Abstract
Background: Our previous findings showed that miR-33 expressed abnormally in clinical specimens of melanoma, but the exact molecular mechanism has not been elucidated. Object: To determine miR-33's roles in melanoma and confirm whether HIF-1α is a direct target gene of miR-33a. Methods: First miR-33a/b expression levels were detected in HM, WM35, WM451, A375 and SK-MEL-1. Then lentiviral vectors were constructed to intervene miR-33a expression in melanoma cells. Cell proliferation, invasion and metastasis were detected. A375 cells mice model was performed to test the tumorigenesis of melanoma in vivo. Finally the dual reporter gene assay was carried out to confirm whether HIF-1α is a direct target gene of miR-33a. Results: MiR-33a/b exhibited a lower expression in WM35, WM451, A375 and SK-MEL-1 of the metastatic skin melanoma cell lines than that in HM. Then inhibition of miR-33a expression in WM35 and WM451 cell lines could promote cell proliferation, invasion and metastasis. Conversely, increased expression of miR-33a in A375 cells could inhibit cellproliferation, invasion and metastasis. In vivo tests also confirmed that overexpression of miR-33a in A375 cells significantly inhibited melanoma tumorigenesis. Finally, we confirmed that HIF-1α is a direct target gene of miR-33a. Conclusion: The newly identified miR-33a/HIF-1α axis might provide a new strategy for the treatment of melanoma.
Abbreviations
| miR-33a/b | = | microRNA 33a/b |
| HIF-1α | = | hypoxia inducible factor 1, α subunit |
Introduction
Malignant melanoma causes about 80% deaths related to skin cancer, and the incidence increases significantly in recent years. In addition, a familial aggregation appears in malignant melanoma. Melanoma progresses rapidly and is easy to transfer. The metastatic phenotype of malignant melanoma is one of the main reasons for cancer-related deaths. It has been reported that tumor invasion and metastasis is closely related to epithelial-mesenchymal transition, tumor microenvironment changes, angiogenesis and lymph angiogenesis.Citation1 Therefore, understanding the underlying molecular mechanism involved in the process of invasion and metastasis of melanoma is crucial.
Recent studies have shown that there are many microRNAs aberrantly expressing in melanoma, and microRNAs as oncogenes or tumor suppressor genes are able to be involved in the development and progression of melanoma, and play an important function in tumor cell proliferation and apoptosis, metabolism and immune, as well as invasion and metastasis.Citation2 To inhibit proliferation, invasion and metastasis of melanoma, miR-34a and miR-137 are targeted to regulate c-met,Citation3,4 miR-18b is shown to target MDM2-p53 signaling pathway.Citation5 And miR-7-5p partially inhibits IRS-2 expression and Akt signaling pathway.Citation6 Conversely,miR-195 can be targeted to regulate WEE1,Citation7 miR-182regulates MATF-M and FOXO3,Citation8 miR-193b can down-regulate STMN1 expression, and miR-376a and miR-376c control the expression of IGF1R,Citation9 in order to promote proliferation, invasion and metastasis of melanoma.Citation10 Up to now, the special molecular mechanism by which microRNA in melanoma affects the tumor invasion and metastasis is still unclear.
Our previous studies have shown abnormal expression of miR-33, let-7b and miR-199a in microRNA microarray from clinical specimens of melanoma. It has been verified that let-7b and miR-199a can suppress proliferation of melanoma cells.Citation11 In the present study, miR-33a/b was lower in the metastatic melanoma cells than that in the normal melanocytes. In vitro tests exhibited that miR-33a could inhibit proliferation, invasion and metastasis of melanoma cells. In vivo experiment was further confirmed that miR-33a had anti-tumor growth and anti-metastatic effects of melanoma cells. These findings indicate that targeting miR-33a may be a new therapeutic strategy for treatment of melanoma.
Results
Low expression of miR-33a/b in human metastatic melanoma cells
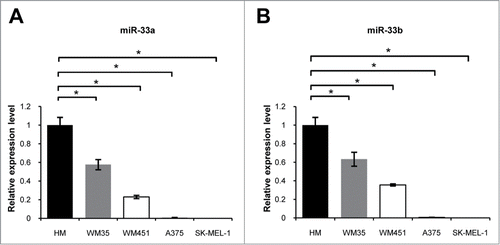
In order to verify microRNA microarray results of previous clinical studies, we made real-time PCR detection of miR-33a/b expression in primary melanocytes (HM) and melanoma cell lines with different metastatic potentials. The results showed that the expressions of miR-33a and miR-33b were all higher in primary melanocytes (HM) than in melanoma cell lines WM35, WM451, A375 and SK-MEL-1 (P < 0.05, ). These findings indicate that miR-33a/b may be tightly associated with the metastatic phenotype of malignant melanoma. Because of a same expression trend for miR-33a and miR-33b in HM and melanoma cell lines, we only selected miR-33a for in-depth study.
Figure 1. miR-33a/b was downregulated in metastatic melanoma cell lines: real-time PCR assay was used to detect miR-33a / b expression in melanoma cell lines with different metastatic potentials. (A) miR-33a in WM35, WM451, A375 and SK-MEL-1 was significantly reduced, normalized with that in HM, respectively (*P < 0.05). (B) miR-33b in WM35, WM451, A375 and SK-MEL-1 was significantly reduced, normalized with that in HM, respectively (*P < 0.05).
miR-33a inhibits the HIF-1α expression
To further explore the biological functions of miR-33a in melanoma cells, we cloned the mir-33a-sponge to bind to pLVX-IRES-ZsGreen1 plasmid so as to construct pYr-LVX-miR-33a-sponge lentiviral expression vector (Fig. S1A) with down-regulated miR-33a expression. After transfection, the expression of miR-33a in WM35 and WM451 cells was 49%–55%. After transfection with pYr-LVX-pri-miR-33a lentivirus vector (Fig. S1B) overexpressing miR-33a constructed using the above-mentioned method, miR-33a expression in transfected A375 cells could be increased by about 12 folds (). The results showed that we successfully obtained melanoma cell lines which stably up-regulated or downregulated miR-33a.
Figure 2. miR-33a was down-regulated or overexpressed in melanoma cells. (A) Real-time PCR detection of the relative expression level of miR-33a in pYr-LVX-miR-33a-sponge-transfected WM35 and WM451 as compared with that after transfection with the blank vector (*P < 0.05). Real-time PCR detect the miR-33a relative expressionin pYr-LVX-pri-miR-33a-transfected as compared with that in empty vector-transfected A375 (*P < 0.05). (B) When miR-33a was downregulated, HIF-1α protein level in WM35 and WM451 cells was significantly increased; when miR-33a overexpressed, HIF-1α protein level in A375 cells was significantly reduced. (C) Relative expression levels of HIF-1α protein in corresponding cells, normalized with that in WM35 cells (*P <0 .05).
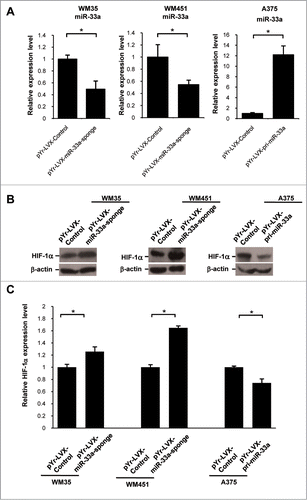
We firstly detected miR-33a effects on HIF-1α protein expression using western blot assay. The results showed that after down-regulation of miR-33a, the protein level of HIF-1α in WM35 cells was remarkably increased to be (1.26 ± 0.08) times that of the negative control group(P < 0.05), while the protein level of HIF-1α in WM451 cells was significantly increased to be (1. 65 ± 0.03) times that of the negative control group (P < 0.05). However, the protein level of HIF-1α in A375 cells overexpressing miR-33a was significantly lower than that of the negative control group (P < 0.05, ).
miR-33a inhibits the proliferation of human melanoma cells
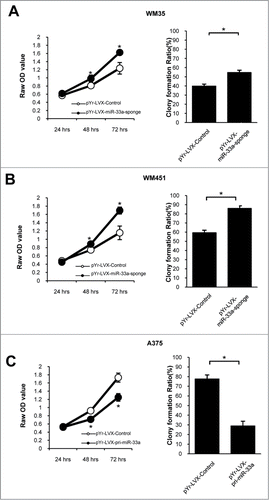
We examined the miR-33a effects on melanoma cell proliferation, and found that down-regulating miR-33a expression could raise proliferative ability of WM35 and WM451 cells. Colony-formation efficiency in WM35 cells was (54.6 ± 0.26) % in pYr-LVX-miR-33a-sponge group and (39.8 ± 0.23) % in negative control group (P < 0.05), while the colony-formation efficiency in WM451 cells was (85.9 ± 0.28) % and (59.4 ± 0.28) %, respectively (P < 0.05, ). On the contrary, overexpression of miR-33a could reduce the proliferative ability of A375 cells. The colony-formation efficiency in A375 cells was (41.6 ± 0.48) % in the pYr-LVX-pri-miR-33a group and (77.5 ± 0.43) % in the negative control group (P < 0.05, ). The MTT assays also have similar results. Thus, miR-33a could significantly inhibit the proliferation of human melanoma cells.
Figure 3. miR-33a suppresses melanoma cell proliferation. MTT assay and colony formation assay were performed for miR-33a effects on cell proliferation and clone formation. In colony formation test, there were 1000 cells per well. (A) WM35 cells transfected with pYr-LVX-miR-33a-sponge had a stronger proliferation ability than those transfected with blank vector, and the colony formation efficiency also increased after transfection with pYr-LVX-miR-33a-sponge (*P < 0.05). (B) The proliferative ability and colony formation efficiency were significantly higher in pYr-LVX-miR-33a-sponge-transfected WM451 cells than the blank vector group (*P < 0.05). (C) The proliferative ability and colony formation efficiency of pYr-LVX-pri-miR-33a-transfected A375 cells was significantly lower than those transfected with the blank vector (*P < 0.05).
miR-33a inhibits the invasion of human melanoma cells
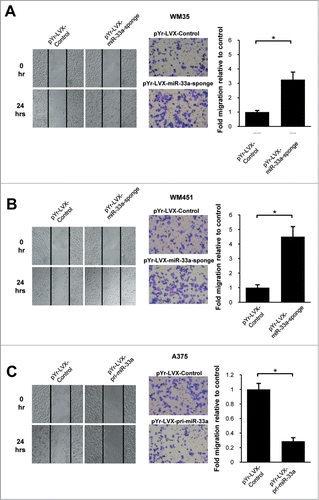
The miR-33a had a lower expression in the high-metastatic melanoma cell lines, suggesting that miR-33a may be associated with metastatic phenotype of malignant melanoma. Our experimental findings in scratch tests showed that downregulation of miR-33a could increase the invasion and metastasis of WM35 and WM451 cells (P < 0.05; ). In contrast, after overexpression of miR-33a, A375 cells appeared to have a reduction in invasion of tumor cells (P < 0.05; ). In the Transwell experiments, after down-regulating miR-33a, the number of WM35 and WM451 cells penetrating the Transwell chamber was (3.26 ± 0.52) folds and (4.50 ± 0.70) folds that of the negative control group (P<0.05; ); while the number of A375 cells penetrating the Transwell chamber after overexpressing miR-33a was only (0.29 ± 0.05)% of that in the negative control group (P < 0.05; ). In summary, miR-33a could significantly inhibit the invasion of human melanoma cells.
Figure 4. miR-33a inhibits melanoma cell migration. Cell scratch test and Transwell assay were used to detect miR-33a effect on melanoma cell migration. (A) WM35 cells transfected with pYr-LVX-miR-33a-sponge had a higher cell migration ability and more cells passing through the Transwell chamber as compared with those transfected with blank vector (*P < 0.05). (B) WM451 cells transfected with pYr-LVX-miR-33a-sponge had a higher cell migration ability and more cells passing through the Transwell chamber as compared with those transfected with blank vector (*P < 0.05). (C) The cell migration ability of pYr-LVX-pri-miR-33a-transfected A375 cells was significantly decreased as compared with the blank vector group, as well as the number of cells passing through the Transwell chamber significantly reduced (*P < 0.05).
Anti-tumor growth and anti-metastatic effects of miR-33a of human melanoma in nude mice
We further validated the effects of upregulated miR-33a in A375 cells on the growth of human melanoma xenograft formation in nude mice. Nude mice were subcutaneously inoculated human melanoma A375 cells blank vector and pYr-LVX-pri-miR-33a vector, respectively. After 30 days, the mice were sacrificed to remove and weigh tumors. The average tumor weight in the blank vector group was (0.37 ± 0.09) g, which was significantly higher than that in the pYr-LVX-pri-miR-33a group (0.16 ± 0.04) g. The average tumor volume in the blank vector group was (258.4 ± 153) mm3, which was significantly higher than that in the pYr-LVX-pri-miR-33a group (89.3 ± 66.82)mm3 (P < 0.05; ).
Figure 5. miR-33a reduces in vivo growth and metastasis of A375 xenografts. miR-33a effects on in vivo growth of A375 xenografts in nude mice were detected through subcutaneous tumor implantation. (A) Left figure shows tumor formation in nude mice of the blank vector group and pYr-LVX-pir-miR-33a group. Right figure shows the excision tumor of A375 xenografts in the blank vector and pYr-LVX-pir-miR-33a groups (*P < 0.05). (B) This figure show the tumor volume of excision tumor by the formula V(mm3) = 0.5 × a × b2(a: maximum length to diameter, b: maximum transverse diameter) (*P < 0.05). (C) This figure show the tumor weight of excision tumor (*P < 0.05). (D) Mice were injected i.v. with melanoma cells to determine their metastatic activity. The lung tissue of mice in pYr-LVX-pir-miR-33a group was normal, whereas the lung tissue of mice in the blank vector group remarkably manifested melanoma colonies (*P < 0.05).
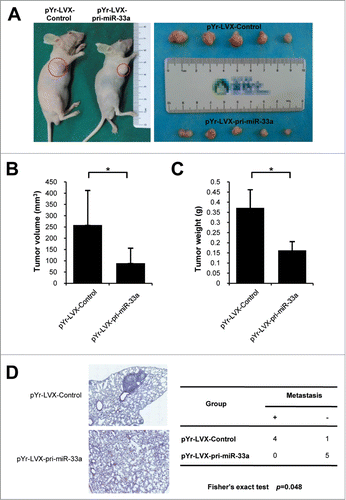
None of mice in the pYr-LVX-pri-miR-33a group (n = 0) developed lung metastasis while 80% of blank vector group (n = 4) developed lung tumor colonies detected in HE stained sections (). Variances were statistically significantly different (P < 0.05).
These results of in vivo experiments suggest that miR-33a could significantly inhibit human melanoma xenograft growth and metastasis in nude mice.
miR-33a directly targets HIF-1α
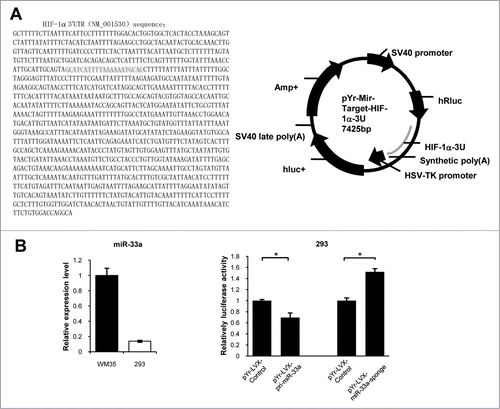
A microRNA is confirmed to be able to simultaneously regulate multiple target genes. In combination with the above experimental results, we used a series of bioinformatics software to predict miR-33a downstream target genes. Interestingly, PicTar (2006) and miRanda (2010) databases both prompt that HIF −1α is most likely to be a target gene of miR-33a.
HIF-1α was remarkably lower at both gene transcription and translation levels in up-regulated miR-33a nude mice group than that in blank vector group (Figs. S2A, B).To confirm whether HIF −1α is the direct target gene of miR-33a, we constructed dual-luciferase reporter plasmids containing HIF-1α 3 'UTR (). Then, 293 cells with high transfection efficiency, in which the miR-33a expression was only about 1/5 as much as that in WM35 cells, were adopted for dual-luciferase experiments to further validate whether HIF-1α is a direct target gene of miR-33a. The results showed that with the reporter plasmid containing HIF-1α 3 'UTR had the dual luciferase activity decreased to be (69.6 ± 0.08) % of that in the control group, when 293 cells overexpressed miR-33a (P < 0.05). When endogenous miR-33a expression was down-regulated in 293 cells, the dual luciferase activity was (1.52 ± 0.06) times that of the control group (P < 0.05; ). In summary, miR-33a could be directly targeted to regulate HIF-1α.
Figure 6. miR-33a directly targets HIF-1α. (A) pYr-Mir-Target-HIF-1α-3U luciferase report plasmid profile. The left is HIF-1α 3'UTR (NM_001530) sequence inserted into pYr-Mir-Target-HIF-1-3U dual luciferase reporter plasmid. Red area is the predicted loci which hsa-miR-33a binds to. The right is pYr-Mir-Target-HIF-1-3U dual luciferase reporter plasmid sketch. (B) Real-time PCR detection of miR-33a relative expression levels in WM35 and 293 cells. When miR-33a overexpressed in 293 cells, the dual luciferase activity decreased; while when endogenous miR-33a was down-regulated in 293 cells, the dual luciferase activity increased (*P < 0.05).
As HIF-1α is known to be a core gene for affecting the biological behavior of malignant melanoma,Citation12 we hypothesized that miR-33a abnormally expressed in melanoma could exert biological functions in inhibition of tumor cell proliferation, metastasis and invasion at least through partially targeting HIF-1α.
Discussion
MiR-33a and miR-33b are co-transferred with their own host genes, SREBP-2 and SREBP-1, respectively, which play important roles in cholesterol uptake and synthesis, fatty acid metabolism, insulin signaling pathway and glycoL-metabolism.Citation13-15 Inhibiting miR-33a and miR-33b is also considered as an effective way to reduce the risk of cardiovascular disease.Citation16 We have found miR-33 expressed abnormally in clinical specimens of melanoma. So it may act as an inhibitor or promoter in melanoma. But there are no evidences about their roles in melanoma.
In this study, we first detected miR-33a/b expression in metastatic human melanoma cells was relatively low. As miR-33a has a similar expression trend of miR-33b, we chose miR-33a for the subsequent experiments. MiR-33a can regulate Pim-1, CDK6 and CCND1, thereby affect in tumor cell proliferation and cell cycle progression.Citation17,18 TTF-1 is capable of binding to the promoter region SREBF2, thus indirectly affecting HMGA2 gene expression regulated by miR-33a, and inhibiting cancer cell invasion and metastasis.Citation19 MiR-33a can also target PTHrP so as to inhibit bone metastasis.Citation20 It is reported recently that low expression of miR-33a in the plasma is highly closely related to the high risk of melanoma recurrence, suggesting that miR-33a may function as a tumor suppressor gene.Citation21
We overexpressed miR-33a in A375, which has lower miR-33a expression, also inhibited it in WM35 and WM451, which have higher miR-33a expression. In vitro experiments of the 3 cell lines demonstrated that miR-33a can inhibit the proliferation, invasion and metastasis of human melanoma cells. In nude mice experiments, miR-33a was shown to inhibit the both growth and metastasis of melanoma xenografts. These results are more consistent with previous literature, and further provide a new experimental basis for miR-33a to function as a tumor suppressor gene in the occurrence and development of melanoma.
MicroRNA in melanoma cells can affect malignant phenotypes through directly targeting transcription factors. It is reported that miR-137, miR-205 and miR-9 can target EZH2, ZEB2, and NF-κB1-Snail1 pathway, respectively, to reduce E-cadherin expression, while miR-125b can control the incidence of melanoma and invasion through direct inhibition of c-Jun expression.Citation5,22-24 Very interestingly, in this study we used bioinformatics software to successfully predict and confirm that HIF-1α is a very important transcription factor and functions as a direct target gene of miR-33a.
HIF-1α is the important factor to exacerbate the conversion of normal melanocytes into melanoma.Citation25,26 HIF-1α activity in melanoma is even increased in normal oxygen levels, which is helpful for tumor cells to invade the adjacent tissues and to get adequate blood supply in hypoxic microenvironment.Citation27,28 HIF-1α overexpression in primary melanoma and its metastases is highly positively correlated to the severity of malignancy and metastasis.Citation28,29 It is reported that HIF-1α can activate CXCR4, PDK1 to promote the invasion and metastasis of melanoma cells.Citation30,31 In addition, bcl-2 plays an important role in promotion of melanoma angiogenesis.Citation31,32
After overexpression or inhibition of miR-33a, we tested HIF-1α expression and found it had an opposite trend of miR-33a, which highly suggested it may be a target gene of miR-33a. To further confirm that HIF-1α is the direct target of miR-33a, we performed dual luciferase reporter assay which testify that miR-33a has a direct binding at HIF-1α 3'-UTR. Also, in the nude mouse model, A375 xenografts show inhibition of HIF-1α in tissue after miR-33a overexpressed. Thus, in melanoma, miR-33a can function as a tumor suppressor gene to inhibit tumor cell proliferation, invasion and metastasis in part to target HIF-1α at least. In the next study we will investigate miR-33a-targeted HIF-1α downstream signaling pathways.
In activation of HIF-1α/PDK3 signaling axis can lead to mitochondrial oxidative metabolism of melanoma, which is a new therapeutic means for tumor metastasis.Citation33 C-1311 is a better inhibitor for HIF-1α pathway inhibitor to exhibit better anti-tumor effects.Citation34 MicroRNA interference technique can effectively inhibit the progress of melanoma invasion.35 Therefore, targeting the miR-33a/HIF-1α signaling axis is likely to provide a new strategy for the clinical treatment of melanoma and inhibition of tumor invasion and metastasis.
Materials and Methods
Cell culture
Human primary melanocyte (HM) was purchased from XinYu Biological Technology (Shanghai, China). The human melanoma cell line WM35 that was derived from a primary cutaneous melanoma, has no metastasis potential. A375, WM451 and SK-MEL-1 were derived from metastases of malignant melanomas. A375 was obtained from the Xiangya Medical School Cell Center of Central South University, (The Cell Center of Xiangya Medical School of CSU, China) WM35, WM451 and SK-MEL-1 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). A375, SK-MEL-1 were maintained in RPMI-1640 medium and 10% fetal calf serum, WM35, WM451 were maintained in Dulbecco's modified Eagle's medium (DMEM) and 10% fetal calf serum at 37˚C with 5% CO2 in humidified atmosphere.
RNA isolation and reverse transcription
miR-33a primer was designed and synthesized by Changsha Yingrun biotechnology. Total cellular RNA were isolated from cultured cells using Trizol reagents according to the manufacturer's instructions (Invitrogen), cDNAs were generated by reverse transcriptase reaction performed in 20 μl reaction volume, containing total cellular RNA 1-5μg, RT Primer Mix 2 μl, 10 × RT Buffer 2 μl, RNase inhibitor 1 μl, dNTPs (2.5mM) 4 μl, reverse transcriptase 1μl and ddH2O. The reaction mixture was incubated at 70°C for 10 min, transcribed at 42°C for 1h, inactivated at 70°C for 15min and preserve at −20°C.
Analyze miR-33a expression by quantitative PCR
miRNA cDNA template 2 μl, 2×SYBRGreenMix 12.5 μl, miR-33a (200nM) forward and reverse primers 0.5μl and U6 RT Primer Mix(200nM) 1μl in a total of 25μl were applied to the following PCR program: 10min 95°C (initial denaturation); temperature transition up to 95°C for 20s, 62°C for 30s and 72°C for 30s, repeated for 40 times (amplification). The PCR reaction was evaluated by melting curve analysis.
Grouping, plasmid and cell transfection
Experiments were divided into the following groups:WM35-pYr-LVX, WM35-pYr-LVX-miR-33a-sponge, WM451-pYr-LVX, WM451-pYr-LVX-miR-33a-sponge, A375-pYr-LVX, A375-pYr-LVX-pri-miR-33a.pLVX-IRES-ZsGreen1(No. 632187) was bought from Clontech Company, pLP1, pLP2, pLP/VSVG vector and slow virus vector were bought from Invitrogen Company. The sequence of pri-mir33a and miR33a-sponge (shown in Fig. S1) was synthesized by Yingrun Company (Changsha, China) and was combined to the vectors and sequencing in Genscript Company (Nanjing, China). Then transfect melanoma cells through lentivirus method, real-time PCR and fluorescence microscope to confirm the efficiency of transfection.
MTT mapping cell growth curve
Cell proliferation was determined by MTT ((3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide) assay. The 96-well plates were inoculated with 10,000 cells per well. MTT assays were performed 24 h to 72 h as follows: 10 μl MTT solution was added to each well and cells were incubated for 4 h, then the supernate was carefully discarded to terminate the reaction. Next 100 μl DMSO was added to each well, and samples were oscillated for 10 min to fully dissolve any crystals. Absorption was measured at 490 nm.
Soft-agar colony formation assay
To analyze growth rates of cells stably transfected with pYr-LVX-pri-miR-33a and pYr-LVX-miR-33a-sponge in soft agar, 6-well plates were coated with 1.5ml base agar, DMEM or RPMI-1640 medium, and 10% fetal calf serum (FCS). Cells were trypsinized and counted, 1×103 cells/well. Cells were incubated at 37°C in a humidified incubator for 2 weeks. The clones were fixed with 5ml methanol for 15min and stained by Giemsa staining for 10–30 min, then slow water wash to dry in the air. Clones were counted under a dissecting microscope. Colony formation rate = the clone number/cell number ×100%
Cell motility assay using Transwell
To investigate the effect of over-expression or down-expression of miR-33a on cell migration, cells (5×104 cells per well) in serum-free media were plated in the top chamber of non-coated polyethylene terephthalate membranes (6-well insert; 8 mm pore size; Becton Dickinson, Franklin Lakes, NJ, USA). The bottom chamber contained 500μl chemotactic factor. The cells were incubated for 24 h at 37°C and cells that did not pass through the membrane pores were removed. Migrated cells were fixed by pre-cooling methanol for 30min, wood staining for 1 min, gradient of ethanol dehydration 1 min (80%, 95%, 100%), xylene transparent for 2–3 min and counted in 5 random fields (400×). Repeat the experiment 2 times. Cells were than analyzed for migration.
Xenograft animal models
Male BALB/C-nu/nu (4–6 weeks) were purchased from animal lab of the Third Xiangya Hospital of Central South University and maintained under specific pathogen-free condition. To test the tumourigenic capacity and growth kinetics, group of 5 nude mice were injected subcutaneously in the dorsal flank with 2 × 106 cells (A375-pYr-LVX, A375-pYr-LVX-pir-miR-33a).Tumor volume was calculated by using the formula V(mm3) = 0.5 × a × bCitation2 (a maximum length to diameter, b maximum transverse diameter). Nude mice were sacrificed on 30 days after tumor implantation. Test the tumor weight by electronic scale with high accuracy (unit: g). According our present method of evaluating metastasis of the cells, suspended cells (1 × 106/0.2 mL) were injected into the lateral tail vein of nude mice. They were sacrificed 42 days after inoculation or when they became moribund, and the lungs were excised and fixed in 4% buffered formaldehyde for 24 h and rinsed with phosphate buffered saline. The tissues were embedded in paraffin and stained with haematoxylin and eosin (H.E.) for histologic examination.Citation36
Western blot assay for verifying HIF-1α protein expression
The HIF-1a antibody was bought from CST Company. Cells were processed for protein extraction and western blotting using standard procedures. The protein concentration of each sample was determined using the BCA method. 20μg of protein supernatant was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto nitrocellulose membranes. Membranes were blocked for 1.5 h and incubated with mouse anti-HIF-1α antibody (1:1000), or mouse anti-β-actin antibody (1:10000). All membranes were incubated at 4°C overnight. Membranes were washed 3 times with PBST then incubated for 1 h at room temperature in anti-mouse secondary antibody (1:10000). ECL chemiluminescence was used to reveal proteins and Bandscan software (Glyko, USA) was used to scan the gray value of Western blot bands to calculate the relative content of protein, protein relative content = gray value of protein bands/gray value of β-actin bands.
Dual luciferase reporter assays
Luciferase reporter assays were performed using the pYr-Mir-Target-HIF1a-3U report vector. The 293 cell was transfected with pYr-LVX-miR-33a-sponge vector or pYr-LVX-pri-miR-33a vector, then co-transfected with pYr-MirTarget-HIF1a-3U report vector′-UTR. Cells were incubated with transfection reagent/DNA complexes for 3 h. Firefly and Renilla luciferase activities were then evaluated using the Dual-Luciferase Reporter Assay System (Promega), where Renilla luciferase activity was normalized to firefly luciferase activity.
Statistical analysis
All values are expressed as Mean ± Sd. Differences between the groups were compared using the unpaired 2-tailed t-test in SPSS software (SPSS, Chicago, IL, USA). In vivo analysis was performed using Mann–Whitney U-test for significance. P <0.05 was considered statistically significant.
Conclusion
The newly identified miR-33a/HIF-1α axis might provide a new strategy for the treatment of melanoma.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
supplementary_file.zip
Download Zip (1.6 MB)Funding
This work was supported by National Natural Science Foundation of China (Grant No. 81372140, 81301688, 81272192, 81171882, 81071645); Ph.D. Programs Foundation of Ministry of Education of China (No. 20130162110050 and 20130162120093); Natural Science Foundation of Hunan Province (Grant No.13JJ4028); Technology Project of Hunan Province(2012SK3229); Project of the Department of Science and Technology of Hunan Province (No. 2013FJ6003); Program for New Century Excellent Talents in University (NCET-11-0527) ; Fundamental Research Funds for the Central Universities(2011JQ028); Post-doctoral Foundation of Central South University (No. 131425) ; 125 Talent Project of the Third Xiangya Hospital of Central South University and Hunan Provincial Innovation Foundation for Postgraduate.
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144:646-74; PMID:21376230; http://dx.doi.org/10.1016/j.cell.2011.02.013
- Segura MF, Greenwald HS, Hanniford D, Osman I, Hernando E. MicroRNA and cutaneous melanoma: from discovery to prognosis and therapy. Carcinogenesis 2012; 33:1823-32; PMID:22693259; http://dx.doi.org/10.1093/carcin/bgs205
- Dong F, Lou D. MicroRNA-34b/c suppresses uveal melanoma cell proliferation and migration through multiple targets. Mol Vis 2012; 18:537-46; PMID:22419847
- Luo C, Tetteh PW, Merz PR, Dickes E, Abukiwan A, Hotz-Wagenblatt A, Holland-Cunz S, Sinnberg T, Schittek B, Schadendorf D, et al. miR-137 inhibits the invasion of melanoma cells through downregulation of multiple oncogenic target genes. J Invest Dermatol 2013; 133:768-75; PMID:23151846; http://dx.doi.org/10.1038/jid.2012.357
- Dar AA, Majid S, Rittsteuer C, de Semir D, Bezrookove V, Tong S, Nosrati M, Sagebiel R, Miller JR 3rd, Kashani-Sabet M. The role of miR-18b in MDM2-p53 pathway signaling and melanoma progression. J Natl Cancer Inst 2013; 105:433-42; PMID:23365201; http://dx.doi.org/10.1093/jnci/djt003
- Giles KM, Brown RA, Epis MR, Kalinowski FC, Leedman PJ. miRNA-7-5p inhibits melanoma cell migration and invasion. Biochem Biophys Res Commun 2013; 430:706-10; PMID:23206698; http://dx.doi.org/10.1016/j.bbrc.2012.11.086
- Bhattacharya A, Schmitz U, Wolkenhauer O, Schonherr M, Raatz Y, Kunz M. Regulation of cell cycle checkpoint kinase WEE1 by miR-195 in malignant melanoma. Oncogene 2013; 32:3175-83; PMID:22847610; http://dx.doi.org/10.1038/onc.2012.324
- Segura MF, Hanniford D, Menendez S, Reavie L, Zou X, Alvarez-Diaz S, Zakrzewski J, Blochin E, Rose A, Bogunovic D, et al. Aberrant miR-182 expression promotes melanoma metastasis by repressing FOXO3 and microphthalmia-associated transcription factor. Proc Natl Acad Sci U S A 2009; 106:1814-9; PMID:19188590; http://dx.doi.org/10.1073/pnas.0808263106
- Zehavi L, Avraham R, Barzilai A, Bar-Ilan D, Navon R, Sidi Y, Avni D, Leibowitz-Amit R. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol cancer 2012; 11:44; PMID:22747855; http://dx.doi.org/10.1186/1476-4598-11-44
- Chen J, Abi-Daoud M, Wang A, Yang X, Zhang X, Feilotter HE, Tron VA. Stathmin 1 is a potential novel oncogene in melanoma. Oncogene 2013; 32:1330-7; PMID:22665054; http://dx.doi.org/10.1038/onc.2012.141
- Xu D, Tan J, Zhou M, Jiang B, Xie H, Nie X, Xia K, Zhou J. Let-7b and microRNA-199a inhibit the proliferation of B16F10 melanoma cells. Oncol Lett 2012; 4:941-6; PMID:23162627
- Hanna SC, Krishnan B, Bailey ST, Moschos SJ, Kuan PF, Shimamura T, Osborne LD, Siegel MB, Duncan LM, O'Brien ET 3rd, et al. HIF1alpha and HIF2alpha independently activate SRC to promote melanoma metastases. J Clin Invest 2013; 123:2078-93; PMID:23563312; http://dx.doi.org/10.1172/JCI66715
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010; 328:1566-9; PMID:20466882; http://dx.doi.org/10.1126/science.1189123
- Davalos A, Goedeke L, Smibert P, Ramirez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC, et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci U S A 2011; 108:9232-7; PMID:21576456; http://dx.doi.org/10.1073/pnas.1102281108
- Ramirez CM, Goedeke L, Rotllan N, Yoon JH, Cirera-Salinas D, Mattison JA, Suárez Y, de Cabo R, Gorospe M, Fernández-Hernando C, et al. MicroRNA 33 regulates glucose metabolism. Mol Cell Biol 2013; 33:2891-902; PMID:23716591; http://dx.doi.org/10.1128/MCB.00016-13
- Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 2011; 478:404-7; PMID:22012398; http://dx.doi.org/10.1038/nature10486
- Cirera-Salinas D, Pauta M, Allen RM, Salerno AG, Ramirez CM, Chamorro-Jorganes A, Wanschel AC, Lasuncion MA, Morales-Ruiz M, Suarez Y, et al. Mir-33 regulates cell proliferation and cell cycle progression. Cell Cycle 2012; 11:922-33; PMID:22333591; http://dx.doi.org/10.4161/cc.11.5.19421
- Thomas M, Lange-Grunweller K, Weirauch U, Gutsch D, Aigner A, Grunweller A, Hartmann RK. The proto-oncogene Pim-1 is a target of miR-33a. Oncogene 2012; 31:918-28; PMID:21743487; http://dx.doi.org/10.1038/onc.2011.278
- Rice SJ, Lai SC, Wood LW, Helsley KR, Runkle EA, Winslow MM, Mu D. MicroRNA-33a mediates the regulation of high mobility group AT-hook 2 gene (HMGA2) by thyroid transcription factor 1 (TTF-1/NKX2-1). J Biol Chem 2013; 288:16348-60; PMID:23625920; http://dx.doi.org/10.1074/jbc.M113.474643
- Kuo PL, Liao SH, Hung JY, Huang MS, Hsu YL. MicroRNA-33a functions as a bone metastasis suppressor in lung cancer by targeting parathyroid hormone related protein. Biochimica et biophysica acta 2013; 1830:3756-66; PMID:23458685; http://dx.doi.org/10.1016/j.bbagen.2013.02.022
- Friedman EB, Shang S, de Miera EV, Fog JU, Teilum MW, Ma MW, Berman RS, Shapiro RL, Pavlick AC, Hernando E, et al. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med 2012; 10:155; PMID:22857597; http://dx.doi.org/10.1186/1479-5876-10-155
- Liu S, Tetzlaff MT, Liu A, Liegl-Atzwanger B, Guo J, Xu X. Loss of microRNA-205 expression is associated with melanoma progression. Lab Invest 2012; 92:1084-96; PMID:22525428; http://dx.doi.org/10.1038/labinvest.2012.62
- Liu S, Kumar SM, Lu H, Liu A, Yang R, Pushparajan A, Guo W, Xu X. MicroRNA-9 up-regulates E-cadherin through inhibition of NF-kappaB1-Snail1 pathway in melanoma. J Pathol 2012; 226:61-72; PMID:22131135; http://dx.doi.org/10.1002/path.2964
- Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff AK. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene 2013; 32:2984-91; PMID:22797068; http://dx.doi.org/10.1038/onc.2012.307
- Nys K, Maes H, Dudek AM, Agostinis P. Uncovering the role of hypoxia inducible factor-1alpha in skin carcinogenesis. Biochimica et biophysica acta 2011; 1816:1-12; PMID:21338656
- Bedogni B, Welford SM, Cassarino DS, Nickoloff BJ, Giaccia AJ, Powell MB. The hypoxic microenvironment of the skin contributes to Akt-mediated melanocyte transformation. Cancer Cell 2005; 8:443-54; PMID:16338658; http://dx.doi.org/10.1016/j.ccr.2005.11.005
- Kuphal S, Winklmeier A, Warnecke C, Bosserhoff AK. Constitutive HIF-1 activity in malignant melanoma. Eur J Cancer 2010; 46:1159-69; PMID:20185296; http://dx.doi.org/10.1016/j.ejca.2010.01.031
- Sun B, Zhang D, Zhang S, Zhang W, Guo H, Zhao X. Hypoxia influences vasculogenic mimicry channel formation and tumor invasion-related protein expression in melanoma. Cancer Lett 2007; 249:188-97; PMID:16997457; http://dx.doi.org/10.1016/j.canlet.2006.08.016
- Giatromanolaki A, Sivridis E, Kouskoukis C, Gatter KC, Harris AL, Koukourakis MI. Hypoxia-inducible factors 1alpha and 2alpha are related to vascular endothelial growth factor expression and a poorer prognosis in nodular malignant melanomas of the skin. Melanoma Res 2003; 13:493-501; PMID:14512791; http://dx.doi.org/10.1097/00008390-200310000-00008
- Victor N, Ivy A, Jiang BH, Agani FH. Involvement of HIF-1 in invasion of Mum2B uveal melanoma cells. Clin Exp Metastasis 2006; 23:87-96; PMID:16826425; http://dx.doi.org/10.1007/s10585-006-9024-z
- Cheli Y, Giuliano S, Fenouille N, Allegra M, Hofman V, Hofman P, Bahadoran P, Lacour JP, Tartare-Deckert S, Bertolotto C, et al. Hypoxia and MITF control metastatic behaviour in mouse and human melanoma cells. Oncogene 2012; 31:2461-70; PMID:21996743; http://dx.doi.org/10.1038/onc.2011.425
- Iervolino A, Trisciuoglio D, Ribatti D, Candiloro A, Biroccio A, Zupi G, Del Bufalo D. Bcl-2 overexpression in human melanoma cells increases angiogenesis through VEGF mRNA stabilization and HIF-1-mediated transcriptional activity. FASEB J 2002; 16:1453-5; PMID:12205045
- Kluza J, Corazao-Rozas P, Touil Y, Jendoubi M, Maire C, Guerreschi P, Jonneaux A, Ballot C, Balayssac S, Valable S, et al. Inactivation of the HIF-1alpha/PDK3 signaling axis drives melanoma toward mitochondrial oxidative metabolism and potentiates the therapeutic activity of pro-oxidants. Cancer Res 2012; 72:5035-47; PMID:22865452; http://dx.doi.org/10.1158/0008-5472.CAN-12-0979
- Paradziej-Lukowicz J, Skwarska A, Peszynska-Sularz G, Brillowska-Dabrowska A, Konopa J. Anticancer imidazoacridinone C-1311 inhibits hypoxia-inducible factor-1alpha (HIF-1alpha), vascular endothelial growth factor (VEGF) and angiogenesis. Cancer Biol Ther 2011; 12:586-97; PMID:21775820; http://dx.doi.org/10.4161/cbt.12.7.15980
- Huynh C, Segura MF, Gaziel-Sovran A, Menendez S, Darvishian F, Chiriboga L, Levin B, Meruelo D, Osman I, Zavadil J, et al. Efficient in vivo microRNA targeting of liver metastasis. Oncogene 2011; 30:1481-8; PMID:21102518; http://dx.doi.org/10.1038/onc.2010.523
- Chen X, Lin J, Kanekura T, Su J, Lin W, Xie H, Wu Y, Li J, Chen M, Chang J. A small interfering CD147-targeting RNA inhibited the proliferation, invasiveness, and metastatic activity of malignant melanoma. Cancer Res 2006; 66:11323-30; PMID:17145878; http://dx.doi.org/10.1158/0008-5472.CAN-06-1536
