Abstract
Colorectal cancer (CRC) is one of the most common malignancies worldwide. The prognosis for this cancer is poor, and the development of novel biomarkers, particularly non-invasive surrogate biomarkers, is urgently needed. Recent studies have demonstrated that microRNAs (miRNAs) are stably detectable in the blood and can serve as useful biomarkers for various types of cancer. In this study, the miR-183 expression levels were found to be significantly overexpressed in plasma samples from CRC patients compared with controls, and the postoperative plasma miR-183 levels were significantly reduced compared with the preoperative levels. The value of the area under the receiver operating characteristic (ROC) curve obtained for miR-183 was 0.829, which was higher than those for carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9). High plasma miR-183 expression was significantly associated with lymph node metastasis, distant metastasis, higher pTNM stage (III-IV), and tumor recurrence. CRC patients with elevated miR-183 expression in plasma displayed shorter disease-free survival (DFS) and lower overall survival (OS). More importantly, plasma miR-183 was independently correlated with tumor recurrence and a lower OS. Collectively, our results suggested that the elevated miR-183 in the plasma could be a promising biomarker for predicting the risk of tumor recurrence and poor survival in CRC patients.
Abbreviations
| CRC | = | colorectal cancer |
| MiRNAs | = | microRNAs |
| CEA | = | carcinoembryonic antigen |
| CA19–9 | = | carbohydrate antigen 19–9 |
| PCR | = | polymerase chain reaction |
| pTNM | = | pathological tumor-node-metastasis |
| HR | = | hazard ratio |
| CI | = | confidence interval |
| ROC | = | receiver operating characteristic |
| AUC | = | area under curve |
| OS | = | overall survival |
| DFS | = | disease-free survival |
Introduction
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in the world, with over 1.2 million new cases and more than 600,000 cancer-related deaths in 2008.Citation1 The overall 5-year relative survival of CRC patients is 65.0%, but this rate varies depending on the stage distribution. Approximately 20% of diagnoses are made during the metastatic stage. Unfortunately, metastatic CRC (mCRC) has a very poor prognosis, with a median survival of less than 2 yCitation2 Even after curative resection and multimodal therapy, the majority of patients eventually develop local or distant recurrent disease. Hence, markers that predict inherently poor prognoses or recurrence risk would be helpful with regard to treatment decisions.
MicroRNAs (miRNAs) are a class of untranslated single-stranded RNA molecules that negatively regulate target genes by translational repression or degradation of the complementary mRNAs.Citation3 The aberrant expression of miRNAs in specific tissues may be related to the process of malignant organ transformation and cancer developmentCitation4,5. In addition, studies have shown that unique miRNA expression profiles in cancer tissues may contribute to the diagnosis of malignancies and the prediction of prognosis.Citation6,7 In recent years, an increasing number of studies has demonstrated that tumor‑derived miRNAs are present in the plasma or serum in remarkably stable forms that are resistant to endogenous ribonuclease activity. These findings highlight the potential of plasma/serum miRNA–based assays as accurate methods for the diagnosis and prognosis of human cancer.Citation8,9
The miRNA-183 family is highly conserved and includes 3 members, miR-96, miR-182, and miR-183.Citation10 These miRNAs have potentially oncogenic roles during carcinogenesis and have been shown to regulate cancer development and progression.Citation11,12 According to previous reports, members of the miRNA-183 family are highly expressed in CRC tissues compared with normal colorectal tissues, and miR-183 is the most highly up-regulated member of this family.Citation13–16 Additionally, a previous study showed that overexpression of miR-183 in tumors was significantly correlated with advanced clinical stage, lymph node and distant metastases, and poor prognosis of CRC.Citation17 Therefore, we hypothesized that the expression levels of miR-183 in the plasma could be useful clinical biomarkers in CRC patients.
In the present study, we evaluated the expression levels of plasma miR-183 in CRC patients and healthy individuals to determine whether there was a correlation between its expression and the clinical outcomes of CRC patients. We also compared the diagnostic sensitivity and specificity of plasma miR-183 with those of conventional CRC biomarkers, such as carcinoembryonic antigen (CEA) and carbohydrate antigen 19–9 (CA19–9) in serum.
Results
Expression of miR-183 in primary CRC tissues and CRC cell lines
To verify the previous reports of high miR-183 expression in primary CRC, we used real-time PCR to evaluate miR-183 expression in 8 paired CRC tissues and adjacent non-cancerous tissues. In addition, the expression levels of miR-183 in 5 human CRC cell lines, Caco-2,CW-2,LoVo,HCT 116, SW480, and the normal human colon cell line FHC, were also analyzed by real-time PCR. As shown in , the miR-183 expression levels were significantly higher in CRC tissues and all examined CRC cell lines than in normal colorectal tissues (all P < 0.001). Furthermore, the level of miR-183 expression was lower in the normal colon cell line FHC than in any of the examined CRC cell lines.
Figure 1. The expression levels of miR-183 in primary colorectal cancer (CRC) tissues and CRC cell lines. (A) The expression levels of miR-183 were significantly higher in CRC tissues than in paired normal colorectal tissues (P <0.001; Wilcoxon t-test). (B) The expression levels of miR-183 were significantly higher in CRC cell lines than in the normal colon cell line FHC and normal colorectal tissues (P <0.001; Mann-Whitney U-test). RNU6B (U6) was used as an internal control.

Up-regulation of miR-183 in CRC plasma samples
We evaluated the plasma expression level of miR-183 in a total of 118 CRC patients and 61 healthy controls by real-time PCR. As shown in , the relative miR-183 expression level in plasma from CRC patients was 0.019 ±0 .008, which was significantly higher than the value obtained for the healthy controls (0.011 ±0 .004; P < 0.0001). The 95% confidence intervals (95% CIs) were [0.018–0.020] and [0.010–0.012] in the CRC and healthy control groups, respectively.
Figure 2. The miR-183 expression levels in plasma simples. (A) The miR-183 expression levels in plasma were significantly higher in colorectal cancer (CRC) patients (n = 118 ) than in healthy controls (n = 61 ) (P < 0.0001; Mann-Whitney U-test). (B) The miR-183 expression levels in the postoperative samples were significantly lower than those in the preoperative samples (p = 0 .001; Wilcoxon t-test). In three cases, the plasma miR-183 expression levels were re-elevated when patients developed postoperative recurrence. RNU6B (U6) was used as an internal control.
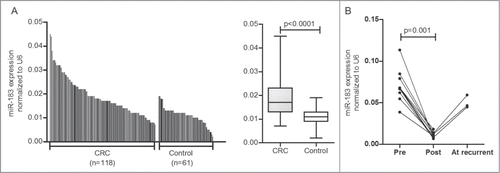
We next compared the miR-183 expression levels in paired plasma samples from 11 CRC patients before and 1 month after surgery. Results showed that the miR-183 expression levels in the postoperative samples were significantly lower than those in the preoperative samples (p = 0 .001). Moreover, the plasma miR-183 expression levels were found to be re-elevated in 3 patients who developed postoperative recurrence of solitary liver metastasis during follow-up ().
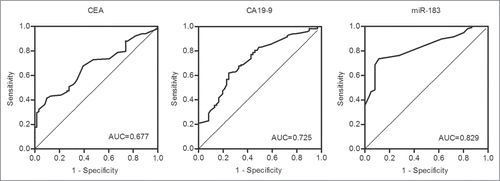
In addition, we assessed miR-183 expression in the plasma as a tool for clinical diagnosis in comparison to other CRC biomarkers, such as CEA and CA19–9. shows the receiver operating characteristic (ROC) curves that were obtained for this analysis. The ROC curve analysis revealed diagnostic sensitivities of 73.7, 41.5, and 61.9% for miR-183, CEA, and CA19–9, respectively, and diagnostic specificities of 88.5, 90.2, and 75.4%, for miR-183, CEA, and CA19–9, respectively. The area under the curve (AUC) values obtained for miR-183, CEA, and CA19–9 were 0.829 (95%CI: 0.770–0.888), 0.677 (95%CI: 0.599–0.755), and 0.725 (95%CI: 0.647–0.802), respectively.
Correlation of the plasma miR-183 levels with the clinicopathological features of CRC
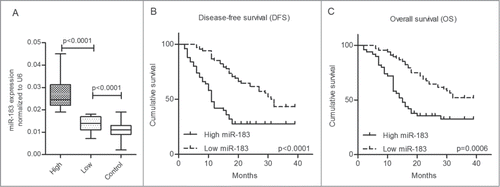
In this study, the CRC patients were separated into 2 groups (high- and low-expression) using the mean miR-183 expression level of the 118 patients (2-ΔCt =0.019) as the threshold. Using this approach, we found that the miR-183 level in the plasma was up-regulated in 42.4% (50/118) of the CRC patients compared with only 3.3% (2/61) of the healthy controls (P < 0.001). Box plots of the miR-183 levels in the controls and in the low-expression and high-expression categories are shown in , and the mean values in these 3 groups were 0.011 ±0 .004, 0.014 ±0 .003 and 0.026 ±0 .006, respectively. There were significant differences between the controls and the patients with low plasma miR-183 expression (P < 0.0001) and between the patients with low and high levels of expression (P < 0.0001).
Figure 4. Survival analysis of 118 colorectal cancer (CRC) patients grouped by their mean level of plasma miR-183. (A) The levels of plasma miR-183 in the high-expression and low-expression and controls groups. (B-C) Kaplan–Meier curves of 118 CRC patients showing the disease-free survival (DFS) and the overall survival (OS). The CRC patients with high miR-183 expression in the plasma displayed shorter DFS (P < 0.0001; log-rank test) and lower OS (p = 0 .0006; log-rank test).
The relationship between the plasma miR-183 expression level and the clinicopathological factors of the CRC patients is shown in . The results revealed that the miR-183 expression levels in the plasma samples from the CRC patients were not significantly correlated with gender, age, histology, tumor depth, or CEA level. The associations between the miR-183 expression level in the plasma and lymph node metastasis (p = 0 .048), distant metastasis (p = 0 .004) and pTNM stage (p = 0 .043) were significant. More importantly, elevated miR-183 expression in the plasma was significantly correlated with tumor recurrence (p = 0 .003).
Table 1. The relationship between plasma miR-183 expression and the clinicopathological characteristics
Plasma miR-183 is prognostic and can be used as a predictor of tumor recurrence in CRC
According to the log-rank test, CRC patients with high miR-183 expression in the plasma displayed shorter DFS (P <0.0001) and lower OS (p = 0 .0006). The Kaplan–Meier cumulative survival curves for 118 CRC patients are presented in . The Cox regression analysis was used to determine whether plasma miR-183 expression can predict CRC prognosis (). In the univariate analysis, a high T stage (T3/4; p = 0 .01), lymph node metastasis (P < 0.0001), distant metastasis (P < 0.0001), high pTNM stage (stage III/IV; P < 0.0001) and high levels of miR-183 in the plasma (P=0 .001) were significantly associated with overall survival. These significant factors (P < 0.05) based on the above univariate analysis were included in a multivariate analysis. The results showed that distant metastasis (p = 0 .013), high pTNM stage (p = 0 .009) and high plasma miR-183 expression (p = 0 .021) were independent prognostic markers for predicting poorer overall survival in CRC patients, with hazard ratios of 2.055 (95% CI 1.164–3.629), 5.218 (95% CI 1.517–17.952) and 1.831 (95% CI 1.098–3.054), respectively. In addition, we also utilized the Cox regression analysis to determine whether plasma miR-183 can serve as a predictor of tumor recurrence after curative surgery (). Univariate analysis showed that high T stage (T3/4; P < 0.0001), lymph node metastasis (P < 0.0001), distant metastasis (P <0.0001), high pTNM stage (stage III/IV; P < 0.0001), high levels of CEA (≥ 3.5; p = 0 .034) and high levels of miR-183 in plasma (P < 0.0001) were significantly associated with tumor recurrence. Additionally, the multivariate analysis revealed that high pTNM stage (p = 0 .003) and high plasma miR-183 expression (p = 0.001) were independent predictors of tumor recurrence in CRC patients, with hazard ratios of 11.782 (95% CI 2.259–61.436) and 3.448 (95% CI 1.670–7.120), respectively.
Table 2. Univariate and multivariate analyses with regard to overall survival
Table 3. Univariate and multivariate analyses with regard to tumor recurrence
Expression of miR-183 in hematocytes
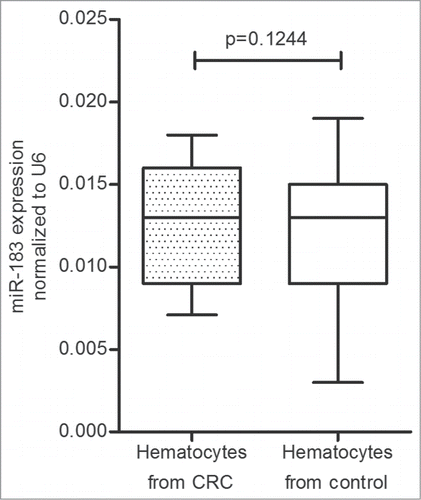
After isolating plasma from the blood samples by centrifugation, we also used real-time PCR to evaluate the miR-183 expression levels in the rest of the peripheral blood, which was mainly composed of the hematocytes, such as red blood cells, white blood cells, and platelets. The result revealed that there was no significant difference in the miR-183 expression levels between CRC patients (0.013 ± 0.003) and healthy controls (0.012 ± 0.004; p = 0 .1244). The 95% CIs were [0.012–0.013] and [0.011–0.013] in the CRC and healthy control groups, respectively ().
Figure 5. The miR-183 expression levels in hematocytes collected from 118 colorectal cancer (CRC) patients and 61 healthy controls. There was no significant difference in the miR-183 expression levels between CRC patients and healthy controls (p = 0 .1244; Mann-Whitney U-test). RNU6B (U6) was used as an internal control.
Discussion
In recent years, the survival rates in individuals with CRC have increased substantially, possibly as a result of early diagnosis and improved treatment.Citation18 However, because of the absence of typical symptoms or signs, and the lack of a sufficiently sensitive and specific biomarker, CRC continues to have an extremely poor prognosis. The most important prognostic factors for CRC remain tumor stage and curative surgery; however, these postoperative factors are not routinely available, which severely limits their clinical application. Thus, the development of novel biomarkers is urgently needed, especially noninvasive surrogate biomarkers, which can yield an improved prognosis of CRC. In the present study, we found that the plasma miR-183 levels may be more valuable than CEA and CA19–9 for CRC diagnosis. More importantly, elevated miR-183 levels in the plasma were significantly associated with tumor recurrence and a poor prognosis of CRC.
MiRNAs are a class of small non-coding endogenous RNA molecules that regulate gene expression at the post-transcriptional level and play important roles in the development and progression of various malignancies.Citation4,19 In recent years, accumulating reports have demonstrated that measuring levels of circulating miRNAs in the peripheral blood is feasible for clinical application. Even in the RNase-rich environment of the blood, miRNAs have been shown to be remarkably stable.Citation8 In addition, miRNA levels were independent of the subject's age and sexCitation20, and both serum and plasma samples were suitable for investigation of miRNAs as blood-based biomarkers.Citation8 In fact, many reports have indicated that circulating miRNAs have the potential to be used as molecular markers in the early diagnosis and prognosis of various tumors,Citation21,22 such as leukemia,Citation23 lung cancerCitation24, breast cancer,Citation25 prostate cancer,Citation26 esophageal cancer,Citation27 gastric cancerCitation28, and colorectal cancer.Citation29
MiR-183 is a member of the miRNA-183 family, which is an evolutionarily conserved miRNA cluster (miR-183, miR-96, and miR-182) located in the same chromosomal region (7q32.2)Citation10. Several studies have demonstrated that the members of the miRNA-183 family are directly involved in human cancer-related processes, such as cellular differentiation, tumorigenesis, proliferation, apoptosis and metabolism.Citation11,12 It was shown that miR-183 was highly expressed in CRC and some other types of cancerCitation16, but it was down-regulated in osteosarcomaCitation30 and inconsistently expressed in breast tumor tissues and the corresponding normal tissues.Citation31 A previous study has revealed an miRNA network composed of miR-183–EGR1–PTEN and has shown that the up-regulation of miR-183 can reduce EGR1 and PTEN protein expression in CRC cells.Citation32 EGR1 and PTEN are known to be key tumor suppressors, and their loss of expression leads to decreases in cell migration and invasion.Citation33-35 Zhou T et al.Citation17 recently reported that the increased expression of miR-183 in tumors was related to advanced clinical stage, lymph node and distant metastases, and poor prognosis of CRC. Therefore, we hypothesized that the quantitative detection of miR-183 in the plasma could be a useful clinical biomarker in CRC patients.
In the present study, we confirmed that miR-183 was significantly overexpressed in CRC tissues and cell lines, which is consistent with the results of previous studies.Citation13,15,17 Additionally, we demonstrated that miR-183 was significantly overexpressed in the plasma in CRC patients compared with controls. Based on the ROC curve analysis, the plasma level of miR-183 had a clinically satisfactory degree of sensitivity and specificity with an AUC of 0.829, which was higher than the AUC values obtained with conventional biomarkers for CRC, such as CEA and CA19–9. Furthermore, in eleven cases, the preoperative levels of miR-183 in the plasma were found to be significantly reduced compared with the postoperative levels, and the plasma miR-183 levels were re-elevated at recurrence after surgery in 3 patients, suggesting the plasma miR-183 levels may be useful for evaluating the tumor dynamics of CRC.
Although many studies have evaluated the diagnostic potential of circulating miRNAs in CRC patients, few have explored their prognostic potential.Citation36-39 In the present study, our findings revealed that the miR-183 expression in plasma was significantly associated with lymph node metastasis, distant metastasis, pTNM stage, and tumor recurrence. In addition, overexpression of miR-183 in plasma was significantly associated with tumor recurrence and poor survival in CRC patients.
A previous study demonstrated that some specific circulating miRNAs may originate from peripheral blood cells, and plasma miRNA biomarker levels may be substantially influenced.Citation40 Therefore, we compared the miR-183 levels in hematocytes between CRC patients and healthy controls. We found that there was no significant difference in the miR-183 levels between these 2 groups. Although further studies are needed to obtain a deeper understanding of the circulating miRNAs, including investigations of their source and mechanism of action, our results suggest that the level of miR-183 in hematocytes may not be a useful biomarker for CRC.
In summary, the current study demonstrated the up-regulation of miR-183 in plasma from CRC patients. The plasma levels of miR-183 may be more valuable than the levels of CEA and CA19–9 for CRC diagnosis, and miR-183 levels may be useful for evaluating the tumor dynamics of CRC. More importantly, elevated expression of miR-183 in plasma could be a promising biomarker to predict the risk of tumor recurrence and poor survival in CRC patients. Future studies with larger sample sizes and longer patient follow-up times should be carried out to confirm the present findings.
Methods
Study population and samples
Between September 2011 and July 2013, a total of 118 blood samples from consecutive CRC patients (mean age 60.9 ±7 .9 years; range 42–78 years) were collected within 1 week of resection in the Department of Oncology Surgery of the First Affiliated Hospital of Xi'an Jiaotong University. The diagnoses were confirmed by histopathology. In addition, blood samples from 61 healthy individuals with no history of malignant disease (mean age 55.2 ± 7.3 years; range 33–71 years) were collected at the Medical Center of the First Affiliated Hospital and used as controls. We also collected paired blood samples from 11 patients with stage II-III CRC before and 1 month after curative resection. Sixteen tissue samples (8 CRC tissues, 8 adjacent non-cancerous colorectal tissues) were snap frozen at the time of surgery and stored at −80°C for further analysis. Patients treated with radiotherapy or chemotherapy before surgery were not included in this study. The CRC patients were treated according to the same therapeutic strategy, i.e., complete tumor resection with negative margins (R0 resection). After surgery, patients with advanced-stage cancer received 5-fluorouracil-based chemotherapy. Tumor stage and histological grade were recorded using the classification guidelines of the American Joint Committee on Cancer (AJCC).Citation41 This study was approved by the Ethics Committee at the First Affiliated Hospital of Xi’an Jiaotong University. All of the specimens were collected after written informed consent was obtained from each patient.
Collection of survival data from gastric cancer patients
After surgery, all of the CRC patients were followed up regularly by our clinicians for more than 36 months or until death. The last follow-up was conducted in May 2014. Overall survival (OS) was measured as the time elapsed between surgery and death or the date of the last follow-up. Disease-free survival (DFS) was measured as the time from surgery to the date of an event or the last follow-up, where the “event” represented recurrence, metastasis, or death from any cause.
Cell lines and cell culture
Five human CRC cell lines, Caco-2,CW-2,LoVo,HCT 116,and SW480, were obtained from the Cell Bank of the Chinese Academy of Sciences, and the normal colon cell line FHC was obtained from the American Type Culture Collection (ATCC). Caco-2 cells were cultured in MEM, CW-2 cells in RPMI-1640, LoVo cells in F12-K, HCT-116 cells in McCoy's 5a, SW480 cells in Leibovitz's L-15 and FHC cells in DMEM:F12. All media were purchased from Sigma Company and supplemented with 10% fetal bovine serum (FBS; Trace Scientific). All of the cell lines were incubated at 37° in a humidified atmosphere containing 5% carbon dioxide.
Plasma preparation, RNA extraction and quantitative real-time RT-PCR
To isolate plasma from the blood samples, peripheral blood (3 ml) was collected in EDTA-K2 anti-coagulant tubes and transferred to the laboratory within 30 min for processing. Plasma samples were obtained by centrifuging the peripheral blood at 3,000 rpm for 20 min and at 12,000 rpm for 10 min at 4°C. The plasma was aliquoted and stored in fresh tubes at −80°C before the extraction of RNA. Total RNA containing small RNA was extracted from plasma or cultured cells or tissues using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. The concentrations of total RNA in the samples were quantified using a NanoDrop ND-1000 spectrophotometer (Thermo Scientific). The reverse transcription and real-time PCR of miRNA were performed using the miDETECT A TrackTM miRNA qRT-PCR kit (RiboBio). Real-time PCR was performed in a 20-μl reaction mixture containing 2 μl cDNA template (200 ng), 10 μl 2× SYBR Green Mix, 0.5 μl 200 nM forward and reverse primers, and 6 μl nuclease-free water. PCR amplification was performed with an iQ5 system (Bio-Rad) in accordance with the manufacturer's instructions, and RNU6B (U6) was used as an internal control. The forward primers used for miR-183 and U6 and a 3' universal reverse primer were purchased from RiboBio. MiRNA expression was defined based on the threshold cycle (Ct), and the relative expression levels were calculated by the 2-ΔCt method (ΔCt=CtmiR-183–CtU6). Each sample was analyzed in triplicate, and the experiment was repeated at least twice. All PCR experiments were performed by the same investigator, who was blinded for the clinical samples.
Conventional tumor biomarker detection
The serum CEA and CA19–9 levels in 118 CRC patients and 61 healthy individuals were determined using electrochemiluminescence (ECL) on a Cobas e-602 analyzer (Roche).
Statistical analysis
The Mann–Whitney U-test was performed to compare the plasma miRNA expression levels between the CRC patients and the healthy control patients. The Wilcoxon t-test was used to compare the paired plasma samples obtained from the pre- and postoperative patients and the paired tissue samples obtained from primary CRC and matched adjacent non-cancerous colorectal tissues. The Chi-squared test was used to evaluate the correlations between the serum miRNA expression levels and the clinicopathological factors. The CRC biomarkers (plasma miRNA-183, serum CEA, and CA19–9) were assessed by the ROC curve analysis; the sensitivity, specificity, and AUC were determined. The Cox proportional hazard regression test was used to estimate the univariate and multivariate hazard ratios for recurrence and prognosis. The Kaplan–Meier method was used to estimate the distribution of the survival curves; log-rank tests were used to compare the distributions between the groups. All tests were 2-sided, and the significance level was set at P<0.05. SPSS Statistics 20.0 for Windows (IBM) and Graph Pad Prism 5 (GraphPad Software) were used for the data analyses.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Dr. Shemin Lu for his excellent technical assistance with some of the experiments.
Funding
The present study was supported by grants from the National Natural Scientific Foundation of China (#81370069 to K.L.).
References
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA: Cancer J Clin 2011; 61:69-90; PMID:21296855
- Kraus S, Nabiochtchikov I, Shapira S, Arber N. Recent advances in personalized colorectal cancer research. Cancer Letters 2014; 347:15-21; PMID:24491406; http://dx.doi.org/10.1016/j.canlet.2014.01.025
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116:281-97; PMID:14744438; http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA. MicroRNA expression profiles classify human cancers. Nature 2005; 435:834-8; PMID:15944708; http://dx.doi.org/10.1038/nature03702
- Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer 2006; 6:857-66; PMID:17060945; http://dx.doi.org/10.1038/nrc1997
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006; 9:189-98; PMID:16530703; http://dx.doi.org/10.1016/j.ccr.2006.01.025
- Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res 2005; 65:7065-70; PMID:16103053; http://dx.doi.org/10.1158/0008-5472.CAN-05-1783.
- Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci 2008; 105:10513-8; PMID:18663219; http://dx.doi.org/10.1073/pnas.0804549105
- Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, Guo J, Zhang Y, Chen J, Guo X. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 2008; 18:997-1006; PMID:18766170; http://dx.doi.org/10.1038/cr.2008.282
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol Dev 2008; 10:106-13; PMID:18184361; http://dx.doi.org/10.1111/j.1525-142X.2007.00217.x
- Abraham D, Jackson N, Gundara JS, Zhao J, Gill AJ, Delbridge L, Robinson BG, Sidhu SB. MicroRNA profiling of sporadic and hereditary medullary thyroid cancer identifies predictors of nodal metastasis, prognosis, and potential therapeutic targets. Clin Cancer Res 2011; 17:4772-81; PMID:21622722; http://dx.doi.org/10.1158/1078-0432.CCR-11-0242
- Xu X, Dong Z, Li Y, Yang Y, Yuan Z, Qu X, Kong B. The upregulation of signal transducer and activator of transcription 5-dependent microRNA-182 and microRNA-96 promotes ovarian cancer cell proliferation by targeting forkhead box O3 upon leptin stimulation. Int J Biochem Cell Biol 2013; 45:536-45; PMID:23262295; http://dx.doi.org/10.1016/j.biocel.2012.12.010
- Sarver AL, French AJ, Borralho PM, Thayanithy V, Oberg AL, Silverstein KA, Morlan BW, Riska SM, Boardman LA, Cunningham JM. Human colon cancer profiles show differential microRNA expression depending on mismatch repair status and are characteristic of undifferentiated proliferative states. BMC Cancer 2009; 9:401; PMID:19922656; http://dx.doi.org/10.1186/1471-2407-9-401
- Motoyama K, Inoue H, Takatsuno Y, Tanaka F, Mimori K, Uetake H, Sugihara K, Mori M. Over-and under-expressed microRNAs in human colorectal cancer. Int J Oncol 2009; 34:1069-75; PMID:19287964
- Earle JS, Luthra R, Romans A, Abraham R, Ensor J, Yao H, Hamilton SR. Association of microRNA expression with microsatellite instability status in colorectal adenocarcinoma. J Mol Diagn 2010; 12:433-40; PMID:20413677; http://dx.doi.org/10.2353/jmoldx.2010.090154
- Zhang Q-H, Sun H-M, Zheng R-Z, Li Y-C, Zhang Q, Cheng P, Tang Z-H, Huang F. Meta-analysis of microRNA-183 family expression in human cancer studies comparing cancer tissues with noncancerous tissues. Gene 2013; 527:26-32; PMID:23791657; http://dx.doi.org/10.1016/j.gene.2013.06.006
- Zhou T, Zhang G-J, Zhou H, Xiao H-X, Li Y. Overexpression of microRNA-183 in human colorectal cancer and its clinical significance. Eur J Gastroenterol Hepatol 2014; 26:229-33; PMID:24150523; http://dx.doi.org/10.1097/MEG.0000000000000002
- Winawer SJ, Zauber AG, Fletcher RH, Stillman JS, O'Brien MJ, Levin B, Smith RA, Lieberman DA, Burt RW, Levin TR. Guidelines for colonoscopy surveillance after polypectomy: a consensus update by the us multi-society task force on colorectal cancer and the american cancer society*,†. CA: Cancer J Clin 2006; 56:143-59; PMID:16737947; http://dx.doi.org/10.3322/canjclin.56.3.143
- Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet 2009; 10:704-14; PMID:19763153; http://dx.doi.org/10.1038/nrg2634
- Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee M-LT, Schmittgen TD. Detection of microRNA expression in human peripheral blood microvesicles. PloS one 2008; 3:e3694; PMID:19002258; http://dx.doi.org/10.1371/journal.pone.0003694
- Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 2012; 32:326-48; PMID:22383180; http://dx.doi.org/10.1002/med.20215
- Madhavan D, Cuk K, Burwinkel B, Yang R. Cancer diagnosis and prognosis decoded by blood-based circulating microRNA signatures. Front Gen 2013; 4:116; PMID:23802013; http://dx.doi.org/10.3389/fgene.2013.00116
- Zhi F, Cao X, Xie X, Wang B, Dong W, Gu W, Ling Y, Wang R, Yang Y, Liu Y. Identification of circulating micrornas as potential biomarkers for detecting acute myeloid leukemia. PloS one 2013; 8:e56718; PMID:23437222; http://dx.doi.org/10.1371/journal.pone.0056718
- Yu H, Jiang L, Sun C, Guo L, Lin M, Huang J, Zhu L. Decreased circulating miR-375: a potential biomarker for patients with non-small-cell lung cancer. Gene 2014; 534:60-5; PMID:24404590; http://dx.doi.org/10.1016/j.gene.2013.10.024
- Sochor M, Basova P, Pesta M, Dusilkova N, Bartos J, Burda P, Pospisil V, Stopka T. Oncogenic MicroRNAs: miR-155, miR-19a, miR-181b, and miR-24 enable monitoring of early breast cancer in serum. BMC Cancer 2014; 14:448; PMID:24938880; http://dx.doi.org/10.1186/1471-2407-14-448
- Mahn R, Heukamp LC, Rogenhofer S, von Ruecker A, Müller SC, Ellinger J. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology 2011; 77:1265. e9-. e16; PMID:NOT_FOUND; http://dx.doi.org/10.1016/j.urology.2011.01.020
- Komatsu S, Ichikawa D, Takeshita H, Tsujiura M, Morimura R, Nagata H, Kosuga T, Iitaka D, Konishi H, Shiozaki A. Circulating microRNAs in plasma of patients with oesophageal squamous cell carcinoma. British J Cancer 2011; 105:104-11; PMID:21673684; http://dx.doi.org/10.1038/bjc.2011.198
- Zhu X, Lv M, Wang H, Guan W. Identification of circulating MicroRNAs as novel potential biomarkers for gastric cancer detection: a systematic review and meta-analysis. Dig Dis Sci 2014; 59:911-9; PMID:24337687; http://dx.doi.org/10.1007/s10620-013-2970-9
- Fesler A, Jiang J, Zhai H, Ju J. Circulating microRNA Testing for the Early Diagnosis and Follow-up of Colorectal Cancer Patients. Mol Diagn Ther 2014; 18:303-8; PMID:24566942; http://dx.doi.org/10.1007/s40291-014-0089-0
- Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang X, Wang L. Down-regulation of miR-183 promotes migration and invasion of osteosarcoma by targeting Ezrin. Am J Pathol 2012; 180:2440-51; PMID:22525461; http://dx.doi.org/10.1016/j.ajpath.2012.02.023
- Lowery AJ, Miller N, Dwyer RM, Kerin MJ. Dysregulated miR-183 inhibits migration in breast cancer cells. BMC Cancer 2010; 10:502; PMID:20858276; http://dx.doi.org/10.1186/1471-2407-10-502
- Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res 2010; 70:9570-80; PMID:21118966; http://dx.doi.org/10.1158/0008-5472.CAN-10-2074
- Liu C, Adamson E, Mercola D. Transcription factor EGR-1 suppresses the growth and transformation of human HT-1080 fibrosarcoma cells by induction of transforming growth factor beta 1. Proc Natl Acad Sci 1996; 93:11831-6; PMID:NOT_FOUND; http://dx.doi.org/10.1073/pnas.93.21.11831
- Baron V, Adamson ED, Calogero A, Ragona G, Mercola D. The transcription factor Egr1 is a direct regulator of multiple tumor suppressors including TGFβ1, PTEN, p53, and fibronectin. Cancer Gene ther 2005; 13:115-24; PMID:16138117; http://dx.doi.org/10.1038/sj.cgt.7700896
- Krones-Herzig A, Mittal S, Yule K, Liang H, English C, Urcis R, Soni T, Adamson ED, Mercola D. Early growth response 1 acts as a tumor suppressor in vivo and in vitro via regulation of p53. Cancer Res 2005; 65:5133-43; PMID:15958557; http://dx.doi.org/10.1158/0008-5472.CAN-04-3742
- Pfütze K, Luo X, Burwinkel B. MicroRNA signatures as biomarkers of colorectal cancer. MicroRNAs Med 2013:329-42; http://dx.doi.org/10.1002/9781118300312.ch19
- Pu XX, Huang Gl, Guo HQ, Guo CC, Li H, Ye S, Ling S, Jiang L, Tian Y, Lin TY. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol 2010; 25:1674-80; PMID:20880178; http://dx.doi.org/10.1111/j.1440-1746.2010.06417.x
- Cheng H, Zhang L, Cogdell DE, Zheng H, Schetter AJ, Nykter M, Harris CC, Chen K, Hamilton SR, Zhang W. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PloS one 2011; 6:e17745; PMID:21445232; http://dx.doi.org/10.1371/journal.pone.0017745
- Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg 2014; 259:735-43; PMID:23982750; http://dx.doi.org/10.1097/SLA.0b013e3182a6909d
- Pritchard CC, Kroh E, Wood B, Arroyo JD, Dougherty KJ, Miyaji MM, Tait JF, Tewari M. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Preven Res 2012; 5:492-7; PMID:22158052; http://dx.doi.org/10.1158/1940-6207.CAPR-11-0370
- Edge SBBD, Compton CC, Fritz AG, Greene FL, Trotti A, editors, ed. AJCC cancer staging manual (7th ed). New York: NY: Springer, 2010.