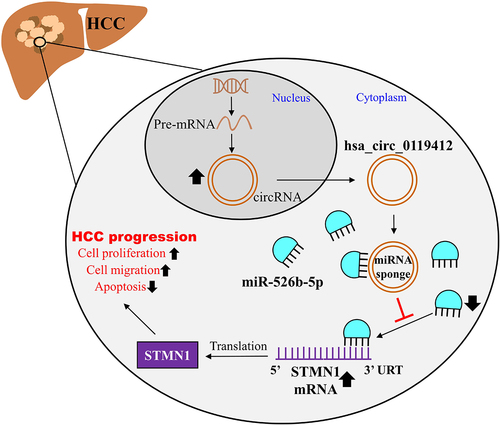
ABSTRACT
Hepatocellular carcinoma (HCC) is always deemed a deadly malignancy worldwide. Non-coding RNAs, including circRNAs, are becoming more widely recognized as essential regulators of the malignant development of HCC. Thus, we elaborated the regulating role of hsa_circ_0119412 in HCC advancement. The qRT-PCR was done to estimate the expressions of hsa_circ_0119412, miR-526b-5p, and Stathmin 1 (STMN1) in HCC (clinical samples and cell lines), and immunoblotting was used to detect STMN1 protein level in HCC cell lines. The stability of the circRNA was checked by processing with ribonuclease R. The proliferative potential of HCC cells was examined via the CCK-8 assay and the migratory potential by the wound healing assay. Immunoblotting was done to examine Bax and Bcl-2 (apoptosis-related proteins). Luciferase and RIP assays were employed to establish the direct interactions among miR-526b-5p and hsa_circ 0119412/STMN1. In vivo tumor growth was measured by doing a xenograft tumor experiment. In the tissues of HCC patients and cell lines derived from HCC cells, hsa_circ_0119412 was distinctly over-expressed. Knocking down hsa_circ_0119412 impeded proliferation and migration while inducing apoptosis in HCC cells. Moreover, silencing hsa_circ_0119412 diminished tumor weight and volume in vivo. Interestingly, miR-526b-5p inhibition partially restored the anti-tumor effects of silencing hsa_circ_0119412. STMN1 expression was also abundant in HCC, suggesting that it play a tumor-promoting role. Mechanistically, hsa_circ_0119412 sponged miR-526b-5p, resulting in STMN1 upregulation and thus facilitating the progression of HCC. In conclusion, this study reveals that hsa_circ_0119412 knockdown attenuates the progression of HCC by targeting miR-526b-5p/STMN1 axis.
Introduction
In 2020, liver cancer was ranked sixth in terms of incidence and third in terms of mortality worldwideCitation1. Hepatocellular carcinoma (HCC) accounts for approximately 75–85% of all liver cancers, followed by intrahepatic cholangiocarcinoma (about 10–15%) and a few other rare types. In China, because of the frequent infection of chronic HBV and continual exposure to aflatoxin, HCC is posing an increasing threat to people’s healthCitation2. Due to recurrence and metastasis, the prognosis for patients with advanced HCC has been unsatisfactory over the past decadesCitation3. Hence, it is imperative to uncover the key regulators in HCC progression, which may provide new biomarkers for developing the tumor immune targeting drugs to improve the prognosis of HCC.
Circular RNAs (circRNAs) having a covalently closed continuous loop are incredibly stable compared to linear RNAs, and have significant biological functions in tumorigenesisCitation4. For instance, circ_PIP5K1A sponges miR-493-5p to facilitate cisplatin resistance and malignancy in non-small cell lung cancer (NSCLC) ultimately enhancing ROCK1 expressionCitation5. In HCC, circFAM13B enhances the proliferation of HCC via targeting miR-212/E2F5 axis and activating the P53 pathwayCitation6. Another circRNA, UBAP2, has been shown to sponge miR-1294 to accelerate tumorigenesis in HCC via enhancing c-Myc expressionCitation7. According to a recent study, hsa_circ_0119412 was expressed at high levels in gastric tissues as well as cells, and played a part in the progression of malignant characteristics of gastric cancer cellsCitation8. However, there are no more studies to clarify the functional role and potential regulating mechanisms of hsa_circ_0119412 in HCC.
CircRNAs have consistently been deemed to regulate cancer progression by serving as microRNA (miRNAs) sponges to regulate the target genes of miRNAsCitation9. MiR-526b-5p, an miRNA, has been reported to play the key roles in different cancers by interacting different circRNAs and targeting different genes. For examples, hsa_circ_0085539 was proved to target miR-526b-5p to enhance SERP1 expression, thereby promoting osteosarcoma progressionCitation10. In cervical cancer, circ_0002762 could accelerate the tumorigenesis by sponging miR-526b-5p to upregulate HK2Citation11. In HCC, miR-526b-5p was reported to interact circUGGT2 or circEIF3I, thereby playing the inhibitory role in HCC developmentCitation12,Citation13. The studies on miR-526b-5p in HCC suggest that miR-526b-5p can interact with different circRNAs. However, the relationship between hsa_circ_0119412 and miR-526b-5p has not been identified.
Stathmin1 (STMN1), located on chromosome 1p36.11, has eight exons. It encodes a ubiquitous cytosolic phosphoprotein belonging to the stathmin family of genes and plays an essential role in the cellular environment aimed at an intracellular relay for integrating regulatory signals. Numerous researchers have confirmed that the dysregulation of the STMN1 gene contributed to tumor progression in lung squamous cell carcinomaCitation14, breast cancerCitation15, and pancreatic cancerCitation16. STMN1 has been found to be elevated in HCC, which has been linked to the occurrence and advancement of cancerCitation17,Citation18. However, the upstream regulators of STMN1 involving circRNA/miRNA axis in HCC have not been investigated.
Our study aimed to validate the hsa_circ_0119412/miR-526b-5p/STMN1 interactome and investigate its impact on HCC. By a series of in vivo and in vitro experiments, this study found that hsa_circ_0119412 was upregulated in HCC, and hsa_circ_0119412 knockdown could suppress the HCC malignancy. Moreover, this study also revealed that hsa_circ_0119412 could interact with miR-526b-5p to regulate STMN1 expression in HCC. Our findings may enrich the regulatory mechanism of HCC progression, potentially pointing to a novel therapeutic target.
Materials and methods
Clinical specimens
The study was authorized by the Ethics Committee of our hospital. HCC tissue and nearby healthy liver tissue were acquired from the HCC patients (n = 45) in our hospital. Samples of only those patients who did not receive radiation or chemotherapy before the surgery were included in the investigations. For further experimental use, samples were transferred to liquid nitrogen as soon as they were excised and stored in it. All HCC patients enrolled in the study signed consent forms.
Cell culture
The human normal hepatocyte cell line THLE-2 and the HCC cell line SNU-182 were obtained from American Type Culture Collection (USA). The other two HCC cell lines (Huh7 and Hep 3B) were sourced from China Cell Bank of Academy of Sciences (China). All cell lines mentioned above were cultured in DMEM medium along with 10% FBS (both from Gibco, USA) at 37°C in a surrounding with 5% CO2.
Quantitative reverse transcription PCR (qRT-PCR) assay
The TRIzol reagent (Invitrogen, USA) was utilized to purify whole RNA. Subsequently, 1 μg RNA was prepared to carry out the reverse transcription procedure using the FastKing RT Kit (with gDNase) (Invitrogen, USA). The Roche 480 detection system was then used to detect amplified cDNA by a SYBR Green Kit (Vazyme, China) with the following protocol: 95°C for 10 minutes, 40 amplification cycles of 95°C for 15 s and 60°C for 30 s, and 72°C for 30 s. All primer sequences used in this study are presented in . The 2−△△t method was employed in calculating the relative expression levels (normalized to either GAPDH or U6).
Table 1. Real-time PCR primer synthesis list.
Detection of hsa_circ_0119412 localization and stability
In order to verify the localization of hsa_circ_0119412, the Cytoplasmic and Nuclear RNA Purification Kit (Invitrogen, USA) was carried out per supplier’s instructions. This was followed by qRT-PCR assay to assess the expression of hsa_circ_0119412 in the nuclear and cytoplasmic fractions. For internal controls, GAPDH (in cytoplasmic fraction) or U6 (in nuclear fraction) were used.
For stability, Ribonuclease R (RNase R) enzyme was adopted to digest the whole RNA (at 37°C for 1 hour), and then qRT-PCR was done to estimate the expression levels of hsa_circ_0119412 and its linear gene PER2.
Cell transfection
siRNA specific for hsa_circ_0119412 (si-circ) and STMN1 (si-STMN1), miR-526b-5p mimic and inhibitor, along with their corresponding negative controls (NCs), were purchased from Genepharma (China). For transfection, the cells (1 × 105 cells per 100 μL culturing medium) were inoculated in 96-well plates and cultured for a period of 24 hours at 37°C in incubators containing 5% CO2. Then, Lipofectamine 2000 Reagent (Invitrogen, USA) was appended to 96-well plates in compliance with the steps provided by the distributor to facilitate the cellular transfection of the vector constructs (stated above). The cells were collected after 48 hours, followed by qRT-PCR to check the transfection efficiency.
Cell counting kit-8 (CCK-8) assay
Briefly, 48 hours post-transfection, cells (20000 cells/mL) were plated into 96-well plates. Next, these were cultivated at 37°C in containers supplied with 5% CO2 for four time points (0, 24, 48 and 72 hours). After each specified time point, cells were administered with CCK-8 reagent (10 μL, Dojindo, Japan). Subsequently, an additional incubation of 2 hours at 37°C was given, followed by measuring the absorbance of each well at a 450 nm wave length with a microplate reader (ThermoFisher, USA).
Dual-luciferase reporter assay
CircInteractome (https://circinteractome.nia.nih.gov) was utilized to predict the binding sequences between miR-526b-5p and hsa_circ_0119412, whereas TargetScan (www.targetscan.org/vert_71/) was employed to detect the possible binding sequences miR-526b-5p and STMN1. According to the predicted sequences, the pGL3 vector (Promega, USA) was deployed to subclone the hsa_circ_0119412/STMN1 3’-UTR wild type (WT, with biding sites) or mutant (MUT, without binding sites) sequences. After that, co-transfection of WT or MUT plasmids constructed above, and miR-526b-5p mimic or its NC into HCC cells was carried out. Then, the cells were harvested 48 hours post co-transfection, and luciferase signals were assessed by employing a Dual Luciferase Reporter Gene Assay Kit (Promega, USA).
Wound healing assay
The transfected cells (1 × 106) were plated into 6-well plates and allowed to reach 100% confluence. Then, with a sterile pipette tip (200 μL), a wound was scratched. Following this, the wells were cleaned with PBS (washed thrice) to clear the subtracted cells, and thereafter cultured in a medium which was serum-free. The images were taken at 0 h and 24 hours for assessing the cell migration using ImageJ software.
RNA immunoprecipitation (RIP) assay
RIP assay was performed by using a RNA-binding protein immunoprecipitation kit (Millipore, USA). Briefly, with the help of RIP lysis buffer (comprising RNase- and protease-inhibitor), the cells were lysed. Next, the process of incubating magnetic beads (labeled with AGO2/IgG antibody) with cell lysates was done overnight (O/N) at 4°C. Next day, the magnetic beads were isolated, cleaned by washing, and digested in a buffer containing Proteinase K. Finally, the RNA was extracted from the immunoprecipitates, and hsa_circ_0119412 and miR-526b-5p enrichment on the beads containing AGO2 was measured by qRT-PCR.
Immunoblotting
Cell lysate was prepared by using a Lysis Buffer (Beyotime, Shanghai, China), and the extracted proteins were separated via SDS-PAGE. These proteins were then transferred onto PVDF membrane, blocked in 5% nonfat milk at room temperature (RT) for 1 hour, and incubated with the primary antibodies overnight at 4°C. The list of primary antibodies (purchased from either Abcam, USA or Beyotime, China) are as follows: Bax (1:5000, ab270742), Bcl-2 (1:3000, ab32124), STMN1 (1:5000, ab52630) and GAPDH (1:5000, AF5009). Next day, the membrane was treated at RT for 1 hour with the secondary antibody (HRP-conjugated, 1:5000, A0192, Beyotime, China). Then, image of the protein bands was acquired by utilizing an ECL kit (Pierce, USA), and quantitated by densitometry via ImageJ software.
Tumor xenograft model
The xenograft assay was approved by the Ethics Committee of our hospital. Nude mice (strain: BALB/c, gender: ♀, age: ~4 weeks, and weight: ~28 g) were procured from the Nanjing Qinglongshan Experimental Animal Factory, and housed in an environment free of pathogens. Huh7 cells stably transfected with lentiviral vector hsa_circ_0119412 (sh-circ_0119412) or sh-NC (RiboBio, Guangzhou, China) were injected (1 × 107 cells) subcutaneously into the flank of mice (n = 5/group). Every week, the length (l) and width (w) of the tumor was measured with the help of calipers to calculate the tumor volume. All animals were sacrificed after 5 weeks and tumor weights were obtained after excising them.
Statistical analysis
GraphPad prism 7.0 software was used to carry out all the statistical analyses. Each assay was done thrice independently, and the results were presented in the form of mean ± SD. To compare two groups, the student’s t test was used, and to compare multiple groups, one-way ANOVA was used, followed by Tukey post hoc analysis. The correlation among miR-526b-5p and hsa_circ_0119412/STMN1 expressions was investigated using Spearman’s correlation analysis. P-value less than 0.05 was considered indicative of a significant difference.
Results
Hsa_circ_0119412 was remarkably upregulated in HCC
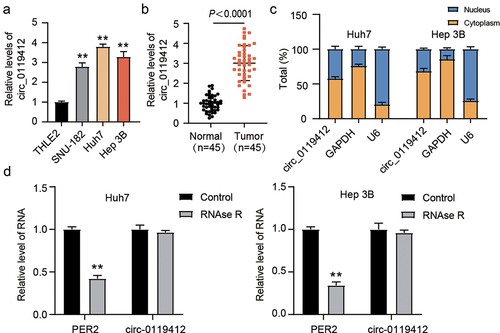
Firstly, we determined the hsa_circ_0119412 expression levels in THLE-2 and three cell lines (SNU-182, Huh7 and Hep 3B) of HCC by qRT-PCR assay. It was seen that hsa_circ_0119412 was significantly upregulated in HCC cell lines as compared to THLE-2 (). Since hsa_circ_0119412’s expression was comparatively high in Huh7 and Hep 3B, they were chosen for the subsequent study. Consistent with the trend in cell lines, hsa_circ_0119412 was also predominantly expressed in HCC tissues (). Besides, in both Huh7 and Hep 3B, the cytoplasm was the main site of hsa_circ_0119412 localization (). Furthermore, RNase R exposure reduced linear transcript PER2 expression but was not able to change hsa_circ_0119412 expression in both Huh7 and Hep 3B cells (). The outcomes above reveal that hsa_circ_0119412 is upregulated in HCC, has a high stability, and is primarily found in the cytoplasm. Various studies have shown that circRNAs, which are mostly found in the cytoplasm, can act as miRNA sponges, regulating miRNA expression levels. Hence, in the following study, we looked into the possible sponged miRNAs of hsa_circ_0119412 as well as the detailed regulatory mechanisms.
Figure 1. Hsa_circ_0119412 was upregulated in HCC. (a) expression of hsa_circ_0119412 was evaluated via qRT-PCR in THLE2 and three HCC (SNU-182, Huh7 and Hep 3B) cell lines. (b) expression of hsa_circ_0119412 was evaluated in adjacent normal and HCC tissues by qRT-PCR assay. (c) localization of hsa_circ_0119412 expression was assessed by qRT-PCR assay in the cytoplasmic and nuclear fractions. (d) the qRT-PCR was done to measure hsa_circ_0119412 and PER2 expressions in the presence or absence of RNase R. **P < .01 vs. THLE2/Control.
Hsa_circ_0119412 silencing suppressed the progression of HCC
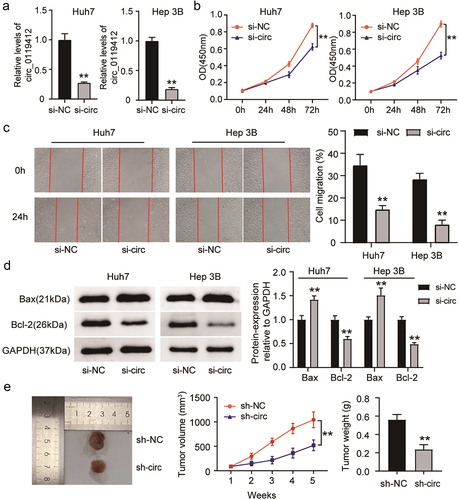
To illustrate the role of hsa_circ_0119412, it was silenced in Huh7 and Hep 3B cells by treating with si-circ_0119412 (si-circ). The experimental results confirmed that the levels of hsa_circ_0119412 expression dramatically reduced when transfected with si-circ, indicating that it was successfully knocked down (). The proliferation of HCC cells decreased after hsa_circ_0119412 silencing (), and their ability to migrate was remarkably reduced (). Additionally, the relative protein level of Bax in hsa_circ_0119412-silenced cells was dramatically higher as compared to the NC group, whereas the opposite trend was observed for Bcl-2, indicating that si-circ induced apoptosis in HCC cells (). We also checked the impact of silencing hsa_circ_0119412 on HCC tumor growth in nude mice. As shown in the image, after hsa_circ_0119412 depletion, the xenograft volume and weight were remarkably lower as compared to control group (). These observations illustrated that hsa_circ_0119412 knockdown restrained proliferation, migration and induced apoptosis in HCC.
Figure 2. Hsa_circ_0119412 knockdown suppressed HCC progression. (a) Huh7 and Hep 3B were transfected with si-circ_0119412 or si-NC, and transfection efficiency was measured by qRT-PCR. (b) cell proliferation in these above mentioned transfected cells was determined using CCK-8 assay. (c) the migration in these above mentioned transfected cells was assessed using wound healing assay. (d) the levels of apoptotic molecules (Bax and Bcl-2) in transfected Huh7 and Hep 3B cells were determined by immunoblotting. (e) the volume of the tumor xenograft was measured every week for 5 consecutive weeks, and after 5 weeks, the tumor was excised, imaged, and weighed (n = 5). **P < .01 vs. si-NC/sh-NC.
Hsa_circ_0119412 binds to miR-526b-5p
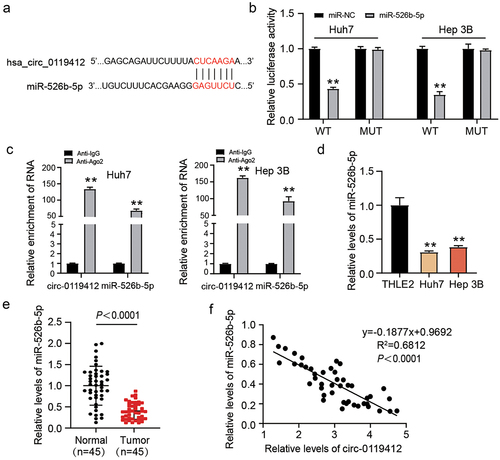
To identify the downstream miRNA targeted by hsa_circ_0119412 in HCC, we used an online website called circInteractome, finding the binding sites between miR-526b-5p and hsa_circ_0119412 (). Notably, miR-526b-5p overexpression strikingly lowered the luciferase activity of hsa_circ_0119412-WT, while that of hsa_circ_0119412-MUT stayed the same (). RIP experiment revealed that both hsa_circ_0119412 as well as miR-526b-5p were remarkably abundant in the AGO2 group (). Furthermore, in HCC cell lines, miR-526b-5p expression was significantly lower as compared to THLE-2 cell line (). Similarly, the tissues of patients with HCC had remarkably low miR-526b-5p expression as compared to the respective controls (). In tumor tissues, Spearman correlation analysis illustrated a negative association among the expressions of miR-526b-5p and hsa_circ_0119412 (). The experimental results of this section illustrated that miR-526b-5p was downstream target mRNA of hsa_circ_0119412.
Figure 3. Hsa_circ_0119412 directly targeted miR-526b-5p. (a) circInteractome predicted the complementary binding sequences for miR-526b-5p in hsa_circ_0119412. (b) detection of relative luciferase activity by co-transfecting the cells with either miR-526b-5p mimic or miR-NC along with either circ_0119412-WT or MUT. (c) the interaction of miR-526b-5p and hsa_circ_0119412 was examined using RIP assay. (d) miR-526b-5p expression was assessed in THLE2 and HCC cells (Huh7 and Hep 3B) via qRT-PCR. (e) miR-526b-5p level was evaluated in adjacent normal and HCC tissues via qRT-PCR assay. (f) correlation between hsa_circ_0119412 and miR-526b-5p in cohort of HCC tissues was analyzed through Spearman correlation analysis.
Hsa_circ_0119412 depletion repressed HCC malignant development via targeting miR-526b-5p
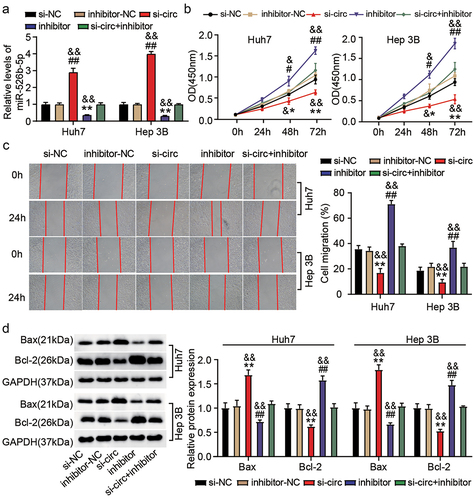
We further explored whether hsa_circ_0119412 regulated HCC progression via miR-526b-5p by performing rescue experiments. For this purpose, transfection was executed in Huh7 and Hep 3B cells with (i) si-circ_0119412 (si-circ) (ii) inhibitor (for miR-526b-5p) (iii) si-circ+inhibitor, and corresponding NCs (si-NC and inhibitor-NC). We observed that miR-526b-5p expression was enhanced in hsa_circ_0119412-silenced cells and low in miR-526b-5p inhibitor transfected cells. Moreover, silencing hsa_circ_0119412 reversed the effect of miR-526b-5p inhibitor to some extent by restoring its expression in the cells (). Functional experiments uncovered that si-circ significantly decreased the proliferative as well as migratory abilities of HCC cells, whereas miR-526b-5p inhibitor had the inverse effect. As anticipated, the suppressive impact of hsa_circ_0119412 with low expression on HCC cells was reversed by downregulating miR-526b-5p . Consistently, the decreased Bcl-2 and increased Bax protein levels induced by si-circ were overturned by inhibiting miR-526b-5p (). These results manifested that hsa_circ_0119412/miR-526b-5p pathway took part in the progression of HCC.
Figure 4. Hsa_circ_0119412 depletion repressed HCC progression via targeting miR-526b-5p. Transfection was carried out in Huh7 and Hep 3B cells with (i) si-circ_0119412 (si-circ) (ii) inhibitor (for miR-526b-5p) (iii) si-circ+inhibitor, and corresponding NCs. (a) MiR-526b-5p level was measured by qRT-PCR in the transfected cells. (b) cell proliferation in various transfected groups was determined by CCK-8 assay. (c) the changes of migratory capacity in different transfected groups were assessed by wound healing assay. (d) the protein levels of apoptotic molecules as determined in different groups by immunoblotting. **P < .05, **P < .01 vs. si-NC, #P <.05, ##P <.01 vs. inhibitor-NC and &P <.05, &&P <.01 vs. si-circ+inhibitor.
STMN1 was a target of miR-526b-5p
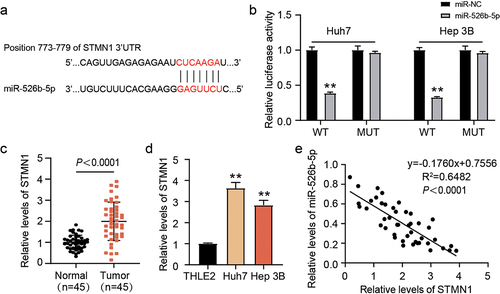
We aimed to further exploit the target of miR-526b-5p and the bioinformatics analysis predicted STMN1 to be its downstream target (). As expected, forced miR-526b-5p expression remarkably lowered the luciferase activity of STMN1-WT, while that of STMN1-MUT remained unaffected (). Moreover, STMN1 was clearly upregulated in tumor tissues () and HCC cell lines (). Further, a negative association was ascertained among the expressions of miR-526b-5p and STMN1 in the cohort tumor of tissues (). These observations implied that STMN1 was a target of miR-526b-5p.
Figure 5. miR-526b-5p target STMN1. (a) TargetScan predicted the binding sites among miR-526b-5p and STMN1. (b) detection of relative luciferase activity by co-transfecting the cells with either miR-526b-5p mimic or miR-NC along with either STMN1 3’-UTR-WT or MUT. (c) STMN1 expression was evaluated by qRT-PCR in adjacent normal and HCC tissues. (d) the expression level of STMN1 was evaluated by qRT-PCR in THLE2 and HCC cells (Huh7 and Hep 3B). (e) correlation among the expressions of STMN1 and miR-526b-5p in cohort of HCC tissues was analyzed through Spearman correlation analysis.
MiR-526b-5p depletion counteracted the influence of STMN1 inhibition on HCC
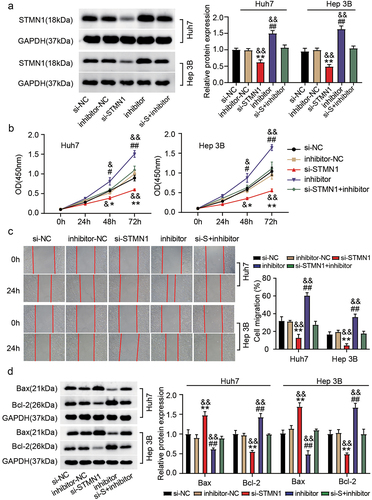
In this part, rescue experiments were launched to elaborate the role of STMN1 in hsa_circ_0119412/miR-526b-5p – regulated progression of HCC. Transfection was done in Huh7 and Hep 3B cells with (i) si-STMN1 (ii) inhibitor (for miR-526b-5p) (iii) si-STMN1+inhibitor, and corresponding NCs (si-NC and inhibitor-NC). Immunoblotting revealed that the miR-526b-5p inhibitor recovered the lower SMTN1 protein levels which was induced by STMN1 knockdown in the cells (). In vitro assays revealed that silencing STMN1 decreased the proliferative and migratory ability of Huh7 and Hep 3B cells, and it was reversed by inhibiting miR-526b-5p . As expected, the decreased Bcl-2 and increased Bax protein level caused by STMN1 knockdown were reversed when miR-526b-5p was inhibited (). Altogether, these observations implicated that miR-526b-5p targeting STMN1 could influence the malignancy of HCC cells.
Figure 6. MiR-526-5p depletion counteracted the effect of STMN1 inhibition on HCC. Transfection was done in Huh7 and Hep 3B cells with (i) si-STMN1 (ii) inhibitor (for miR-526b-5p) (iii) si-STMN1+inhibitor, and corresponding NCs. (a) relative protein level of STMN1 in various transfected groups was determined by immunoblotting. (b) cell proliferation was determined by CCK-8 assay in various transfected groups. (c) the changes of migratory capacity were assessed by wound healing assay in different transfected groups. (d) the protein levels of apoptotic molecules as determined by immunoblotting in different transfected groups. **P < .05, **P < .01 vs. si-NC, #P <.05, ##P <.01 vs. inhibitor-NC and &P <.05, &&P <.01 vs. si-STMN1+inhibitor.
Discussion
Liver cancer is one of the most prevalent types of malignant cancers, with HCC accounting for the vast majority of cases. After surgery, the estimated 5 year survival rate for patients with HCC is just 36.9%Citation19. This study verified that hsa_circ_0119412 was elevated in HCC. Silencing hsa_circ_0119412 obstructed cellular proliferation and migration, and elicited cellular apoptosis in HCC, and reduced the tumorous growth in vivo. As for the regulatory mechanism of hsa_circ_0119412 in HCC, this study confirmed that hsa_circ_0119412 could target miR-526b-5p to enhance STMN1 expression, thereby playing regulatory role in HCC progression ().
Mounting evidence implies that circRNAs take part in the multistep cancerous progressionCitation20. For instance, it was reported that circMRPS35 facilitates malignant phenotypes and chemoresistance in HCCCitation21. Xie et al. revealed that CircFAM13B accelerates the proliferation of HCC cells by sponging miR-212, thus upregulating E2F5 expressionCitation6. CircEPB41L2, another circRNA, exhibited tumor-suppressive role in HCC via sponging miR-590-5pCitation22. CircRNF180 was also found to be a tumor-suppressor in HCCCitation23. These studies suggest that different circRNAs play different roles in HCC progression. In this study, we found a circRNA hsa_circ_0119412 that was upregulated in HCC. According to the reports from the previous studies, hsa_circ_0119412 was proved to be elevated in gastric cancer, and silencing hsa_circ_0119412 could inhibit the malignancy of gastric cancer cell by targeting miR-1298-5p/ZBED3 axisCitation8. Lv et al.Citation24 discovered that hsa_circ_0119412 was also upregulated in cervical cancer, and it targets miR-217/AGR2 axis to facilitate the malignant phenotypes of cervical cancer cells. As for HCC, there is no study to reveal the function of hsa_circ_0119412. In this work, we for the first time observed the high expression of hsa_circ_0119412 in HCC, and hsa_circ_0119412 knockdown slowed down the HCC cell malignant progression both in vitro and in vivo, implying that it plays an oncogenic part in HCC. Moreover, we found that hsa_circ_0119412 could regulate miR-526b-5p/STMN1 axis in HCC, which was different from the previous studies in gastric cancer and cervical cancerCitation8,Citation24.
Numerous reports have illustrated that circRNAs can competitively sponge miRNAs to influence mRNA levels, which are then involved in carcinogenic or anti-cancer processesCitation25. Yu et al. suggested that circRNA UBAP2 can sponge miR-1294 to promote tumorigenesis in HCCCitation7. Another study found that circSP3 accelerated HCC growth by sponging miRNA-198Citation26. In this study, we used circInteractome database, luciferase and RIP assays to confirm that miR-526b-5p could be sponged by hsa_circ_0119412 in HCC. As for miR-526b-5p, its tumor-suppressive roles have been suggested in HCC. According to the study from Kong Q et al., miR-526b-5p was proved to be sponged by circUGGT2 and regulate its target gene RAB1A, thereby inhibiting HCC developmentCitation12. Another study from Liu Y et al., confirmed that miR-526b-5p interacting with circEIF3I and target gene HGF attenuates the HCC tumor growthCitation13. The previous studies indicated that there were multiple circRNAs and mRNAs in HCC to interact with miR-526b-5p. Here, our results proved a novel regulatory mechanism on miR-526b-5p in HCC, showing that hsa_circ_0119412 could sponge miR-526b-5p to regulate the target gene STMN1 of miR-526b-5p in HCC. Moreover, due to their targeting relationship, miR-526b-5p inhibition restored the suppressive effects of hsa_circ_0119412 or STMN1 silencing on HCC cell malignancy.
STMN1 was found to be abnormally expressed in tumors, resulting in the malignant phenotypes of several malignancies, including lung squamous cell carcinomaCitation14, pancreatic-Citation16, liver-Citation27, and breast-cancerCitation15. Therefore, STMN1 inhibition could be crucial target for cancer therapy. In HCC, STMN1 overexpression was proved to be correlated to early recurrence and unfavorable prognosis in HCC patientsCitation28. In vitro and in vivo, STMN1 knockdown could inhibit cell proliferation, migration, and tumor growthCitation18. Consistent with the previous studies, this study also found the upregulation of STMN1 in HCC, and STMN1 knockdown also inhibited the HCC cell proliferation and migration, and induce cell apoptosis. However, our study further explored the upstream regulators of STMN1 in HCC because no study did this. By TargetScan and luciferase assay, STMN1 was proved to be a target gene of miR-526b-5p. The results of this study revealed that hsa_circ_0119412 could sponge miR-526b-5p to enhance STMN1, thereby playing the postive role in HCC progression.
Despite the above-mentioned findings, the study still has certain limitations. Firstly, hsa_circ_0119412 exerts its roles via complicated competing endogenous RNA (ceRNA) network, and we only observed the significance of hsa_circ_0119412/miR-526b-5p/STMN1 axis. Therefore, several potential miRNAs and mRNAs targeted by hsa_circ_0119412 in HCC advancement should be investigated. Second, the findings should be validated in large clinical sample sizes to increase their credibility.
Conclusion
To summarize, this study for the first time confirms that hsa_circ_0119412 can regulate HCC progression by targeting miR-526b-5p/STMN1 axis. These results shed light on the mechanism of hsa_circ_0119412 for HCC and position it as a promising candidate for future research into HCC diagnosis and prognosis.
Authors’ contributions
SW and JLZ performed the experiments and data analysis. TT and HCZ conceived and designed the study. JH and XLK made the acquisition of data. SW and JLZ did the analysis and interpretation of data. All authors read and approved the manuscript.
Data availability of statement
All data generated or analyzed during this study are included in this article.
Consent to participate
Every patient signed a written informed consent form.
Ethics approval
The Ethics Committee of the First Affiliated Hospital of Chengdu Medical College (Chengdu, China) has approved the present study. Clinical tissue samples were processed in strict accordance with the ethical standards of the Declaration of Helsinki. All of the patients provided written informed consent.
The animal experiments in the current work were conducted in compliance with the ARRIVE guidelines and was permitted by the Ethics Committee of the First Affiliated Hospital of Chengdu Medical College.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
Notes on contributors
Song Wu
Song Wu, received Master degree from Southwest Medical University in 2012. Now, he's an attending at the First Affiliated Hospital of Chengdu Medical College. His research focuses on the pathogenesis of hepatobiliary, Pancreatic and vascular diseases.
Tao Tang
Tao Tang, received Master degree from Chongqing Medical University in 2014. Now, he's an attending at the First Affiliated Hospital of Chengdu Medical College. His research focus on the Mechanism of angiogenesis in hepatocellular carcinoma.
Hongchi Zhou
Hongchi Zhou, received bachelor's degree from the Chengdu Medical College in 2018. Now, he is a physician at the First Affiliated Hospital of Chengdu Medical College,His research focus on the mechanism of tumor neovascularization.
Jing Huang
Jing Huang, received Master degree from Chongqing Medical University in 2013. Now, he's an attending at the First Affiliated Hospital of Chengdu Medical College. His research focus on the Pathogenesis of liver cancer.
Xiaoliang Kang
Xiaoliang Kang, received Master degree from Qingdao University Medical College in 2017. Now, he's a physician in the First Affiliated Hospital of Chengdu Medical College. His research focus on the Mechanism of liver cancer recurrence.
Junli Zhang
Junli Zhang, received Master degree from the Chengdu Medical College in 2016. Now, she is a physician at the First Affiliated Hospital of Chengdu Medical College.her research focus on Pathophysiology of liver cancer recurrence.
References
- Erratum: global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2020;70(4):313. doi:10.3322/caac.21609
- Anwanwan D, Singh SK, Singh S, Saikam V, Singh R. Challenges in liver cancer and possible treatment approaches. Biochimica Et Biophysica Acta (BBA) - Reviews On Cancer. 2020;1873(1):188314. doi:10.1016/j.bbcan.2019.188314.
- He Y, Lu M, Che J, Chu Q, Zhang P, Chen Y. Biomarkers and future perspectives for hepatocellular carcinoma immunotherapy. Front Oncol. 2021;11:716844. doi:10.3389/fonc.2021.716844.
- Shen H, Liu B, Xu J, Zhang B, Wang Y, Shi L, Cai X. Circular RNAs: characteristics, biogenesis, mechanisms and functions in liver cancer. J Hematol Oncol. 2021;14:134. doi:10.1186/s13045-021-01145-8.
- Feng N, Guo Z, Wu X, Tian Y, Li Y, Geng Y, Yu Y. Circ_PIP5K1A regulates cisplatin resistance and malignant progression in non-small cell lung cancer cells and xenograft murine model via depending on miR-493-5p/ROCK1 axis. Respir Res. 2021;22:248. doi:10.1186/s12931-021-01840-7.
- Xie Y, Hang X, Xu W, Gu J, Zhang Y, Wang J, Zhang X, Cao X, Zhan J, Wang J, et al. CircFAM13B promotes the proliferation of hepatocellular carcinoma by sponging miR-212, upregulating E2F5 expression and activating the P53 pathway. Cancer Cell Int. 2021;21(1):410. doi:10.1186/s12935-021-02120-6.
- Yu MC, Ding GY, Ma P, Chen YD, Zhu XD, Cai JB, Shen Y-H, Zhou J, Fan J, Sun H-C, et al. CircRNA UBAP2 serves as a sponge of miR-1294 to increase tumorigenesis in hepatocellular carcinoma through regulating c-myc expression. Carcinogenesis. 2021;42(10):1293–11. doi:10.1093/carcin/bgab068.
- Huang T, Wang Y, Li M, Wang W, Qi Z, Li J. Circular RNA hsa_circ_0119412 contributes to tumorigenesis of gastric cancer via the regulation of the miR-1298-5p/zinc finger BED-type containing 3 (ZBED3) axis. Bioengineered. 2022;13(3):5827–5842. doi:10.1080/21655979.2022.2036406.
- Zhong Y, Du Y, Yang X, Mo Y, Fan C, Xiong F, Ren D, Ye X, Li C, Wang Y, et al. Circular RNAs function as ceRnas to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8.
- Liu W, Wang D, Wang X, Liu P, Yan M. hsa_circ_0085539 promotes osteosarcoma progression by regulating miR-526b-5p and SERP1. Mol Ther Oncolytics. 2020;19:163–177. doi:10.1016/j.omto.2020.09.009.
- Zhao N, Hu L, Chen H. Circ_0002762 accelerates glycolysis metabolism to promote cervical cancer progression via the miR-526b-5p/HK2 axis. Gynecol Obstet Invest. 2022;87(6):352–363. doi:10.1159/000526700.
- Kong Q, Fan Q, Ma X, Li J, Ma R. CircRNA circUGGT2 contributes to hepatocellular carcinoma development via regulation of the miR-526b-5p/RAB1A axis. Cancer Manag Res. 2020;12:10229–10241. doi:10.2147/CMAR.S263985.
- Liu Y, Xiao X, Wang J, Wang Y, Yu Y. Silencing CircEIF3I/miR-526b-5p axis epigenetically targets HGF/c-met signal to hinder the malignant growth, metastasis and angiogenesis of hepatocellular carcinoma. Biochem Genet. 2023;61(1):48–68. doi:10.1007/s10528-022-10239-y.
- Bao P, Yokobori T, Altan B, Iijima M, Azuma Y, Onozato R, Yajima T, Watanabe A, Mogi A, Shimizu K, et al. High STMN1 expression is associated with cancer progression and chemo-resistance in lung squamous cell carcinoma. Ann Surg Oncol. 2017;24(13):4017–4024. doi:10.1245/s10434-017-6083-0.
- Obayashi S, Horiguchi J, Higuchi T, Katayama A, Handa T, Altan B, Bai T, Bao P, Bao H, Yokobori T, et al. Stathmin1 expression is associated with aggressive phenotypes and cancer stem cell marker expression in breast cancer patients. Int J Oncol. 2017;51(3):781–790. doi:10.3892/ijo.2017.4085.
- Watanabe A, Araki K, Yokobori T, Altan B, Ishii N, Tsukagoshi M, Kubo N, Saito F, Suzuki H, Kuwano H, et al. Stathmin 1 promotes the proliferation and malignant transformation of pancreatic intraductal papillary mucinous neoplasms. Oncol Lett. 2017;13(3):1783–1788. doi:10.3892/ol.2017.5603.
- Liu X, Xie S, Zhang J, Kang Y. Long noncoding RNA XIST contributes to cervical cancer development through targeting miR-889-3p/SIX1 axis. Cancer Biother Radiopharm. 2020;35(9):640–649. doi:10.1089/cbr.2019.3318.
- Zhang R, Gao X, Zuo J, Hu B, Yang J, Zhao J, Chen J. STMN1 upregulation mediates hepatocellular carcinoma and hepatic stellate cell crosstalk to aggravate cancer by triggering the MET pathway. Cancer Sci. 2020;111:406–417. doi:10.1111/cas.14262.
- Miao L, Zhang Z, Ren Z, Li Y. Application of immunotherapy in hepatocellular carcinoma. Front Oncol. 2021;11:699060. doi:10.3389/fonc.2021.699060.
- Huang Z, Xia H, Liu S, Zhao X, He R, Wang Z, Shi W, Chen W, Kang P, Su Z, et al. The mechanism and clinical significance of Circular RNAs in hepatocellular carcinoma. Front Oncol. 2021;11:714665. doi:10.3389/fonc.2021.714665.
- Li P, Song R, Yin F, Liu M, Liu H, Ma S, Jia X, Lu X, Zhong Y, Yu L, et al. circMRPS35 promotes malignant progression and cisplatin resistance in hepatocellular carcinoma. Mol Ther. 2022;30(1):431–447. doi:10.1016/j.ymthe.2021.08.027.
- Cao H, Quan S, Zhang L, Chen Y, Jiao G. BMPR2 expression level is correlated with low immune infiltration and predicts metastasis and poor survival in osteosarcoma. Oncol Lett. 2021;21(5):391. doi:10.3892/ol.2021.12652.
- Zeng X, Tan C, Mo M, Qin X, Ma X, Huang K, Wang X, Liang W, Yang L. CircRNA profiling identifies circRNF180 as a tumor suppressor in hepatocellular carcinoma. Epigenomics. 2021;13:513–530. doi:10.2217/epi-2020-0385.
- Lv Y, Wang M, Chen M, Wang D, Luo M, Zeng Q. hsa_circ_0119412 overexpression promotes cervical cancer progression by targeting miR-217 to upregulate anterior gradient 2. J Clin Lab Anal. 2022;36(4):e24236. doi:10.1002/jcla.24236.
- Li K, Yao T, Zhang Y, Li W, Wang Z. NEAT1 as a competing endogenous RNA in tumorigenesis of various cancers: role, mechanism and therapeutic potential. Int J Biol Sci. 2021;17(13):3428–3440. doi:10.7150/ijbs.62728.
- Zhao X, Dong W, Luo G, Xie J, Liu J, Yu F. Silencing of hsa_circ_0009035 suppresses cervical cancer progression and enhances radiosensitivity through MicroRNA 889-3p-dependent regulation of HOXB7. Mol Cell Biol. 2021;41(6):e0063120. doi:10.1128/MCB.00631-20.
- Liu YP, Pan LL, Kong CC. Stathmin 1 promotes the progression of liver cancer through interacting with YAP1. Eur Rev Med Pharmacol Sci. 2020;24(13):7335–7344. doi:10.26355/eurrev_202007_21900.
- Hsieh SY, Huang SF, Yu MC, Yeh TS, Chen TC, Lin YJ, Chang C-J, Sung C-M, Lee Y-L, Hsu C-Y, et al. Stathmin1 overexpression associated with polyploidy, tumor-cell invasion, early recurrence, and poor prognosis in human hepatoma. Mol Carcinog. 2010;49(5):476–487. doi:10.1002/mc.20627.