CA from the bacterium Sulfurihydrogenibium azorense. Bioorg Med Chem Lett 2012;22:7142–5.
- Vullo D, De Luca V, Scozzafava A, et al. The first activation study of a bacterial carbonic anhydrase (CA). The thermostable alpha-CA from Sulfurihydrogenibium yellowstonense YO3AOP1 is highly activated by amino acids and amines. Bioorg Med Chem Lett 2012;22:6324–7.
- Russo ME, Olivieri G, Capasso C, et al. Kinetic study of a novel thermo-stable alpha-carbonic anhydrase for biomimetic CO2 capture. Enzyme Microb Technol 2013;53:271–7.
- Migliardini F, De Luca V, Carginale V, et al. Biomimetic CO2 capture using a highly thermostable bacterial alpha-carbonic anhydrase immobilized on a polyurethane foam. J Enzyme Inhib Med Chem 2014;29:146–50.
- Abdelrahim MYM, Martins CF, Neves LA, et al. Supported ionic liquid membranes immobilized with carbonic anhydrases for CO2 transport at high temperatures. J Membr Sci 2017;528:225–30.
- Perfetto R, Del Prete S, Vullo D, et al. Production and covalent immobilisation of the recombinant bacterial carbonic anhydrase (SspCA) onto magnetic nanoparticles. J Enzyme Inhib Med Chem 2017;32:759–66.
- Del Prete S, Perfetto R, Rossi M, et al. A one-step procedure for immobilising the thermostable carbonic anhydrase (SspCA) on the surface membrane of Escherichia coli. J Enzyme Inhib Med Chem 2017;32:1120–8.
- Merlo R, Del Prete S, Valenti A, et al. An AGT-based protein-tag system for the labelling and surface immobilization of enzymes on E. coli outer membrane. J Enzyme Inhib Med Chem 2019;34:490–9.
- Capasso C, De Luca V, Carginale V, et al. Biochemical properties of a novel and highly thermostable bacterial alpha-carbonic anhydrase from Sulfurihydrogenibium yellowstonense YO3AOP1. J Enzyme Inhib Med Chem 2012;27:892–7.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227:680–5.
- Del Prete S, De Luca V, Supuran CT, Capasso C. Protonography, a technique applicable for the analysis of eta-carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:920–4.
- Del Prete S, De Luca V, Iandolo E, et al. Protonography, a powerful tool for analyzing the activity and the oligomeric state of the gamma-carbonic anhydrase identified in the genome of Porphyromonas gingivalis. Bioorg Med Chem 2015;23:3747–50.
- De Luca V, Del Prete S, Su puran CT, Capasso C. Protonography, a new technique for the analysis of carbonic anhydrase activity. J Enzyme Inhib Med Chem 2015;30:277–82.
- Vettone A, Serpe M, Hidalgo A, et al. A novel thermostable protein-tag: optimization of the Sulfolobus solfataricus DNAalkyl-transferase by protein engineering. Extremophiles 2016;20:13.
- Tieleman DP, Berendsen HJ. A molecular dynamics study of the pores formed by Escherichia coli OmpF porin in a fully hydrated palmitoyloleoylphosphatidylcholine bilayer. Biophys J 1998;74:2786–801.
