ABSTRACT
Hepatocellular carcinoma (HCC) is one of the most common types of malignant tumors with increasing incidence rates and high mortality rates. The currently available methods for treating HCC include surgery, radiotherapy or chemotherapy, but all of them have limitations. Therefore, developing novel therapeutic methods for HCC is in great need. Here, in this study, we found that tanshinone I, a small molecule compound, inhibited the proliferation of HCC cells in a dose-dependent manner. We also observed that Tanshinone I destabilized genomes by inhibiting both NHEJ and HR repair pathways, which are responsible for repairing DNA double strand breaks (DSBs). Mechanistically, this compound suppressed the expression of 53BP1, and the recruitment of RPA2 to DNA damage sites. Importantly, we demonstrated that combining Tanshinone I with radiotherapy exhibited better therapeutic potential for treating HCC.
Introduction
As the most common type of malignant liver tumors, hepatocellular carcinoma (HCC) ranks fourth in incidence rates and second in mortality rates among all types of cancers in 2022.Citation1 A number of factors such as the difficulty in early diagnosis and the lack of effective therapeutic methods lead to the high mortality. Surgical treatment is suitable for early stage HCC, but not for patients with local advanced and distant metastasis,Citation2,Citation3 while the response of these pateints with late stage HCC to radiotherapy and chemotherapy is sometimes poor. The patients may develop radio- or chemo-resistance, and these therapeutic methods may have severe side effects,Citation4,Citation5 so radiotherapy and chemotherapy are now mainly used for adjuvant therapy in treating HCC.Citation6,Citation7 Therefore, finding new methods to sentisize HCC to radiotherapy or chemotherapy may benefit the HCC patients.
Although the etiology and pathogenesis of HCC are not fully clear, it shares several common hallmarks of malignant tumors: persistent proliferative signaling, immune evasion, genomic instability, and high burden of mutations. Among them, the accumulation of DNA damage is a very major one.Citation8 Numerous endogenous or exogenous factors may cause DNA damage to hepatocytes such as endogenously generated reactive oxygen species (ROS), collapsed DNA replication forks, exogenous toxic chemical substances, ionizing radiation.Citation9,Citation10
In response to different types of damage, cells evolved different repair pathways to fix them. Among all types of DNA damage, DNA double-strand breaks (DSBs) are considered the most serious form. The defects in DSB repair pathways, homologous recombination (HR) and Non-homologous end joining (NHEJ)Citation11–13 may lead to the rise in genomic instability, resulting in apoptosis, cellular senescence or even tumorigenesis. Interestingly, our previous work demonstrated that both DSB repair pathways are up-regulated in HCC tissues than in normal adjacent livers.Citation14 Importantly, it was shown that targeting the two pathways by inhibiting PARP1 and DNA-PKcs enzymatic acitivities synergistically suppressed HCC growth using both mouse models and human PDX models.Citation14 In addition, the XRCC4-like factor XLF activates the NHEJ pathway in hepatoma cells, leading to radio-resistance.Citation14,Citation15 These pieces of evidence suggest that the survival of HCC is inextricably linked to the ability of DSB repair.
Tanshinone I is one of the most popular traditional Chinese herbs extracted from Salvia miltiorrhiz, also known as Dan-shen. Tanshinone I has many functions including anti-bacterial, anti-inflammation, promoting blood circulation, removing blood stasis, and promoting wound healing.Citation16 A number of reports also indiate that Tanshinone I has certain therapeutic effects on mastitis, arthritis and cancers.Citation17–19 As reproted, Tanshinone I may induce apoptosis and autophagy via different pathways to attenuate the malignant biological properties of ovarian and breast cancers.Citation16 However, whether Tanshinone I has any effects in HCC and whether the Tanshinone I inhibits the DSB repair ability to reduce the survival of HCC have not been characterized.
Here, we demonstrated that Tanshinone I disrupts genome stability by inhibiting DSB repair. Further mechanistic studies indicate that Tanshinone I impairs the expression of 53BP1 at transcription level, and inhibits the recruitment of RPA2 to DNA damage sites upon X-ray irradiation. Combining Tanshinone I and X-ray had additional suppressive effect on the survival of HCC cells, indicating a potential therapeutic method for HCC.
Materials and methods
Cell culture
Hep3B and Huh7 cells were cultured in 10-cm plates with DMEM (supplemented with 10% FBS and 1% penicillin/streptomycin). Both Hep3B and Huh7 cell lines were cultured with 5% CO2 at 37°C.
Cell viability assay
1,000 cells were seeded in 96-well plates and cultured in DMEM for 24 hours before being supplemented with Tanshinone I at increasing concentrations. At 24 h, 48 h, 72 h and 96 h post Tanshinone I treatment, the medium was removed and cells were washed with PBS three times. Next, cells were added with 90 μL fresh medium, and 10 μL MTT solution (Solarbio, Cat. # M1020), and cells were then maintained in the incubator for 4 more hours. Then, the medium was aspirated and each well was added with 110 μL Formazan solution, and the plates were shaken slowly for 10 minutes before the absorption was measured using the microplate reader at a wavelength of 490 nm.
Clonogenic assay
300 cells per well were plated in 6-well plates. After 24-hours culture, Tanshinone I was added to the plates at the following concentrations: DMSO, 250 nM, 500 nM, 750 nM, 1 μM, 2 μM. After a 2-week incubation, cells were fixed with formaldehyde for 30 minutes and stained with crystal violet for 30 minutes. Finally, the cells were washed twice with distilled water. Clones with 50 cells were counted.
Analysis of HR/NHEJ repair efficiency
The HR and NHEJ reporter plasmids used in this paper were as previously descibed.Citation20 4 × 104 cells were seeded in 6-cm culture plates, and cultured for 24 h before the cells were pretreated with Tanshinone I and DMSO. After 24 h treatment, the I-SceI digested HR or NHEJ plasmids were purified and transfected into cells using the Lonza 4D system. At 72 h post transfection, cells are collected for analysis on FACS Caliber.
Comet assay
2 × 104 cells were seeded in 6-well plates. After 24 hours, cells were treated with Tanshinone I. On day 3, cells were collected for analysis of genomic stability using a Trevigen alkaline comet assay kit (Cat. number: 4250–050-K,Trevigen, Gaithersburg, MD, USA). Pictures were taken with a confocal microscope (TCS SP8; Leica, Wetzlar, Germany).
Immunofluorescence assay
4,000 cells were seeded on coverslips in 12-well plates. After 24 h, the cells were treated with Tanshinone I for 72 h and irradiated at a dosage of 2 Gy. The cells were collected at different time points and quickly washed twice with 1×PBS. For the RPA2 staining, 1×CSK buffer (NaCl 1 M, PEPES 100 mM, EGTA 10 mM, MgCl2-6 H2O 30 mM, TritonX-100 5%) was also added and incubated for 10 min to remove the cytoplasm. Then the cells were fixed with 4% formaldehyde for 30 min, and quickly washed twice with cold 1×PBS. Then the cells were treated with 0.25% Triton X-100 for 10 min. After washing with 1×PBS three times, the cells were blocked with 2% BSA for 1 hour, and a primary antibody was added, followed by the incubation at 4-degree overnight. Afterwards, the plates were washed with 1×PBS three times. Each wash lasts 5 minutes,followed by the incubation with secondary antibodies at room temperature for 1 h in the dark. Immunofluorescence pictures were also taken on a confocal microscope (TCS SP8; Leica, Wetzlar, Germany).
Quantitative real-time PCR
4 × 104 cells per well were cultured with DMEM medium. Tanshinone I was added at the concentration of 1 μM, the cells were harvested for RNA extraction with an RNA extraction kit (Tiangen, Lot# X0629). The RNA were reverse-transcribed to cDNA with a cDNA reverse transcription kit (Trans, Lot# O11210). Quantitative real-time PCR was performed with a SYBR Green kit (Roche,Lot46660900).
Results
Tanshinone I inhibits the proliferation and destabilizes genomes of HCC cells
The structure of Tanshinone I we used is shown in . To understand the toxic effects of Tanshinone I on HCC cells, we first examined the proliferation in two HCC cell lines, Hep3B and Huh7. Using a clonogenic assay, we observed a significantly decreased proliferation rate in a dose-dependent manner in both of the cell lines ( and S1a,b). Consistently, we observed similar reduction in proliferation rate using the MTT assay ().
Figure 1. Tanshinone I inhibits proliferation of Hep3B cells by destabilizing genomes.
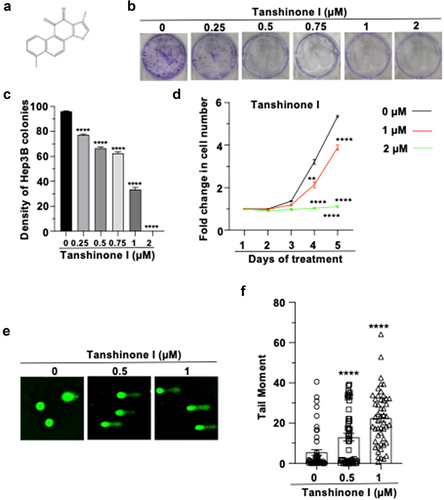
To test whether Tanshinone I inhibits cell proliferation by causing the rise in genomic instability, we performed the alkaline comet assay to measure the changes in genome stability. Indeed, we observed a dose-dependent increase in tail moment in drug-treated Hep3B and Huh7 cells ( and S1c,d). Together, these data suggest that Tanshinone I disrupts the genomic stability, thereby impairing the proliferation of HCC cells.
Tanshinone I inhibits HR repair and NHEJ repair in Hep3B cells
We next sought to determine whether this drug affects the repair of DNA damage. γH2AX is a widely used marker of DSBs.Citation21 We performed immunostaining assay to examine the kinetics of γH2AX clearance at different time points post X-ray irradiation in DMSO or Tanshinone I treated Hep3B cells. We found that the treatment of Tanshinone I significantly reduced the clearance efficiency of γH2AX (). Additionally, a Western blot analysis also indicates that treating cells with Tanshinone I led to the upregulation in the γH2AX level ().
Figure 2. Tanshinone I suppresses the recruitment of γH2AX and inhibits HR and NHEJ repair in Hep3B cells.
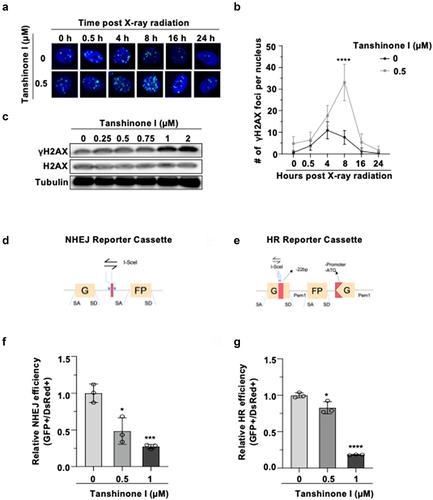
Deficiency in DNA repair pathway increases the genomic instability, and DSBs are the most dangerous type of DNA lesion. To investigate whether Tanshinone I causes genomic instabilty through targeting DSB repair, we tested DSB repair efficiency in Tanshinone I treated cells using our well-established reporter cassettesCitation14 (). The I-SceI digested HR and NHEJ reporters were transfected along with DsRed plasmids, which were used to normalize the transfection efficiency, into Hep3B cells. At 72 h post transfection, the ratio of GFP+ cells versus DsRed+ cells, which were quantified by flow cytometry, was utilized as the measure of DSB repair efficiency. We found that in Tanshinone I treated Hep3B cells both NHEJ () and HR () efficiency was significantly reduced in a dose-dependent manner. The NHEJ efficiency of Hep3B decreased by approximately 70% and the HR efficiency was reduced by nearly 80% at the concentration of 1 μM, indicating that Tanshinone I destabilizes the genome through inhibting NHEJ and HR repair in HCC cells. We also ruled out the possibility that observed decline in both NHEJ and HR was due to the altered cell cycle distribution as we observed that there was no significant change in the number of cells in different cell cycle phases after 72 h treatment of Tanshinone I (Figure S2).
Importantly, by analyzing proliferation rates of Tanshinone I treated HCC cells overexpressing RPA2, a critical HR factor, we found that restoring DNA repair greatly attenuated the cytotoxic effects of Tanshinone I on HCC cells (Figure S3). Taken together, these results indicate that, in addition to its know function of inducing DNA damage, Tanshinone I directly participates in negatively regulating DSB repair.
Tanshinone I reduces the expression of 53BP1 and the recruitment of RPA2
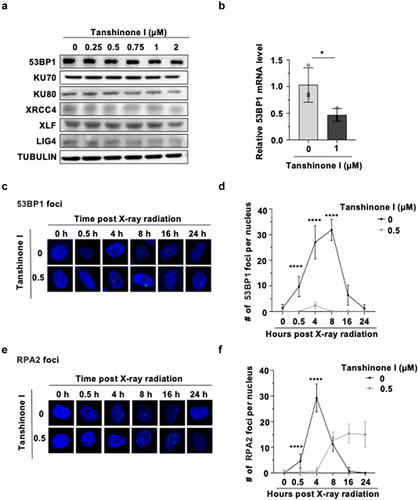
To investigate the underlying molecular mechanisms of down-regulating both NHEJ and HR, we pretreaeted Hep3B cells with DMSO and Tanshinone I for 72 hours, and then examined the expression of key proteins involved in NHEJ and HR pathway. We found that the expression of 53BP1, which is critical to NHEJ pathway,Citation22,Citation23 was significantly reduced in a dose-dependent manner, while we did not observe any statistically significant change in HR proteins between the drug treated and un-treated cells ( and S4). Next, we examined whether the Tanshinone I-mediated decline in 53BP1 expression is through transcriptional mechanisms. Quantitative PCR experiments reveal that the transcription level of 53BP1 was significantly decreased in drug-treated cells (). As a consequence, the recruitment of 53BP1 to DNA damage site was also impaired in cells treated with X-ray irradiation (). We also performed immunostaining experiments with an antibody against RPA2, an essential HR factor that protects the resected single strand DNA from accidental degradation during HR process.Citation24,Citation25 We found that the recruitment of RPA2 to DNA damage sites was also sigificantly reduced in X-ray irradiated HCC cells (). Together, Tanshinone I suppresses both NHEJ and HR through different molecular mechanisms.
Figure 3. Tanshinone I suppresses the mRNA level of 53BP1 and inhibits the recruitment of RPA2 to DNA damage sites.
Tanshinone I sensitizes HCC cells to ionizing radiation
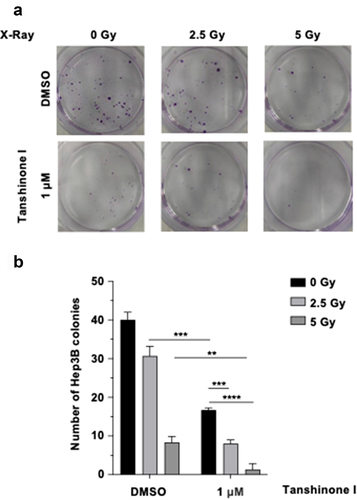
Radiotherapy is one of therapeutic methods in treating HCC cancer. Since Tanshinone I inhibits DSB repair by both NHEJ and HR, we examined whether Tanshinone I sensitizes HCC cells to radiotherapy using a clonogenic assay. We found that combination of X-ray and Tanshinone I led to a 80.00% (2.5 Gy +1 μM) and 96.67% (5 Gy +1 μM) decrease in Hep3B cells, compared with a 23.33% (2.5 Gy) and 79.17% (5 Gy) decrease with single-X-ray treatment and a 58.33% decrease with single-agent Tanshinone I treatment (). These data suggest that combining Tanshinone I with radiotherapy exerts an additional effect of inhibiting the survival of HCC cells, indicating that Tanshinone I holds the potential to be a sensitizer for radiotherapy.
Figure 4. Tanshinone I sensitizes HCC cells to ionizing radiation.
Discussion
Tanshinone I is the traditional Chinese herb, which is one of the fat-soluble monomer components extracted from Salvia miltiorrhiza. In recent years, with the in-depth study of the pharmacology of Tanshinone I, a large amount of evidence has shown that Tanshinone I has a variety of biological activities, including improving the activity of antioxidant enzymes, enhancing the ability to scavenge free radicals, reducing the production of lipid peroxides, inhibiting the proliferation of cancer cells and so on.Citation16–19,Citation26 Our present work adds the suppression of DSB repair by both NHEJ and HR to the function list of Tanshinone I. The newly discovered function also provides the foundation for Tanshinone I application in HCC therapy.
A number of studies have indicated that Tanshinone I affects the expression of a number of proteins, which participate in the pathways of apoptosis, autophagy, mTOR, NRF2. Although further evidence is still needed to clarify how Tanshinone I changes the expression, the previous work demonstated that Tanshinone I directly targets the epigenetic factor EZH2, which is associated with chromatin. Since our work indicates that Tanshinone I negatively regulates transcription of 53BP1, we speculate that the changes in the expression of these proteins in the presence of Tanshinone I might be the consequences of the alteration in mRNA level of these genes. Future work on whether Tanshinone I targets EZH2 to regulate the promoters of these genes would greatly help elucidate the mechanisms.
Although we demonstrated that Tanshinone I suppressed the recruitment of RPA2 to DNA damage sites, at which step Tanshinone I directly inhibits the HR process remains to be determined. Since it does not affect the recruitment of γH2AX to DNA damage sites, Tanshinone I might suppress the chromatin relaxation upon the occurrence of DNA damage, the end resection, or the loading of the RPA2 to resected DNA ends. A further thourough study based on mass spectrometry results would help identify the direct targets of Tanshinone I, which are involved in HR repair.
It is of note that we did not observe the synergistic effect of combining Tanshinone I and radiation on the survival of HCC cells. Theoretically, inhibiting the repair of X-ray induced DNA damage by Tanshinone I would cause synergistic effect on HCC cell survival. We reasoned that the amount of DSBs accounts for only 50% of all types of X-ray-induced DNA damage, and the role of Tanshinone I in alleviating oxidative stress further complicates the expected results. Nevertheless, our data still indicate that treating cells with Tanshinone I at a low concentration may still sensitize HCC cells to radiotherapy. Moreover, it is still possible that combining Tanshinone I with other types of therapeutic method such as PD-1 or PD-L1 antibodies might yield better response.
Ethical statement
No animals or patient samples were used in this study, so the ethical statement is not applicable.
Supplemental Material
Download MS Word (151.3 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated during this study are included in the main text and supplementary information files.
Supplemental material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2023.2229958
Additional information
Funding
Notes on contributors
Zhen Qian
Ms. Zhen Qian, Master Graduate Student, School of Public Health Shanghai Jiao Tong University School of Medicine; Shanghai Key Laboratory of Maternal Fetal Medicine, Clinical and Translational Research Center of Shanghai First Maternity and Infant Hospital, Frontier Science Center for Stem Cell Research, School of Life Sciences and Technology, Tongji University, Shanghai, China.
Nannan Feng
Dr. Nannan Feng, Associate professor, School of Public Health Shanghai Jiao Tong University School of Medicine, Shanghai, China.
Anke Geng
Dr. Anke Geng, Associate research fellow, Shanghai Key Laboratory of Maternal Fetal Medicine, Clinical and Translational Research Center of Shanghai First Maternity and Infant Hospital, Frontier Science Center for Stem Cell Research, School of Life Sciences and Technology, Tongji University, Shanghai, China.
References
- Xia C, Dong X, Li H, Cao M, Sun D, He S, Yang F, Yan X, Zhang S, Li N, et al. Cancer statistics in China and United States, 2022: profiles, trends, and determinants. Chin Med J (Engl). 2022;135(5):584–7. doi:10.1097/CM9.0000000000002108.
- Crocetti L, Bargellini I, Cioni R. Loco-regional treatment of HCC: current status. Clin Radiol. 2017;72(8):626–635. doi:10.1016/j.crad.2017.01.013.
- Kirstein MM, Wirth TC. Multimodal treatment of hepatocellular carcinoma. Internist (Berl). 2020;61(2):164–169. doi:10.1007/s00108-019-00722-x.
- Nia A, Dhanasekaran R. Genomic landscape of HCC. Curr Hepatol Rep. 2020;19(4):448–461. doi:10.1007/s11901-020-00553-7.
- Chen W, Chiang CL, Dawson LA. Efficacy and safety of radiotherapy for primary liver cancer. Chin Clin Oncol. 2021;10(1):9. doi:10.21037/cco-20-89.
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: gLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. A Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660.
- Yang SF, Chang CW, Wei RJ, Shiue YL, Wang SN, Yeh YT. Involvement of DNA damage response pathways in hepatocellular carcinoma. Biomed Res Int. 2014;2014:153867. doi:10.1155/2014/153867.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013.
- Chatterjee N, Walker GC. Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen. 2017;58(5):235–263. doi:10.1002/em.22087.
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73(1):39–85. doi:10.1146/annurev.biochem.73.011303.073723.
- Nambiar M, Raghavan SC. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011;39(14):5813–5825. doi:10.1093/nar/gkr223.
- Zhu C, Mills KD, Ferguson DO, Lee C, Manis J, Fleming J, Gao Y, Morton CC, Alt FW. Unrepaired DNA breaks in p53-deficient cells lead to oncogenic gene amplification subsequent to translocations. Cell. 2002;109(7):811–821. doi:10.1016/S0092-8674(02)00770-5.
- Curtin NJ. DNA repair dysregulation from cancer driver to therapeutic target. Nat Rev Cancer. 2012;12(12):801–817. doi:10.1038/nrc3399.
- Wang C, Tang H, Geng A, Dai B, Zhang H, Sun X, Chen Y, Qiao Z, Zhu H, Yang J, et al. Rational combination therapy for hepatocellular carcinoma with PARP1 and DNA-PK inhibitors. Proc Natl Acad Sci USA. 2020;117(42):26356–26365. doi:10.1073/pnas.2002917117.
- Yang S, Wang XQ. XLF-mediated NHEJ activity in hepatocellular carcinoma therapy resistance. Bmc Cancer. 2017;17(1):344. doi:10.1186/s12885-017-3345-y.
- Zhou J, Jiang YY, Chen H, Wu YC, Zhang L. Tanshinone I attenuates the malignant biological properties of ovarian cancer by inducing apoptosis and autophagy via the inactivation of PI3K/AKT/mTOR pathway. Cell Prolif. 2020;53(2):e12739. doi:10.1111/cpr.12739.
- Wang X, Fan J, Ding X, Sun Y, Cui Z, Liu W. Tanshinone I inhibits IL-1beta-induced apoptosis, inflammation and extracellular matrix degradation in chondrocytes CHON-001 cells and attenuates murine osteoarthritis. Drug Des Devel Ther. 2019;13:3559–3568. doi:10.2147/DDDT.S216596.
- Yang L, Zhou G, Liu J, Song J, Zhang Z, Huang Q, Wei F. Tanshinone I and Tanshinone IIA/B attenuate LPS-induced mastitis via regulating the NF-kappaB. Biomed Pharmacother. 2021;137:111353. doi:10.1016/j.biopha.2021.111353.
- Luo H, Vong CT, Chen H, Gao Y, Lyu P, Qiu L, Zhao M, Liu Q, Cheng Z, Zou J, et al. Naturally occurring anti-cancer compounds: shining from Chinese herbal medicine. Chin Med. 2019;14(1):48. doi:10.1186/s13020-019-0270-9.
- Zhang F, Tang H, Jiang Y, Mao Z. The transcription factor GATA3 is required for homologous recombination repair by regulating CtIP expression. Oncogene. 2017;36(36):5168–5176. doi:10.1038/onc.2017.127.
- Sharma A, Singh K, Almasan A. 2012. Histone H2AX phosphorylation: a marker for DNA damage. Methods Mol Biol. 920:613–626.
- Zimmermann M, de Lange T. 53BP1: pro choice in DNA repair. Trends Cell Biol. 2014;24(2):108–117. doi:10.1016/j.tcb.2013.09.003.
- Setiaputra D, Durocher D. Shieldin – the protector of DNA ends. EMBO Rep. 2019;20(5). doi:10.15252/embr.201847560.
- Cao L, Xu X, Bunting SF, Liu J, Wang RH, Cao LL, Wu JJ, Peng TN, Chen J, Nussenzweig A, et al. A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency. Mol Cell. 2009;35(4):534–541. doi:10.1016/j.molcel.2009.06.037.
- Bunting SF, Callen E, Wong N, Chen HT, Polato F, Gunn A, Bothmer A, Feldhahn N, Fernandez-Capetillo O, Cao L, et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell. 2010;141(2):243–254. doi:10.1016/j.cell.2010.03.012.
- Wang X, Yang Y, Liu X, Gao X. 2020. Pharmacological properties of tanshinones, the natural products from Salvia miltiorrhiza. Adv Pharmacol. 87:43–70.
