Abstract
Colorectal cancer (CRC) is one of the most common cancers worldwide. The molecular mechanisms underlying CRC development involve a multistep process with the accumulation of both genetic and epigenetic changes. To deeply understand CRC tumorigenesis and progression, advances in identification of novel mechanisms and key factors are therefore in an urgent need. Here, we examined the correlation of factor inhibiting HIF-1α (FIH-1) expression with clinicopathological features of CRC. The finding that FIH-1 was not only significantly decreased in tumor tissue but also was significantly correlated with tumor invading depth, lymph node involvement, and metastasis suggested the role of FIH-1 as a tumor suppressor in CRC development. To further support the above hypothesis, we performed both in vitro and in vivo experiments to identify the role of FIH-1 in CRC development. FIH-1 was found to inhibit CRC cell proliferation, migration, invasion, and colony formation in vitro. FIH-1 was also shown to repress LOVO xenograft tumor growth in vivo. To decipher the mechanism, we examined the expression level of HIF-1α and its target genes. We found that FIH-1 was able to inhibit HIF1α mediated transcription of GLUT1 and VEGF in CRC cells. The above observation points to the possibility that loss or decreased expression of FIH-1 gene may lead to a constitutive activation of HIF1α and an alteration of HIF-1 targets such as GLUT-1 and VEGF. These findings highlight the critical role of FIH-1 in CRC and indicate FIH-1 functions as a tumor suppressor in human CRC by repressing HIF1α pathway.
Abbreviations
| FIH-1 | = | Factor inhibiting HIF1α |
| HIF1α | = | Hypoxia-inducible transcription factor 1α |
| VEGF | = | vascular endothelial growth factor |
| GLUT1 | = | glucose transporter 1 |
| CRC | = | colorectal cancer |
| IHC | = | immunohistochemistry |
| TMA | = | tissue microarrays |
Introduction
Colorectal cancer (CRC) is the third most frequent cause of cancer-related death worldwide.Citation1 The molecular mechanisms underlying CRC development involve a multistep process with the accumulation of both genetic and epigenetic changes,Citation2 including alternations in the Hypoxia-inducible transcription factor 1α (HIF-1α) pathway. HIF1α acts as a master regulator of transcriptional response to low oxygen, which is able to respond to a wide range of environmental oxygen concentrations, and control angiogenesis, erythropoiesis, and glycolysis via alterations in at least 300 oxygen-responsive genes.Citation3 Additionally, HIF-1α also plays a pivotal role in maintaining cellular activities even during normoxia.Citation4 According to the Warburg effect, a hallmark of malignant tumors that characterized by increased activity of aerobic glycolysis, HIF-1α is constitutively activated in malignant tumor cells and regulates cellular glycolytic activity.Citation5-7 Furthermore, it was reported that HIF-1α could activate a transcription program that promoted aggressive tumor phenotypes by triggering the expression of critical genes, including vascular endothelial growth factor (VEGF), and glucose transporter 1 (GLUT1).Citation8 As a key regulator of HIF-1α, factor inhibiting HIF-1α (FIH-1) catalyzes an asparagine hydroxylation step that controls the association of HIF-1α transcription factors with CBP/p300 transcriptional co-activators, and reduces the transcriptional activity of HIF-1α.Citation9-11 Recently, several studies identified that FIH-1 was associated with neoplastic progression and might be a candidate tumor suppressor.Citation12-14 However, little is known about the role of FIH-1 in human CRC.
In the previous study, we firstly identified that FIH-1 was downregulated in CRC tissue, compared to the normal colorectal tissue.Citation15 Furthermore, we reported FIH-1 mRNA expression was significantly correlated with tumor T stage status by analysis of The Cancer Genome Atlas (TCGA) sample cohort. In the present study, the correlation of the FIH-1 protein expression level with CRC clinicopathologic features in our sample cohort was examined in support of the tumor suppressor role of FIH-1 in CRC progression. The FIH-1 expression was further shown to control cell proliferation, migration and invasion in CRC cell lines and was found to affect the tumor growth in LOVO xenografts. Together, our data provide strong evidence that aberration of the FIH-1 expression plays a critical role in human CRC tumorigenesis and indicate that FIH-1 inhibits CRC development by repressing HIF-1α pathway.
Results
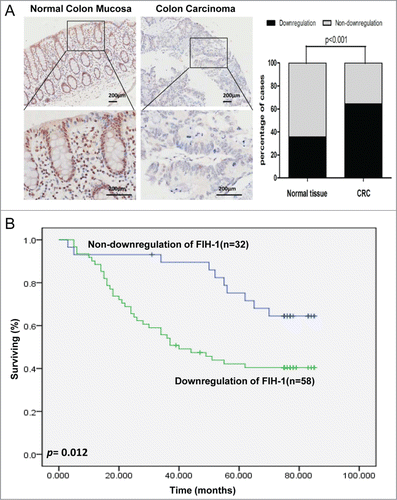
FIH-1 is downregulated in CRC and is a predictor for CRC prognosis
Although we examined the mRNA level of FIH-1 expression in CRC in the previous study, the data about FIH-1 protein expression were unavailable. Furthermore, the data we reported before about the correlation of FIH-1 mRNA expression and tumor T stage status were from TCGA sample cohort, not from Chinese sample cohort of our hospital. To determine FIH-1's role in CRC tumorigenesis, we evaluated FIH-1 protein expression in a large set of 90 colorectal tissue samples from our hospital by IHC analysis on a set of tissue microarrays (TMAs). Using IHC, FIH-1 was shown to localize in both cytoplasm and nucleus of colorectal cells. In the CRC samples, FIH-1 protein expression was significantly downregulated, compared with the adjacent normal samples (P < 0.001, ). This result independently suggested the role of FIH-1 as a tumor suppressor in CRC. In addition, Kaplan-Meier test was employed to analyze the follow-up data and showed that the patients with downregulation of FIH-1 had poorer prognosis than those without downregulation of FIH-1 (p = 0.012, ).
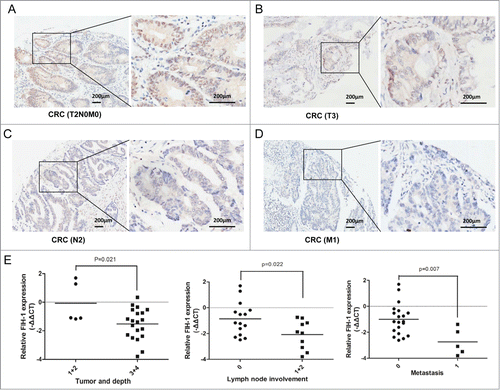
Decreased FIH-1 expression in CRC tissue is associated with disease progression
Given the fact that FIH-1 acts as the key factor in CRC biology, we speculated that FIH-1 expression might play a critical role in suppressing CRC progression. To prove this speculation, we examined the above 90 CRC patient samples with well-annotated clinical information, and analyzed the correlation between FIH-1 protein level and clinicopathology. In addition, we randomly selected 25 pairs of CRC and adjacent normal samples and measured the mRNA level of FIH-1 expression. IHC analysis showed that FIH-1 expression was significantly lower in CRC samples of T(3 + 4) stage (, ), N(1 + 2) stage (, ), and M1 stage (, ) than in CRC samples of T(1 + 2)N0M0 stage (, ). The low mRNA level of FIH-1 expression was also significantly correlated with invading depth, lymph node involvement, and metastasis (). Correlations between the FIH-1 expression level and clinicopathologic characteristics of CRC are summarized in .
Table 1. Clinicopathologic correlation of FIH-1 down-regulation in human CRCs
Figure 2. Decreased FIH-1 expression in CRC tissue is associated with disease progression. (A) FIH-1 expression pattern in T2N0M0 stage CRC sample. (B) FIH-1 expression pattern in T3 stage CRC sample. (C) FIH-1 expression pattern in N2 stage CRC sample. (D) FIH-1 expression pattern in M1 stage CRC sample. (E) Low mRNA level of FIH-1 expression was significantly correlated with CRC invading depth, lymph node involvement, and metastasis.
Increased FIH-1 inhibits CRC cell proliferation, migration, invasion and colony formation in vitro
Thus far we have shown that FIH-1 plays a critical role in both CRC tumorigenesis and progression. We then examined the biological role of FIH-1 in vitro by performing functional assays. First, FIH-1 overexpression plasmids were transfected into LOVO and SW1116 cell lines. We observed an increase of FIH-1 levels. As a consequence, a significant decrease in cell proliferation was observed in both cell lines (). A subsequent wound-healing assay showed that the rate of wound-healing of FIH-1 upregulated cells was significantly slower than that of untreated cells (). At 18h, a wound in LOVO cells was almost closed, while cells treated with FIH-1 overexpression plasmids still showed a noticeable wound. A similar trend was observed in SW1116 cells. Furthermore, upregulation of FIH-1 resulted in significant inhibition of LOVO and SW1116 cell migration and invasion evaluated via cell migration and invasion assays (). Colony formation ability was also inhibited by upregulating FIH-1 in both cell lines (). Taken together, the data suggested that FIH-1 played a tumor suppressor role in CRC development.
Figure 3. Effect of FIH-1 overexpression on proliferation, migration, invasion of LOVO and SW1116 cells. (A) Overexpression of FIH-1 had significant effect on decreasing proliferation rate of both LOVO and SW1116 cell lines. (B) Wound closure was delayed in cells transfected with FIH-1 overexpression plasmidbitorfunctional assays. We 31 can directly regulate FIH-1 expression in both CRC tissue and cell lines. Next we examined th as compared with negative control in 18 h time points in both types of CRC cells. (C) Representative fields of migration (up) or invasion (down) cells on the membrane were on the left. Average migration or invasion cell number per field was on the right. The migration or invasion cell number of LOVO or SW1116 transfected with FIH-1 overexpression plasmidbitorfunctional assays. We 31 can directly regulate FIH-1 expression in both CRC tissue and cell lines. Next we examined th was drastically decreased. (D) The number of clones of LOVO or SW1116 transfected with FIH-1 overexpression plasmid was fewer than that of control cells.
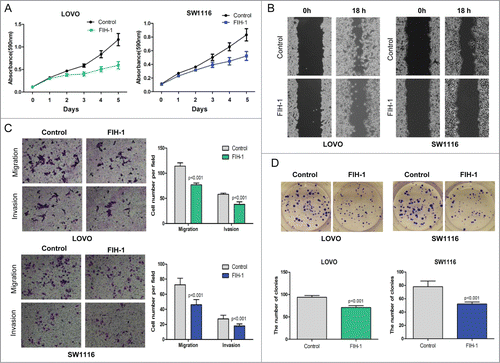
Figure 4. Increased FIH-1 decreases GLUT-1 and VEGF expression levels in LOVO and SW1116. (A) Increased FIH-1 significantly decreased GLUT-1 and VEGF mRNA levels in LOVO cells, but did not affect HIF-1α mRNA level. (B) Increased FIH-1 decreased GLUT-1 and VEGF protein levels in LOVO cells, but did not affect HIF-1α protein level. (C) Increased FIH-1 significantly decreased GLUT-1 and VEGF mRNA levels in SW1116 cells, but did not affect HIF-1α mRNA level. (D) Increased FIH-1 decreased GLUT-1 and VEGF protein levels in SW1116 cells, but did not affect HIF-1α protein level.
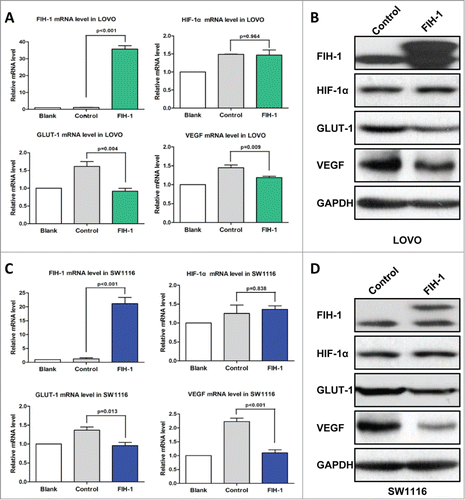
Increased FIH-1 decreases GLUT-1 and VEGF expression levels in LOVO and SW1116
LOVO and SW1116 cells were transiently transfected with plasmid expressing FIH-1, and expression levels of HIF-1α and target genes of HIF-1α, including GLUT-1,Citation16 were examined. First, we found HIF-1α expression was not altered after overexpressing FIH-1. But we observed a decrease in the level of GLUT-1 mRNA (). Furthermore, we also observed a decrease in the level of GLUT-1 protein in both cell lines ().
Apart from GLUT-1, we also examined FIH-1 regulation on VEGF expression, another HIF-1α target gene.Citation17 Like GLUT-1, overexpression of FIH-1 in LOVO and SW1116 cells downregulated VEGF mRNA and protein levels ().
FIH-1 controls the tumor growth of LOVO xenografts in vivo
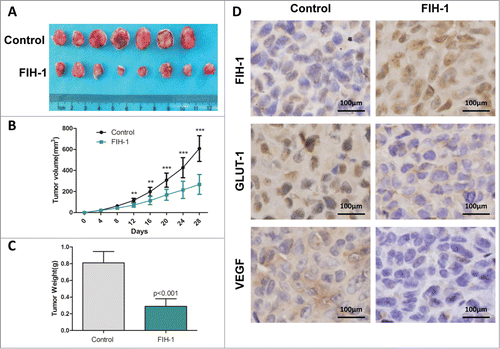
To further investigate whether FIH-1 affects tumor formation in vivo, LOVO cells were stably transfected with LV.FIH-1 and LV.Empty. The cells were then implanted subcutaneously into nude mice and the tumors were monitored every 4 days. At the end of the fourth week, all mice were sacrificed, and the tumor volumes and weights were measured. There was a significant difference in average tumor volume and weight between the 2 groups (). The tumors were larger and heavier in the control group than those in the FIH-1 upregulated group. To further determine whether the growth inhibition was through FIH-1, we performed IHC staining on tumor tissues. We found that the expression level of FIH-1 was increased in the tumors of nude mice injected by FIH-1 upregulated LOVO cells, while the expression levels of GLUT1 and VEGF were decreased (). These data agreed with in vitro results implicating the role of FIH-1 in CRC tumorigenesis and progression.
Figure 5. FIH-1 controls the tumor growth of LOVO xenografts in vivo. (A) Overexpression of FIH-1 strikingly decreased the growth of LOVO cells xenografted in nude mice. (B) The tumors were significantly bigger in control group than those in treated group. (C) The tumors were significantly heavier in control group than those in treated group. (D) The protein expression levels of FIH-1, GLUT-1, and VEGF in the subcutaneous tumor tissues collected from control and treated groups were detected by IHC.
Discussion
HIF-1α regulates the expression of several genes during adaptation to hypoxic conditions and also plays a pivotal role in maintaining cellular activities during normoxia.Citation18 HIF-1α is constitutively activated in malignant tumor cells and its activity or stability is maintained by FIH-1. FIH-1 hydroxylates HIF-1α, blocks the association of HIF-1α with the transcriptional co-activators, and inhibits transcriptional activation.Citation10 Although FIH-1 functions as a transcriptional repressor of HIF target genes and is associated with neoplastic progression, it may have diverse biological functions in different organisms and diseases.Citation19 Liu et al. reported FIH-1 expression decreased the oncogenic potential of head and neck squamous cell carcinoma cells and decreased FIH-1 led to an increased VEGF during head and neck carcinogenesis.Citation20 In Couvelard et al.'s study, FIH-1 was highly expressed in pancreatic endocrine tumors and its expression was correlated with tumor metastases, tumor recurrence, and prognosis.Citation12 Although we reported that FIH-1 was a potential tumor suppressor in human CRC,Citation15 the biological role of FIH-1 in this disease is not well understood.
In the previous study, we firstly identified that FIH-1 was downregulated in CRC tissue, compared to the normal colorectal tissue.Citation15 By analyzing TCGA CRC sample cohort, we found that FIH-1 mRNA expression was significantly correlated with tumor invading depth. However, we didn't analyze the FIH-1 protein levels in CRC, which were not included in TCGA database. In the present study, we examined FIH-1 protein expression in human CRC tissues and found FIH-1 protein level was downregulated in CRC, compared with the adjacent normal samples. Furthermore, CRC with downregulation of FIH-1 had a poor prognosis. These results implicated that FIH-1 might play an important role in CRC progression. To further support this hypothesis, we analyzed the correlation of FIH-1 expression with clinicopathological features of CRC by using our independent data. Both in protein and mRNA expression levels, the downregulation of FIH-1 was significantly correlated with CRC invading depth, lymph node involvement, and metastasis. These data supplement our previous results and indicate that FIH-1 protein may suppress CRC progression and loss of FIH-1 expression is a critical event in human CRC.
In support of the potential tumor suppressor role of the FIH-1 in CRC tumorigenesis and progression, we exogenously upregulated FIH-1 in LOVO and SW1116 cells, and found FIH-1 overexpression was shown to repress CRC cell proliferation, migration, invasion, and colony information. To decipher the mechanism, we examined the expression levels of HIF-1α and its target genes. The most well studied HIF-1α activated growth factors regulate endothelial cell proliferation and blood vessel formation. HIF-1α activates transcription of VEGF and one of its receptors, VEGF receptor 1.Citation8 VEGF is a key angiogenic factor that is secreted by cancer cells and normal cells in response to hypoxia.Citation21 In addition to VEGF, the glucose transporters GLUT1 that mediates cellular glucose uptake has been also shown to be regulated by HIF-1α.Citation22 Studies indicate that increased glycolysis is a normal response to proliferation, and that migrating cells also use this pathway as an energy source.Citation23 In this study, we found HIF-1α expression was not altered after overexpressing FIH-1, but we observed decreased expression of GLUT-1 and VEGF in the level of both mRNA and protein, which underscored FIH-1 ability to inhibit HIF-mediated gene transcription in CRC. Mahon et al. reported that interaction of FIH-1 and VHL could repress HIF-1α transactivation domain function.Citation10 Additionally, Wang et al. found overexpressing FIH-1 could interfere with HIF function and reduce the expression of GLUT-1 and VEGF.Citation24 In light of these observations, we consider that upregulation of FIH-1 may inhibit HIF-1α activation and suppress the HIF-1α pathway in CRC.
To confirm the results of in vitro assays, the LOVO xenograft experiment was performed. The tumors were bigger and heavier in nude mice injected by control cells than in those injected by LOVO cells with upregulated FIH-1. IHC staining results showed that the expression level of FIH-1 protein was increased in tumors of nude mice injected by FIH-1 upregulated LOVO cells, while the expression levels of GLUT1 and VEGF were decreased. These data agreed with in vitro results implicating the role of FIH-1 in CRC tumorigenesis and progression. Overexpression of FIH-1 in LOVO cells repressed HIF-1α activation, caused downregulation of GLUT1 and VEGF, inhibited cellular glucose uptake and angiogenesis, and affected xenografts.
In the present study, one of the limitations is that the experiments under hypoxia are not performed. However, previous reports demonstrated that unlike other hydroxylases, FIH-1 had the ability to function even at substantially low oxygen concentrations.Citation25 Active FIH-1 could also interfere with HIF function at a certain hypoxic stage.Citation25 In this respect, the absence of FIH-1 expression and activity may be a necessary step for HIF function in CRC.Citation24
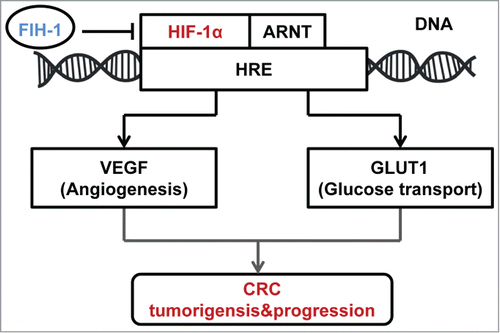
In summary, the results from this study suggest that FIH-1 functions as a tumor suppressor in human CRC development. Loss of FIH-1 expression that affects HIF mediated gene transcription is a crucial event in CRC tumorigenesis and progression, which was not reported in CRC before (). Further in-depth studies are needed to examine FIH-1 regulates transcriptional activation of HIF-1α under hypoxic conditions in CRC. Nevertheless, our data indicate that FIH-1 may play a tumor suppressor role in CRC tumorigenesis and progression through repressing HIF-1α pathway. Further understanding of the pathogenic significance of loss of FIH-1 can contribute to a better diagnosis and treatment of CRC.
Materials and Methods
Patient samples
Written informed consent was obtained from each patient and the investigation was approved by the institutional review board of Zhongshan Hospital, Shanghai Fudan University. Total 90 CRC samples and their paired noncancerous tissues were collected at the time of surgical resection at Zhongshan Hospital. Samples were immediately snap-frozen in liquid nitrogen and stored at −80°C. The patients’ clinicopathological data were also collected.
Cell culture
LOVO and SW1116 human CRC cells were obtained from the Institute of Cell Biology at the Chinese Academy of Sciences. Cell lines were chosen according to the origins and the genomic features.Citation26 LOVO cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS), whereas SW1116 cells were cultured in L-15 medium supplemented with 10% FBS. All cells were cultured in a humidified incubator at 37°C with 5% CO2.
Overexpression of FIH-1
The FIH-1 overexpression plasmid and negative control were purchased from Gene Chem. Cells were seeded at 2 × 10Citation5 per well in 6-well plates and allowed to attach for at least 16 hours. The overexpression plasmid or negative control was transfected using lipofectamine 2000 (Invitrogen). The stable cell lines were established by lentiviral infection.Citation19 LV.FIH-1 and LV.Empty were obtained from GeneChem.
Proliferation assay
Cells were plated in 96-well plates with 1 × 103 cells/well in regular growth medium. Proliferation of cancer cells was measured 5 days after treatment by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay.
Wound healing assay
Cells (1 × 105 per well) were seeded in 6-well plates and grown to confluence. The cells were pretreated with Mitomycin C (0.5 μg/ml), which inhibits cell division, so that the difference in motility was not affected by differences in cell proliferation rates. Then, the cells were wounded with a pipette tip. After being washed with PBS for 3 times, the cells were cultured in full medium and allowed to close the wound for 18 h. Photos of the wound were taken immediately after scratching 18 h later in the same marked location of the dish.
Cell migration and invasion assays
A 24-well transwell plate (8-mm pore size, Corning) was used to measure each cell line's migratory and invasive ability. For the migration assay, 5 × 104 cells were plated in the top chamber lined with a non-coated membrane. For the invasion assay, chamber inserts were coated with 200 mg/ml of Matrigel and dried overnight under sterile conditions. Then, 1 × 105 cells were plated in the top chamber. In both assays, cells were suspended in medium without serum, and medium supplemented with serum was used as a chemo-attractant in the lower chamber. After incubation at 37°C for 24 h, the top chambers were wiped with cotton wool to remove the nonmigratory or noninvasive cells. The invading cells on the underside of the membrane were fixed in 4% Paraformaldehyde for 15 min, airdried, stained in Giemsa (Jiancheng Bioengineering Institute), and counted under a microscope.
Colony formation assay
Colony formation assay in soft agar was carried out according to standard procedures. The cells were mixed with tissue culture media containing 0.6% agar to result in a final agar concentration of 0.4%. Cell suspension 1 mL was immediately plated in 6-well plates coated with 1 mL/well of 0.6% agar in tissue culture media. The colonies were counted in triplicate on day 15 after plating.
RNA extraction and qRT-PCR Assay
Total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. TaqMan real-time PCR assays for FIH-1, HIF-1α, GLUT1, VEGF, and GAPDH were from Takara Bio. All reactions, including the no template controls, were run in triplicate. After the reactions were completed, the CT values were determined by using fixed threshold settings. Data were analyzed using the 2ΔCT method.
Western blotting
Cellular proteins were extracted and separated in SDS-PAGE gels, and western blot analyses were performed according to standard procedures. Western blotting of GAPDH on the same membrane was used as a loading control. The antibodies used were anti-FIH-1 (ab92498, 1:1000), anti-HIF1α (ab51608, 1:1000), anti-GLUT1 (ab652, 1:2000), anti-VEGF (ab1316, 1:1000), and anti-GAPDH (ab37168, 1:5000), from Abcam.
Histological analyses
After mice were sacrificed according to experimental protocol, tumor tissues were isolated, fixed, and embedded in paraffin for histopathological analysis. Immunohistochemistery (IHC) was performed according to standard procedures. The antibodies used were the same as in protein gel blotting. Imaging from tumor tissue was detected with microscope. Compared to the paired normal tissue, downregulation of FIH-1 in CRC tissue was defined by IHC staining. No-downregulation of FIH-1 was defined as no differences of IHC staining between CRC tissue and its paired normal tissue, or upregulation of FIH-1 in CRC tissue.
Immunodeficient mouse xenograft tumor model
All animal experiments were performed according to the regulations of P.R. China and Fudan University, and approved by the animal care and use committee of Fudan University. LOVO cells, which were infected with LV.FIH-1 and LV.Empty, were harvested and injected subcutaneously into the right flank of male nude (nu/nu) mice (2 × 106 viable tumor cells/mouse). The animals were equally divided into control and treated groups (8 mice per group). After subcutaneous implantation of cells, animals were observed daily for tumor growth and subcutaneous tumors were measured on 4, 8, 12, 16, 20, 24, and 28 days. Each tumor volume was calculated according to the following equation: V(mm3) = D2 (mm2) × L (mm)/2, where D and L were the smallest and the largest perpendicular tumor diameters, respectively. The mice were sacrificed at 28 days post-implantation (one mouse of the control group died and there were 7 mice in control group finally); then xenograft tumors were removed for further investigation.
Statistical analysis
All experiments were performed in triplicate. Differences between groups were calculated using Student's t test, Chi-square test, or Fisher's exact test. Additionally, P < 0.05 was selected to indicate a significant difference. These data were analyzed using SPSS version 17.0.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
Project supported by the National Natural Science Foundation of China (81101566) and Scientific Funds of Shanghai Government (12QA1400600, XYQ2011017, 2013SY045, 2013SY054, 11411950500, 11411950501, 13411951600, 201305). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Reference
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin 2013; 63:11-30; PMID:23335087; http://dx.doi.org/10.3322/caac.21166
- Jia Y, Guo M. Epigenetic changes in colorectal cancer. Chin J Cancer. 2013; 32(1):21-30; http://dx.doi.org/10.5732/cjc.011.10245
- Weidemann A, Johnson RS. Biology of HIF-1alpha. Cell Death Differ 2008; 15:621-7; PMID:18259201; http://dx.doi.org/10.1038/cdd.2008.12
- Sakamoto T, Niiya D, Seiki M. Targeting the Warburg effect that arises in tumor cells expressing membrane type-1 matrix metalloproteinase. J Biol Chem 2011; 286:14691-704; PMID:21372132; http://dx.doi.org/10.1074/jbc.M110.188714
- Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Curr Opin Genet Dev 2010; 20:51-6; PMID:19942427; http://dx.doi.org/10.1016/j.gde.2009.10.009
- Wang Y, Li Z, Zhang H, Jin H, Sun L, Dong H, Xu M, Zhao P, Zhang B, Wang J, et al. HIF-1α and HIF-2α correlate with migration and invasion in gastric cancer. Cancer Biol Ther 2010; 10:376-82; PMID:20559021; http://dx.doi.org/10.4161/cbt.10.4.12441
- Hammoudi N, Ahmed KB, Garcia-Prieto C, Huang P. Metabolic alterations in cancer cells and therapeutic implications. Chin J Cancer 2011; 30(8):508-25; PMID:21801600; http://dx.doi.org/10.5732/cjc.011.10267
- Harris AL. Hypoxia–a key regulatory factor in tumour growth. Nat Rev Cancer 2002; 2:38-47; PMID:11902584; http://dx.doi.org/10.1038/nrc704
- Lando D, Peet DJ, Gorman JJ, Whelan DA, Whitelaw ML, Bruick RK. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev 2002; 16:1466-71; PMID:12080085; http://dx.doi.org/10.1101/gad.991402
- Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev 2001; 15:2675-86; PMID:11641274; http://dx.doi.org/10.1101/gad.924501
- Coleman ML, McDonough MA, Hewitson KS, Coles C, Mecinovic J, Edelmann M, Cook KM, Cockman ME, Lancaster DE, Kessler BM, et al. Asparaginyl hydroxylation of the Notch ankyrin repeat domain by factor inhibiting hypoxia-inducible factor. J Biol Chem 2007; 282:24027-38; PMID:17573339; http://dx.doi.org/10.1074/jbc.M704102200
- Couvelard A, Deschamps L, Rebours V, Sauvanet A, Gatter K, Pezzella F, Ruszniewski P, Bedossa P. Overexpression of the oxygen sensors PHD-1, PHD-2, PHD-3, and FIH Is associated with tumor aggressiveness in pancreatic endocrine tumors. Clin Cancer Res 2008; 14:6634-9; PMID:18927305; http://dx.doi.org/10.1158/1078-0432.CCR-07-5258
- Giatromanolaki A, Koukourakis MI, Pezzella F, Turley H, Sivridis E, Bouros D, Bougioukas G, Harris AL, Gatter KC. Expression of prolyl-hydroxylases PHD-1, 2 and 3 and of the asparagine hydroxylase FIH in non-small cell lung cancer relates to an activated HIF pathway. Cancer Lett 2008; 262:87-93; PMID:18187257; http://dx.doi.org/10.1016/j.canlet.2007.11.041
- Morris MR, Maina E, Morgan NV, Gentle D, Astuti D, Moch H, Kishida T, Yao M, Schraml P, Richards FM, et al. Molecular genetic analysis of FIH-1, FH, and SDHB candidate tumour suppressor genes in renal cell carcinoma. J Clin Pathol 2004; 57:706-11; PMID:15220362; http://dx.doi.org/10.1136/jcp.2003.011767
- Chen T, Yao LQ, Shi Q, Ren Z, Ye LC, Xu JM, Zhou PH, Zhong YS. MicroRNA-31 contributes to colorectal cancer development by targeting factor inhibiting HIF-1α (FIH-1). Cancer Biol Ther 2014; 15:516-23; PMID:24521875; http://dx.doi.org/10.4161/cbt.28017
- Greijer AE, van der Groep P, Kemming D, Shvarts A, Semenza GL, Meijer GA, van de Wiel MA, Belien JA, van Diest PJ, van der Wall E. Up-regulation of gene expression by hypoxia is mediated predominantly by hypoxia-inducible factor 1 (HIF-1). J Pathol 2005; 206:291-304; PMID:15906272; http://dx.doi.org/10.1002/path.1778
- Forsythe JA, Jiang BH, Iyer NV, Agani F, Leung SW, Koos RD, Semenza GL. Activation of vascular endothelial growth factor gene transcription by hypoxia-inducible factor 1. Mol Cell Biol 1996; 16:4604-13; PMID:8756616
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, et al. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1. Genes Dev 1998; 12:149-62; PMID:9436976; http://dx.doi.org/10.1101/gad.12.2.149
- Schödel J, Bohr D, Klanke B, Schley G, Schlötzer-Schrehardt U, Warnecke C, Kurtz A, Amann K, Eckardt KU, Willam C. Factor inhibiting HIF limits the expression of hypoxia inducible genes in podocytes and distal tubular cells. Kidney Int 2010; 78:857-67; PMID:20720525; http://dx.doi.org/10.1038/ki.2010.284
- Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY, Wu KJ, Chiou SH, Lin SC, Chang KW. miR-31 ablates expression of the HIF regulatory factor FIH to activate the HIF pathway in head and neck carcinoma. Cancer Res 2010; 70:1635-44; PMID:20145132; http://dx.doi.org/10.1158/0008-5472.CAN-09-2291
- Stacker SA, Achen MG. The VEGF signaling pathway in cancer: the road ahead. Chin J Cancer 2013; 32(6):297-302; PMID:23419196
- Chen C, Pore N, Behrooz A, Ismail-Beigi F, Maity A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J Biol Chem 2001; 276:9519-25; PMID:11120745; http://dx.doi.org/10.1074/jbc.M010144200
- Mazurek S, Boschek CB, Eigenbrodt E. The role of phosphometabolites in cell proliferation, energy metabolism, and tumor therapy. J Bioenerg Biomembr 1997; 29:315-30; PMID:9387092; http://dx.doi.org/10.1023/A:1022490512705
- Wang E, Zhang C, Polavaram N, Liu F, Wu G, Schroeder MA, Lau JS, Mukhopadhyay D, Jiang SW, O'Neill BP, et al. The role of factor inhibiting HIF (FIH-1) in inhibiting HIF-1 transcriptional activity in glioblastoma multiforme. PLoS One 2014; 9:e86102; PMID:24465898; http://dx.doi.org/10.1371/journal.pone.0086102
- Stolze IP, Tian YM, Appelhoff RJ, Turley H, Wykoff CC, Gleadle JM, Ratcliffe PJ. Genetic analysis of the role of the asparaginyl hydroxylase factor inhibiting hypoxia-inducible factor (FIH) in regulating hypoxia-inducible factor (HIF) transcriptional target genes. J Biol Chem 2004; 279:42719-25; PMID:15302861; http://dx.doi.org/10.1074/jbc.M406713200
- Ahmed D, Eide PW, Eilertsen IA, Danielsen SA, Eknæs M, Hektoen M, Lind GE, Lothe RA. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013; 2:e71; PMID:24042735; http://dx.doi.org/10.1038/oncsis.2013.35