Abstract
Triple-negative breast cancer (TNBC) represents a collection of malignant breast tumors that are often aggressive and have an increased risk of metastasis and relapse. Long non-coding RNAs are generally defined as RNA transcripts measuring 200 nucleotides or longer that do not encode for any protein. During the past decade, increasing evidence has shown that lncRNAs play important roles in oncogenesis and tumor suppression; however, the roles of lncRNAs in TNBC are poorly understood. To address this issue, we used Agilent human lncRNA microarray chips and bioinformatics tools, including Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG), to assess lncRNA expression in 3 pairs of TNBC tissues. A dysregulated lncRNA expression profile was identified by microarray and verified by qRT-PCR in 48 pairs of breast cancer subtype tissues. Metastasis is the major cause of cancer-related deaths, including those in TNBC, and the presence of dormant residual disseminated tumor cells (DTC) may be a key factor leading to metastasis. ANKRD30A, a potential target for breast cancer immunotherapy, is currently one of the most used DTC markers. Notably, we found the expression levels of the novel intergenic lncRNA LINC00993 to be associated with the expression levels of ANKRD30A. Furthermore, our qRT-PCR data indicated that the expression of LINC00993 was also associated with the expression of the estrogen receptor. In conclusion, our study identified a set of lncRNAs that were consistently aberrantly expressed in TNBC, and these dysregulated lncRNAs may be involved in the development and/or progression of TNBC.
Introduction
Breast cancer (BC) is a heterogeneous disease with significant molecular variations, both between tumor subtypes and within a single tumor.Citation1 Triple-negative breast cancer (TNBC) is a pathological subtype of BC that responds differently to chemotherapeutic drugs and targeted agents.Citation2 Immunohistochemically, BC tissues that lack detectable levels of the estrogen receptor (ER), progesterone receptor (PR) and human epidermal growth factor receptor 2 (ERBB2/HER2) are categorized as TNBC.Citation2,3 Approximately 12 to 24% of women diagnosed with BC have TNBC.Citation4 Over the past few decades, there have been constant efforts to identify therapeutic targets or early diagnostic biomarkers for this disease; however, clinical management of TNBC is usually less than satisfactory despite the use of standardized treatment regimes.Citation5 Because TNBC patients lack effective therapeutic targets, such as tamoxifen in ER-positive BC and Herceptin in HER2-positive BC, they often have significantly lower rates of disease-free survival (DFS), overall survival (OS) and 5-year survival.Citation6 A study of 906 patients with early-stage invasive BC demonstrated that rates of local recurrence were associated with positive lymph node numbers and TNBC subtype.Citation7
Protein-coding sequence accounts for only a minority (less than 2%) of the human genome; the majority is non-coding sequence such as non-coding RNA (ncRNA).Citation8 ncRNAs can be divided into 2 categories: house-keeping ncRNAs (tRNA, rRNA, etc.) and regulatory ncRNAs (miRNA, lncRNA, piRNA, etc.).Citation9 Long non-coding RNAs (lncRNAs) are regulatory ncRNAs that measure 200 nucleotides (nt) or longer and do not encode any protein.Citation10 Recent reports have shown that lncRNAs are involved in almost all human biological processes, both physical and pathological, and that they act as gene regulators at both the transcriptional and post-transcriptional level by binding to DNA, RNA or proteins.Citation11-13
Recent studies have indicated that altered expression levels of lncRNAs are associated with human diseases, including BC.Citation14-17 For example, it was reported that the lncRNAs H19,Citation18 HOTAIRCitation19-21 and UCA1Citation22 act as oncogenic genes in BC, while GAS5Citation16,23 plays a tumor suppressor role. Additionally, several novel lncRNAs have been associated with drug resistance to standard BC treatment. The discovery of the ARA (Adriamycin Resistance Associated) lncRNA provided novel insights into Adriamycin resistance at the lncRNA level.Citation24 Breast Cancer Antiestrogen Resistance 4 (BCAR4) was found to be related to tamoxifen resistanceCitation25 and could also sensitize BC cells to lapatinib.Citation26 It has also been reported that Colon Cancer Associated Transcript 2 (CCAT2) can downregulate chemosensitivity to 5-FU and up-regulate cell migration in BC cell lines.Citation27
However, whether each BC subtype will have a distinct lncRNA genomic profile remains to be determined. We hypothesize that dysregulated lncRNA expression patterns exist in numerous breast carcinoma subtypes, and that further investigations may help to identify one or more TNBC-associated lncRNAs. In this study, we used Agilent lncRNA microarray chips to explore the association between TNBC and lncRNAs. By comparing the expression profiles of TNBC and non-tumor tissues, we discovered a set of novel lncRNAs that are always aberrantly expressed in TNBC and then validated these findings using qRT-PCR. The results of our study suggest that dysregulated lncRNA may play a role in the developmental cascade of BC, and some key lncRNAs may be critical to this process.
Results
Dysregulated expression profiles of lncRNAs and mRNAs in TNBC
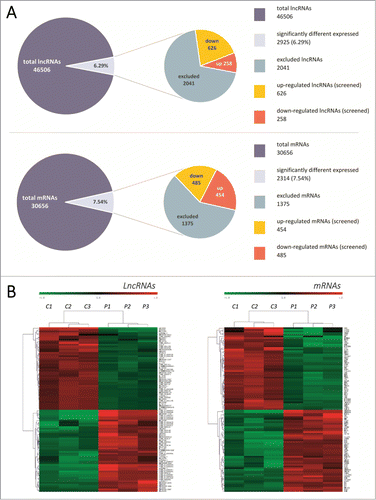
Out of a collection of 46,506 lncRNAs and 30,656 mRNAs probes, the expression of 2925 lncRNAs and 2314 mRNAs was found to be significantly altered () in 3 different pairs of TNBC tissue (fold change >2; p <0.05). Out of the group of RNAs that were up-regulated, lncRNA AK130538 and mRNA HORMAD1 demonstrated the greatest degree of demonstrated up-regulation, with increases of 212.076- and 91.757-fold, respectively; in the down-regulation group, lncRNA TCONS_l2_00002973 and mRNA ANKRD30A demonstrated the greatest degree of down-regulation, with decreases of 298.204- and 275.902-fold, respectively.
Figure 1. (A) Summary of microarray results. Expression levels of 46,506 lncRNAs and 30,656 mRNAs were assessed in 3 pairs of TNBC tissues using Agilent Human lncRNA 4*180K microarrays. Compared with non-tumor tissues, 2925 lncRNAs (6.29%) and 2314 mRNAs (7.54%) had significant changes in expression levels (fold change >2, p < 0.05). A total of 2041 lncRNAs and 1375 mRNAs were excluded due to low expression levels. A total of 884 lncRNAs were then identified from the screen, with 626 up-regulated and 258 down-regulated. (B) Hierarchical clustering map. Hierarchical clustering analysis of the top 100 dysregulated lncRNAs and mRNAs. This clustering map revealed a set of lncRNAs that were often aberrantly expressed in TNBC compared with non-tumor tissues. Each row represents a single lncRNA or mRNA and each column represents one tissue sample. Expression levels of these transcripts are represented in red (elevated expression) or green (reduced expression), indicating expression above and below the median expression levels, respectively. C: carcinoma group, P: paired non-tumor group.
The following data are noteworthy. First, 28 out of the top 30 dysregulated lncRNAs () were found to be down-regulated, with an average fold change of 53.82; this indicated that the down-regulated lncRNAs might have a more significant role in TNBC than the up-regulated group. Secondly, 12 out of the top 15 down-regulated lncRNAs (see supplementary file) are located on chromosome 10, while 4 out of the 5 top downregulated mRNAs (see supplementary file) are also located on chromosome 10, suggesting that transcripts on chromosome 10 have a significant role in the development and/or progression of TNBC. Therefore, we analyzed the chromosomal locations of each of the significantly altered lncRNAs and mRNAs (). We found that 10.56% of the dysregulated lncRNAs and 10.98% of the dysregulated mRNAs mapped to chromosome 1, where as for chromosome 10, these values were 4.17% and 4.49%, respectively. Similar values were found when evaluating dysregulated lncRNAs and mRNAs on each of the chromosomes, which suggested that the expression of protein-coding transcripts could be regulated by lncRNAs, as has been reported previously.Citation28
Figure 2. (A) Chromosomal locations of variably expressed lncRNAs and mRNAs. The X-axis represents the ordinal of the chromosome, and the “NA” stands for non-annotation. The Y-axis represents the number of lncRNAs that expressed differently in TNBC tissues (fold change>2; p < 0.05). Breast cancer associated genes (ESR1, MKI67, PGR, ERBB2, TP53 and BRCA1) are presented according to their respective chromosomal locations. (B) Volcano plots. The negative log of the p value (base 10) was plotted on the Y-axis, and the log of the fold change (base 2) was plotted on the X-axis. The red points on this graph represent lncRNAs that were significantly differently expressed in TNBC tissues (fold change >2 and p <0.05); the gray points represent the remaining lncRNAs (fold change <2 or p >0.05). (C) Scatter plots demonstrating the heterogeneity between samples. A consistent set of lncRNAs was found to be frequently dysregulated in TNBC despite the heterogeneity of the samples. The X-axis represents the logarithmic (base 2) fluorescence signal values of the microarray probes in a specific sample, while the Y-axis represents the equivalent values in a second sample. The size of the red area corresponds to the number of differently expressed lncRNAs; the larger the red area, the more significant the heterogeneity between samples. C: carcinoma group, P: paired non-tumor group.
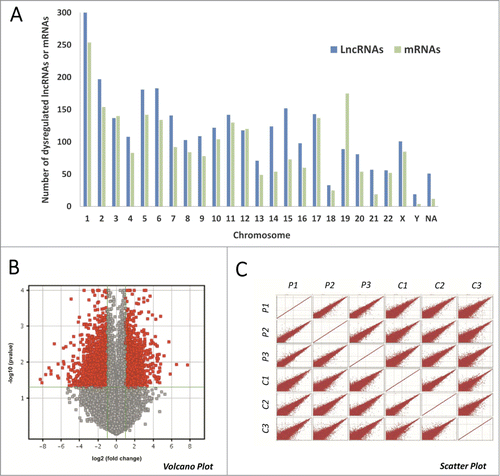
Table 1. The top 30 dysregulated lncRNAs identified by microarrays in 3 pairs of TNBC tissues
Expression of mRNAs of classical BC markers and their correlated lncRNAs in microaray
Five of the classic BC molecular markers that can be used to categorize BC subtypes are ER, PR, p53, KI-67 and ERBB/HER2. Immunohistochemical staining was used to assess the expression of the 5 markers in our collection of tumor samples. We then analyzed their mRNA expression levels in microarray to validate the accuracy of the microarray (see supplementary file). We found that the expression of ER, PR and KI67 were all significantly altered in TNBC (p < 0.05). These results were consistent with the immunochemical results.
To identify TNBC-associated lncRNAs, as well as lncRNAs that are associated with the above-described BC molecular markers, we performed Pearson's correlation coefficient analysis to assess the correlation between the expression levels of dysregulated lncRNAs and mRNAs. The top 400 significantly differently expressed lncRNAs (200 up-regulated and 200 down-regulated), as well as all of the mRNA probes that are present on the Agilent microarray, were chosen as our candidates. A Pearson correlation coefficient value of >0.99 and a p value of <0.05 were chosen as the cutoffs. A total of 22 lncRNAs were found to be co-expressed with the ER mRNAs (), including the most downregulated lncRNA, TCONS_l2_00002973, while none of these 400 lncRNAs was identified to be associated with the other 4 BC subtype markers.
Table 2. Dysregulated lncRNAs associated with ER expressionCorrelation coefficient > 0.99 and p < 0.05 was chosen as the cut-off
Verification of microarray profiling data using qRT-PCR
Considering that lncRNA expression levels are often lower than those of protein-coding genes, we screened lncRNAs for further study using the following filter criteria: 1) we normalized the fluorescence signal values of lncRNA probes using a quantile function and then transformed these values into logarithmic form (base 2), 2) we selected lncRNAs with normalized fluorescence signal values ≥8 in all 3 of the TNBC tissue samples or in all non-tumor tissues, and 3) we selected only lncRNAs with a signal-to-noise ratio of Cy3 that was over 2.6, that is, the parameter of gIsWellAboveBG equaled “P” in the microarray data. A total of 884 lncRNAs were confirmed, including 626 that were upregulated and 258 that were down-regulated. We then chose 8 candidate lncRNAs and used qRT-PCR to analyze their expression patterns in 48 pairs of BC subtype tissues and corresponding non-tumor tissues (). Based on the ER and HER2 expression levels, the 48 pairs of samples were then divided into 4 groups of 12 pairs each.
Figure 3. qRT-PCR verification of 8 candidate lncRNAs in 48 pairs of BC tissues of different subtypes. Expression of the 8 candidate lncRNAs in 24 pairs of (A) ER-negative and (B) ER-positive samples and (C) 48 pairs of BC and corresponding non-tumor tissues. (D) Expression levels of the 8 candidate lncRNAs in TNBC tissues, as measured by microarray and qRT-PCR. The Y-axis represents the relative expression levels of lncRNAs assessed by the 2−ΔCt method and the error bar represents standard deviation. Paired t-tests (2-tailed) were performed to compare the expression levels between carcinoma and non-tumor tissues, and a p value < 0.05 indicated statistical significance. (*p < 0.05, **p < 0.01 and ***p < 0.001).
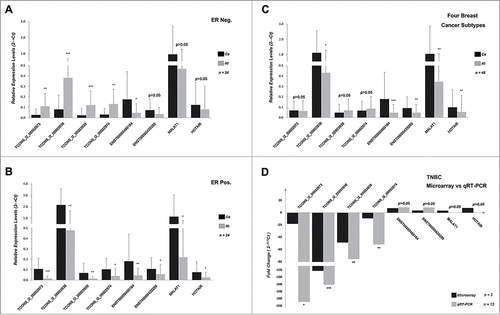
The results confirmed that TCONS_l2_00002973, TCONS_l2_00003938, TCONS_l2_00003939 and TCONS_l2_00002974 were downregulated in TNBC tissue; their expression was associated with the level of ER expression (p < 0.05), whereas the expression levels of the other 4 (ENST00000460164, ENST00000425295, MALAT1 and HOTAIR) were not. None of the 8 lncRNAs had expression levels that could be associated with HER2 expression. We also performed another analysis by dividing the above samples into tumor and non-tumor groups. We found that the expression levels of TCONS_l2_00003938, ENST00000460164, ENST00000425295, MALAT1 and HOTAIR were significantly higher in tumor tissues than non-tumor tissues (p < 0.05), whereas there were no significant differences in the expression levels of the other 3 lncRNAs (p > 0.05). These data suggest that there is a set of lncRNAs that are often dysregulated in BC tissues and that their expression levels can be associated with ER status.
Prediction of dysregulated lncRNAs functional roles using bioinformatics tools
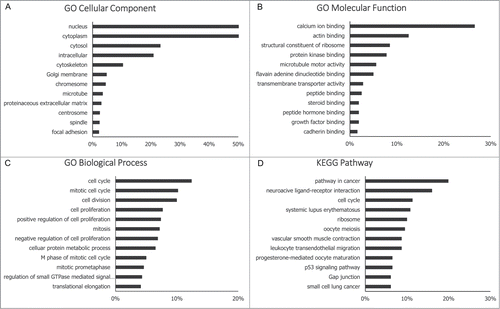
The functional roles of most lncRNAs have not yet been defined, thus their functions can only be indirectly predicted by analyzing the functions of their co-expressed mRNAs. We therefore performed Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis () to explore the functional roles of dysregulated mRNAs. In the GO biological process analysis, the majority of the genes that were assessed were proven to be related to cancer-associated biological behaviors, such as cell cycle control, cell division and cell proliferation. In the KEGG pathway analysis, the majority of dysregulated mRNAs were found to be involved in the pathway in cancer. The details of the GO and KEGG pathway analyses are included in the supplementary file. Collectively, our data suggest that dysregulated mRNAs are involved in the development/progression of TNBC, and that dysregulated novel lncRNAs may have similar functional roles to these mRNAs.
Figure 4. GO and KEGG pathway analysis. Functions of significantly differently expressed mRNAs (fold change>2; p < 0.05) were analyzed by GO and KEGG pathway annotations, and functions of novel lncRNAs were deduced by their co-expressed mRNAs. The GO database includes 3 parts (cellular component, biological process and molecular function) that together describe the genes and gene products across all species (http://www.geneontology.org). The KEGG pathway is a collection of manually drawn signal pathway maps and provides a valuable tool for mapping a specific gene to its corresponding pathway (http://www.genome.jp/kegg/).
LINC00993 and ANKRD30A
Based on above findings, we chose to further investigate the TCONS_l2_00003938 lncRNA and the ANKRD30A mRNA. TCONS_l2_00003938, also known as LINC00993, is a novel intergenic lncRNA located on chromosome 10 (chr10+: 37598113–37635954) that is 652 nucleotides in length. ANKRD30A, an abbreviation of Ankyrin repeat domain 30A, also mapped to chromosome 10 (10p11.21), and is the nearest protein coding gene to LINC00993.
We found a strong association between the expression levels of LINC00993 and ANKRD30A. Our microarray data showed that both LINC00993 and ANKRD30A were significantly down-regulated in TNBC tissues, with 202.33- and 275.90-fold decreases in their expression, respectively. Furthermore, Pearson correlation analysis also indicated a strong association between the expression of LINC00993 and ANKRD30A (correlation coefficient >0.99, p < 0.001).
The ChIPBase database (http://deepbase.sysu.edu.cn/chipbase/index.php) contains high-throughput sequencing data that were generated over 543 ChIP-Seq experiments conducted on a diverse array of tissue samples and cell lines taken from 6 species, including the expression profiles of lncRNAs and mRNAs from 22 different healthy human tissues. After searching this database, we found that both LINC00993 and ANKRD30A are specifically expressed in adipose, breast and testes (see supplementary file). This finding is consistent with previous reports indicating that ANKRD30A is a breast differentiation antigen that is specifically expressed in breast epithelium.
We also found that the expression levels of both ANKRD30A and LINC00993 were associated with ER expression. It has previously been reported that ANKRD30A expression correlated directly with estrogen receptor expression.Citation29 Using qRT-PCR, we found that LINC00993 has significantly different expression patterns in ER-positive and ER-negative breast tumors. Our results indicate that LINC00993 is up-regulated in ER-positive tumors and downregulated in ER-negative tumors compared to non-tumor tissues.
Furthermore, we observed that the expression of LINC00993 might be directly regulated by ANKRD30A. Recent studies have revealed that the dysregulation of the lncRNAs that are known to be associated with human diseases is often due to the aberrant expression of transcription factors.Citation30-32 ANKRD30A is believed to be a putative transcription factor because of its bipartite nuclear localization signal motif and bZIP site (DNA-binding site followed by a leucine zipper motif). ANKRD30A is also the nearest coding gene to LINC00993, being located approximately 70K bp upstream from it.
Collectively, our data indicate a possible association between ER, ANKRD30A and LINC00993. Furthermore, we believe that the aberrant expression of LINC00993 may be related to the oncogenesis and/or development of BC, especially TNBC.
Discussion
Non-coding RNAs constitute the overwhelming majority of the human genome; less than 2 percent of genomic transcripts encode proteins.Citation8,33 This suggests that RNA-based regulatory mechanisms play an important role in physiological and pathological processes. Based on their lengths, ncRNAs can be classified into 2 main groupsCitation34,35: 1) small ncRNAs (miRNA, siRNA and piRNA, etc.), which are shorter than 200 nucleotides and 2) lncRNAs, which are longer than 200 nucleotides and contain no significant open reading frames.
Due to their low levels of expression and poor evolutionary conservation relative to the protein coding regions of the genome, lncRNAs have not been well studied historically.Citation36,37 It has been only recently that their vital roles in nearly every process of life have been demonstrated.Citation11,13,38 Based upon the location of lncRNA transcripts relative to protein coding genes, lncRNAs can be divided into 5 broad categoriesCitation37: 1) sense lncRNAs, which overlap one or more exons of another transcript on the same strand, 2) antisense lncRNAs, which overlap one or more exons of another transcript on the opposite strand, 3) bidirectional lncRNAs, which are located on the opposite strand from a neighboring exon whose transcription is initiated less than 1000 base pairs away, 4) intronic lncRNAs, which are derived wholly from within an intron of another transcript, and 5) intergenic lncRNAs, which lie within the genomic interval between 2 genes.
Increasing evidence highlights that lncRNAs can serve as diagnostic biomarkers and therapeutic targets in solid tumors, including BC; Citation16,22,27 however, their relative expression levels in various subtypes of human BC, particularly the TNBC subtype, remain unknown. In this study, we evaluated the expression levels of lncRNAs and mRNAs in 3 different pairs of TNBC tissue using microarray chips containing 46,506 lncRNA probes and 30,656 mRNA probes. More than 2000 dysregulated lncRNAs were identified, many of which are novel lncRNAs whose functions have not yet been defined. To evaluate whether an association exists between dysregulated lncRNA expression and the clinicopathological characteristics of BC, we used qRT-PCR to quantify the expression levels of 8 candidate lncRNAs in 48 pairs of BC subtype tissues. Bioinformatics analysis was then used to predict the functional roles and target genes of these dysregulated lncRNAs; a variety of databases were searched, including GO, KEGG and ChIPBase.
ER and HER2 are the main prognostic predictors and therapeutic targets for BC. TNBC is a hormone-receptor-negative subtype of BC that can be characterized by the absence of HER2 over-expression. However, it remains unknown whether lncRNAs that are known to be dysregulated in BC are involved in regulating the expression of ER and HER2. Therefore, we analyzed the association between the relative expression levels of dysregulated lncRNAs and ER/HER2 using data generated by qRT-PCR. Of the 8 candidate lncRNAs included in this study, we found that TCONS_l2_00002973, LINC00993, TCONS_l2_00003939 and TCONS_l2_00002974 were associated with the expression of ER; none were associated with HER2 expression.
The exact functions of most lncRNAs are unknown; therefore, their potential functions can only be deduced indirectly by analyzing the functions of co-expressed mRNAs or adjacent genes. We found strong indications that ANKRD30A may be the target of LINC00993. ANKRD30A is a protein coding gene located on chromosome 10 (10p11.21), which has been previously identified as BC antigen NY-BR-1Citation39 or B726P.Citation40 Aberrant expression of ANKRD30A was detected in 14 out of 27 primary and 22 out of 50 lymph-node-metastatic breast tumor tissues by Zehentneret et al. in 2002,Citation41 a finding which was further confirmed by O'Brienet in 44 out of 108 BC tissues and 0 out of 20 non-breast tissues.Citation42 Moreover, expression levels of ANKRD30A have been significantly associated with histologic grade, ER status, and HER2 gene amplification in BC; ANKRD30A was more often detected in well-differentiated, ER-positive and HER2-negative tumors.Citation43 Metastasis is the driving force behind the majority of cancer-related deaths, and because it remains incurable, it is the biggest challenge currently faced by oncologists.Citation44,45 One possible cause of metastasis may be the presence of dormant residual disseminated tumor cells (DTCs) that can evade postoperative adjuvant chemo- or target therapy.Citation45–47 ANKRD30A is currently one of the most used DTC markersCitation48, and one of the potential targets for BC immunotherapy.Citation29,49
LINC00993 is located approximately 70 Kbp downstream from ANKRD30A and is therefore the nearest non-coding RNA to this gene. LINC00993 is a novel intergenic lncRNA and is also known as lncRNA1188–3, TCONS_l2_00003938, lnc-ANKRD30A-2, RP11–20F24.4, OTTHUMG00000017971.1 or ENSG00000235687. In this study, we found for the first time that LINC00993 is both considerably dysregulated in TNBC and associated with ER expression. Furthermore, we also found a strong association between LINC00993 and the BC related gene ANKRD30A. These elements are adjacent to each other on chromosome 10, and both were found to be breast-specific in addition to being down-regulated in ER-negative BC. The structural characteristics of ANKRD30A suggest that it may interact with LINC00993 as a transcription factor; however, additional experiments are needed to elucidate the functions of LINC00993 in vitro and vivo, as well as the nature of its interaction with ANKRD30A.
Taken together, our findings demonstrated that a set of lncRNAs is always dysregulated in TNBC. The results of GO, KEGG pathway and Pearson's correlation analyses indicated that these dysregulated lncRNAs are involved in the oncogenesis and/or development of BC. We then focused on the novel lncRNA LINC00993, which was significantly down-regulated in ER-negative BC, and found that it may interact with the BC-associated gene ANKRD30A. We therefore hypothesize that the LINC00993 may play a key role in BC, especially TNBC. Finally, by identifying a reservoir of candidate lncRNAs that may be important to TNBC pathogenesis, our study provides a foundation for future research into the involvement of lncRNAs in BC.
Materials and Methods
Tissue specimens
A total of 48 pairs of primary breast infiltrating ductal carcinoma tissues and their corresponding non-tumor counterparts were surgically obtained at the First Affiliated Hospital of Chongqing Medical University between November 2013 and August 2014. All cases were diagnosed both clinically and pathologically, and none of the patients had been preoperatively treated with chemotherapy, radiation therapy or endocrine therapy. After surgery, fresh tissues were snap-frozen in liquid nitrogen and then stored at −80°C prior to total RNA extraction. Written informed consent was obtained from all study participants. This study was approved by the Ethics Committee on Human Study of Chongqing Medical University.
Microarray
Three pairs of TNBC tissues and their corresponding non-tumor tissues were used for lncRNA microarray analysis. Total RNA was quantified using the NanoDrop ND-2000 (Thermo Scientific), and RNA integrity was assessed using the Agilent Bioanalyzer 2100 (Agilent Technologies). Sample labeling, microarray hybridization and washing were all performed according to the manufacturer's standard protocols. Briefly, Cyanine-3-CTP-labeled cRNA was hybridized onto the lncRNA microarray chips (Agilent Human lncRNA, 4*180K, Design ID: 042818), which contained 46,506 human lncRNA probes and 30,656 human mRNA probes. The lncRNA microarray analysis was performed by OE Biotech, Shanghai, China.
qRT-PCR
Total RNA was isolated using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and assessed with both the NanoDrop spectrophotometer ND-8000 (NanoDrop Technologies) and by standard denaturing agarose gel electrophoresis. cDNA was synthesized using PrimeScript™ RT Master Mix (Takara Bio, Otsu, Japan) according to the manufacturer's instructions, in a total volume of 10 µL. The 10 µL RT reaction mix was then diluted (˜100ng/ul) with DEPC-Treated Water (Ambion®) and stored at −20°C until further use. Quantitative real-time PCR was performed using SYBR® Premix Ex Taq™ II (Takara Bio, Otsu, Japan) in a 10 µL PCR reaction mixture. GAPDH was used as an internal control. All lncRNA transcript levels were quantified on an ABI 7500 Real-Time PCR system (Applied Biosystems®), and the 2−ΔΔCt method was used to determine the fold change (FC).
Correlation analysis
Pearson's correlation coefficient was calculated to measure the linear correlation of the expression levels of lncRNAs and mRNAs. The top 100 upregulated and top 100 down-regulated lncRNAs were candidates for this calculation. mRNAs with a Pearson's correlation coefficient ≥0.99 and a p value <0.01 were considered significantly correlated.
Data analysis
Feature Extraction software (version10.7.1.1, Agilent Technologies) was used to analyze the microarray images to obtain raw data; Genespring was employed to finish the basic analysis of the raw data. Based on the normalized fluorescence signal values of the lncRNA/mRNA probes, differentially expressed mRNAs and lncRNAs were identified by examining fold changes to their expression levels, as well as by p values calculated via Student's t-test. The threshold that was set for both up- and downregulated genes was a fold change ≥ 2.0 and a p value <0.05. The relative expression of a lncRNA or mRNA detected by qRT-PCR was calculated using the 2 −ΔCt method, and paired t-tests (2-tailed) were performed when appropriate.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental_Data.zip
Download Zip (8.3 MB)Acknowledgment
The technical assistance provided by OE Biotech is greatly appreciated.
Additional information
Funding
References
- Polyak K. Heterogeneity in breast cancer. J Clin Invest 2011; 121:3786-8; PMID:21965334; http://dx.doi.org/10.1172/JCI60534
- Bosch A, Eroles P, Zaragoza R, Vina JR, Lluch A. Triple-negative breast cancer: molecular features, pathogenesis, treatment and current lines of research. Cancer Treat Rev 2010; 36:206-15; PMID:20060649; http://dx.doi.org/10.1016/j.ctrv.2009.12.002
- Jiao Q, Wu A, Shao G, Peng H, Wang M, Ji S, Liu P, Zhang J. The latest progress in research on triple negative breast cancer (TNBC): risk factors, possible therapeutic targets and prognostic markers. J Thorac Dis 2014; 6:1329-35; PMID:25276378; http://dx.doi.org/10.3978/j.issn.2072-1439.2014.08.13
- Rakha EA, Ellis IO. Triple-negative/basal-like breast cancer: review. Pathology 2009; 41:40-7; PMID:19089739; http://dx.doi.org/10.1080/00313020802563510
- Lara-Medina F, Perez-Sanchez V, Saavedra-Perez D, Blake-Cerda M, Arce C, Motola-Kuba D, Villarreal-Garza C, Gonzalez-Angulo AM, Bargallo E, Aguilar JL, et al.. Triple-negative breast cancer in Hispanic patients: high prevalence, poor prognosis, and association with menopausal status, body mass index, and parity. Cancer 2011; 117:3658-69; PMID:21387260; http://dx.doi.org/10.1002/cncr.25961
- Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res 2007; 13:4429-34; PMID:17671126; http://dx.doi.org/10.1158/1078-0432.CCR-06-3045
- Russo AL, Arvold ND, Niemierko A, Wong N, Wong JS, Bellon JR, Punglia RS, Golshan M, Troyan SL, Brock JE, et al.. Margin status and the risk of local recurrence in patients with early-stage breast cancer treated with breast-conserving therapy. Breast Cancer Res Treat 2013; 140:353-61; PMID:23836011; http://dx.doi.org/10.1007/s10549-013-2627-6
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F, et al.. Landscape of transcription in human cells. Nature 2012; 489:101-8; PMID:22955620; http://dx.doi.org/10.1038/nature11233
- Sana J, Faltejskova P, Svoboda M, Slaby O. Novel classes of non-coding RNAs and cancer. J Transl Med 2012; 10:103; PMID:22613733; http://dx.doi.org/10.1186/1479-5876-10-103
- Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23:1494-504; PMID:19571179; http://dx.doi.org/10.1101/gad.1800909
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15:7-21; PMID:24296535; http://dx.doi.org/10.1038/nrg3606
- Hauptman N, Glavac D. Long Non-Coding RNA in Cancer. Int J Mol Sci 2013; 14:4655-69; PMID:23443164; http://dx.doi.org/10.3390/ijms14034655
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861-74; PMID:22094949; http://dx.doi.org/10.1038/nrg3074
- Askarian-Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR, et al.. SNORD-host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 2011; 17:878-91; PMID:21460236; http://dx.doi.org/10.1261/rna.2528811
- Hu P, Yang J, Hou Y, Zhang H, Zeng Z, Zhao L, Yu T, Tang X, Tu G, Cui X, et al.. LncRNA expression signatures of twist-induced epithelial-to-mesenchymal transition in MCF10A cells. Cell Signal 2014; 26:83-93; PMID:24113349; http://dx.doi.org/10.1016/j.cellsig.2013.10.001
- Zhang Z, Zhu Z, Watabe K, Zhang X, Bai C, Xu M, Wu F, Mo YY. Negative regulation of lncRNA GAS5 by miR-21. Cell Death Differ 2013; 20:1558-68; PMID:23933812; http://dx.doi.org/10.1038/cdd.2013.110
- Augoff K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 and its host gene lncRNA LOC554202 are regulated by promoter hypermethylation in triple-negative breast cancer. Mol Cancer 2012; 11:5; PMID:22289355; http://dx.doi.org/10.1186/1476-4598-11-5
- Berteaux N, Lottin S, Monte D, Pinte S, Quatannens B, Coll J, Hondermarck H, Curgy JJ, Dugimont T, Adriaenssens E. H19 mRNA-like noncoding RNA promotes breast cancer cell proliferation through positive control by E2F1. J Biol Chem 2005; 280:29625-36; PMID:15985428; http://dx.doi.org/10.1074/jbc.M504033200
- Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense transcript long noncoding RNA (lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol Biol 2013; 425:3707-22; PMID:23375982; http://dx.doi.org/10.1016/j.jmb.2013.01.022
- Sorensen KP, Thomassen M, Tan Q, Bak M, Cold S, Burton M, Larsen MJ, Kruse TA. Long non-coding RNA HOTAIR is an independent prognostic marker of metastasis in estrogen receptor-positive primary breast cancer. Breast Cancer Res Treat 2013; 142:529-36; PMID:24258260; http://dx.doi.org/10.1007/s10549-013-2776-7
- Gupta R, Shah N, Wang K, Kim J, Horlings H, Wong D, Tsai M, Hung T, Argani P, Rinn J, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010; 464:1071-6; PMID:20393566; http://dx.doi.org/10.1038/nature08975
- Huang J, Zhou N, Watabe K, Lu Z, Wu F, Xu M, Mo YY. Long non-coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014; 5:e1008; PMID:24457952; http://dx.doi.org/10.1038/cddis.2013.541
- Mourtada-Maarabouni M, Pickard M, Hedge V, Farzaneh F, Williams G. GAS5, a non-protein-coding RNA, controls apoptosis and is downregulated in breast cancer. Oncogene 2009; 28:195-208; PMID:18836484; http://dx.doi.org/10.1038/onc.2008.373
- Jiang M, Huang O, Xie Z, Wu S, Zhang X, Shen A, Liu H, Chen X, Wu J, Lou Y, et al.. A novel long non-coding RNA-ARA: adriamycin resistance-associated. Biochem Pharmacol 2014; 87:254-83; PMID:24184505; http://dx.doi.org/10.1016/j.bcp.2013.10.020
- Godinho M, Meijer D, Setyono-Han B, Dorssers LC, van Agthoven T. Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. J Cell Physiol 2011; 226:1741-9; PMID:21506106; http://dx.doi.org/10.1002/jcp.22503
- Godinho MF, Wulfkuhle JD, Look MP, Sieuwerts AM, Sleijfer S, Foekens JA, Petricoin EF 3rd, Dorssers LC, van Agthoven T. BCAR4 induces antioestrogen resistance but sensitises breast cancer to lapatinib. Br J Cancer 2012; 107:947-55; PMID:22892392; http://dx.doi.org/10.1038/bjc.2012.351
- Redis RS, Sieuwerts AM, Look MP, Tudoran O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al.. CCAT2, a novel long non-coding RNA in breast cancer: expression study and clinical correlations. Oncotarget 2013; 4:1748-62; PMID:24077681
- Torring PM, Larsen MJ, Kjeldsen AD, Ousager LB, Tan Q, Brusgaard K. Long non-coding RNA expression profiles in hereditary haemorrhagic telangiectasia. PLoS One 2014; 9:e90272; PMID:24603890; http://dx.doi.org/10.1371/journal.pone.0090272
- Theurillat JP, Zurrer-Hardi U, Varga Z, Storz M, Probst-Hensch NM, Seifert B, Fehr MK, Fink D, Ferrone S, Pestalozzi B, et al.. NY-BR-1 protein expression in breast carcinoma: a mammary gland differentiation antigen as target for cancer immunotherapy. Cancer Immunol Immunother 2007; 56:1723-31; PMID:17410359; http://dx.doi.org/10.1007/s00262-007-0316-1
- Huarte M, Rinn JL. Large non-coding RNAs: missing links in cancer. Hum Mol Genet 2010; 19:R152-61; PMID:20729297; http://dx.doi.org/10.1093/hmg/ddq353
- Spizzo R, Almeida MI, Colombatti A, Calin GA. Long non-coding RNAs and cancer: a new frontier of translational research. Oncogene 2012; 31:4577-87; PMID:22266873; http://dx.doi.org/10.1038/onc.2011.621
- Xu H, He JH, Xiao ZD, Zhang QQ, Chen YQ, Zhou H, Qu LH. Liver-enriched transcription factors regulate microRNA-122 that targets CUTL1 during liver development. Hepatology 2010; 52:1431-42; PMID:20842632; http://dx.doi.org/10.1002/hep.23818
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al.. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447:799-816; PMID:17571346; http://dx.doi.org/10.1038/nature05874
- Mercer T, Dinger M, Mattick J. Long non-coding RNAs: insights into functions. Nat Rev Genet 2009; 10:155-9; PMID:19188922; http://dx.doi.org/10.1038/nrg2521
- Mattick J, Makunin I. Non-coding RNA. Hum Mol Genet 2006; 15 Spec No 1:29; PMID:16651366; http://dx.doi.org/10.1093/hmg/ddl046
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al.. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 2012; 22:577-91; PMID:22110045; http://dx.doi.org/10.1101/gr.133009.111
- Ponting C, Oliver P, Reik W. Evolution and functions of long noncoding RNAs. Cell 2009; 136:629-41; PMID:19239885; http://dx.doi.org/10.1016/j.cell.2009.02.006
- Harries LW. Long non-coding RNAs and human disease. Biochem Soc Trans 2012; 40:902-6; PMID:22817756; http://dx.doi.org/10.1042/BST20120020
- Nissan A, Jager D, Roystacher M, Prus D, Peretz T, Eisenberg I, Freund HR, Scanlan M, Ritter G, Old LJ, et al.. Multimarker RT-PCR assay for the detection of minimal residual disease in sentinel lymph nodes of breast cancer patients. Br J Cancer 2006; 94:681-5; PMID:16495929; http://dx.doi.org/10.1038/sj.bjc.6602992
- Jiang Y, Harlocker SL, Molesh DA, Dillon DC, Stolk JA, Houghton RL, Repasky EA, Badaro R, Reed SG, Xu J. Discovery of differentially expressed genes in human breast cancer using subtracted cDNA libraries and cDNA microarrays. Oncogene 2002; 21:2270-82; PMID:11948410; http://dx.doi.org/10.1038/sj.onc.1205278
- Zehentner BK, Dillon DC, Jiang Y, Xu J, Bennington A, Molesh DA, Zhang X, Reed SG, Persing D, Houghton RL. Application of a multigene reverse transcription-PCR assay for detection of mammaglobin and complementary transcribed genes in breast cancer lymph nodes. Clin Chem 2002; 48:1225-31; PMID:12142378
- O'Brien N, O'Donovan N, Foley D, Hill AD, McDermott E, O'Higgins N, Duffy MJ. Use of a panel of novel genes for differentiating breast cancer from non-breast tissues. Tumour Biol 2007; 28:312-7; PMID:18253069; http://dx.doi.org/10.1159/000115527
- Varga Z, Theurillat JP, Filonenko V, Sasse B, Odermatt B, Jungbluth AA, Chen YT, Old LJ, Knuth A, Jager D, et al.. Preferential nuclear and cytoplasmic NY-BR-1 protein expression in primary breast cancer and lymph node metastases. Clin Cancer Res 2006; 12:2745-51; PMID:16675566; http://dx.doi.org/10.1158/1078-0432.CCR-05-2192
- Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target. Nat Rev Cancer 2010; 10:871-7; PMID:21048784; http://dx.doi.org/10.1038/nrc2933
- Klein CA. Framework models of tumor dormancy from patient-derived observations. Curr Opin Genet Dev 2011; 21:42-9; PMID:21145726; http://dx.doi.org/10.1016/j.gde.2010.10.011
- HADFIELD G. The dormant cancer cell. Br Med J 1954; 2:607-10; PMID:13190204
- Demicheli R, Miceli R, Moliterni A, Zambetti M, Hrushesky WJ, Retsky MW, Valagussa P, Bonadonna G. Breast cancer recurrence dynamics following adjuvant CMF is consistent with tumor dormancy and mastectomy-driven acceleration of the metastatic process. Ann Oncol 2005; 16:1449-57; PMID:15956037; http://dx.doi.org/10.1093/annonc/mdi280
- Lacroix M. Significance, detection and markers of disseminated breast cancer cells. Endocr Relat Cancer 2006; 13:1033-67; PMID:17158753; http://dx.doi.org/10.1677/ERC-06-0001
- Balafoutas D, zur HA, Mayer S, Hirschfeld M, Jaeger M, Denschlag D, Gitsch G, Jungbluth A, Stickeler E. Cancer testis antigens and NY-BR-1 expression in primary breast cancer: prognostic and therapeutic implications. BMC Cancer 2013; 13:271; PMID:23731661; http://dx.doi.org/10.1186/1471-2407-13-271
