ABSTRACT
Background
Bladder cancer is one of the most common malignant tumors of the urinary system, and its incidence is increasing worldwide. However, the underlying mechanisms that trigger migration, invasion and chemotherapy resistance are unclear.
Results
Bioinformatics analysis of bladder cancer cohort indicated that LINC00839 is deregulated in bladder cancer. LINC00839 was validated and highly expressed in bladder cancer patients and cell lines. In addition, LINC00839 induced the migration, invasion and Gemcitabine resistance of bladder cancer cells. We identified that the transcription factor EGR1 directly repressed LINC00839 and thereby suppressed the migration and invasion of bladder cancer cells. Furthermore, LINC00839 interacted with miR-142, which subsequently regulated the expression of SOX5, a well-studied oncogene and targeted by miR-142. In addition, EGR1 served as a suppressive transcription factor of SOX5. Therefore, EGR1 directly or indirectly regulates SOX5 via LINC00839/miR-142 axis. LINC00839 induced Gemcitabine resistance by promoting autophagy.
Conclusions
EGR1, LINC00839/miR-142 and SOX5 form a coherent feed-forward loop that modulates the migration, invasion and Gemcitabine resistance of bladder cancer.
Introduction
Bladder cancer is one of the most common malignant tumors of the urinary system, and its incidence is increasing worldwide.Citation1 Bladder cancer metastasis is a complex process that involves various steps, including invasion, migration, and colonization of cancer cells in distant organs.Citation2 However, the exact molecular mechanisms underlying the bladder cancer metastasis are still unclear. The high incidence of metastasis and poor prognosis of bladder cancer have prompted researchers to investigate the molecular mechanisms underlying its development and progression.
The long non-coding RNA (lncRNA) LINC00839 is located on human chromosome 10q11.21. The function of LINC00839 is largely unknown in bladder cancer.Citation3 To date, limited studies have demonstrated that LINC00839 deregulation is associated with progression of cancers, such as breast cancer,Citation4 hepatocellular carcinoma,Citation5 colorectal cancerCitation3 and lung cancer.Citation6 One previous study showed that LINC00839 may serve as a biomarker of bladder cancer,Citation7 but the underlying molecular mechanism of LINC00839 in the initiation and progression of bladder cancer remains elusive. Therefore, we aim to determine the expression profile and the function of LINC00839 in bladder cancer. LINC00839 can interact with different targeted miRNAs to promote the proliferation, migration and invasion of cancers. LINC00839 promotes the progression of colorectal cancer via activating NRF1.Citation3 In neuroblastoma, LINC00839 induces the tumor progression via targeting miR-454-3p and miR-338-3p.Citation8 In this study, we could show the regulation between LINC00839 and related miRNA in bladder cancer.
miR-142 is a microRNA (miRNA) that can either function as an oncogenic miRNA or a tumor suppressive miRNA.Citation9–12 High expression of miR-142 is associated with the poor prognosis of esophageal squamous cell carcinoma.Citation13 Elevated expression of miR-142 is observed in the tissues derived from colorectal cancer patients.Citation14 However, miR-142 contributes to the chemotherapy sensitivity in breast cancer treatment.Citation15,Citation16 The anti-tumor immunity is enhanced by miR-142 through regulating tumor cell PD-L1 expression.Citation17 Therefore, the function of miR-142 is uncertain, and we aim to clarify the role of miR-142 in bladder cancer in this study.
EGR1, as a transcription factor, displays either oncogenic effect or anti-tumor effect.Citation18 EGR1 promotes epithelial–mesenchymal transition (EMT) by inducing the expression of SNAI1 and SNAI2, which subsequently repress CDH1 expression.Citation19,Citation20 In addition, EGR1 induces hepatocyte growth factor that further increases SNAI1 expression and thereby results in enhanced metastasis.Citation21 However, up-regulation of EGR1 inhibited metastasis of head and neck squamous cell carcinoma by repressing SNAI1 and SNAI2.Citation22
In this study, we determined the expression and function of miR-142, EGR1, LINC00839 in bladder cancer. We characterized that these molecules are involved in bladder cancer progression and chemotherapy resistance. The results of this study could have important implications for the development of novel diagnostic and therapeutic strategies for bladder cancer.
Results
LINC00839 is deregulated in bladder cancer
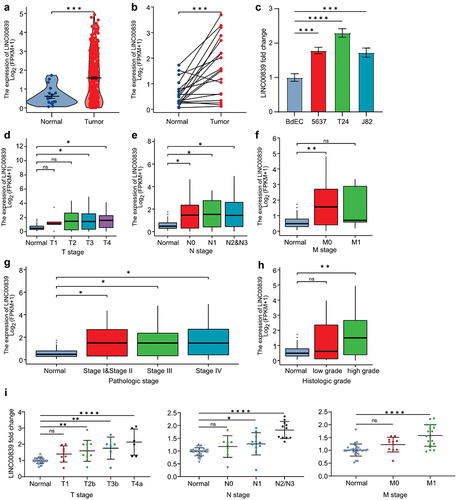
To determine the expression of LINC00839 in bladder cancer, we analyzed the RNA expression profile obtained from The Cancer Genome Atlas Urothelial Bladder Carcinoma (TCGA-BLCA) dataset. The expression of LINC00839 is highly expressed in bladder cancer tissues compared to the normal tissues in the unpaired and paired comparison (). In addition, we evaluated the expression of LINC00839 in three different cell lines derived from bladder cancer patients, including 5637, T24 and J82. All cell lines displayed an increased LINC00839 level compared to the epithelial cell of bladder (), which supports the results of bioinformatics analysis in TCGA-BLCA dataset. Furthermore, we investigated the association between LINC00839 expression and tumor stages. Elevated LINC00839 expression was positively associated with the advanced TNM stages of bladder cancer (). High expression of LINC00839 indicated advanced pathologic stages and histologic grades of bladder cancer (). Next, we performed q-PCR analysis on LINC00839 in tissues derived from 30 bladder cancer patients categorized into different TNM stages (Table S1). Indeed, the expression of LINC00839 was generally higher in the tissues of T1-T4a, N0-N3 and M0-M1 stages compared to the adjacent normal tissue. In addition, the expression of LINC00839 appeared to be significantly higher in the advanced TNM stages compared to early stages (), indicating that the expression of LINC00839 is positively correlated to the advanced bladder cancer stages. Taken together, LINC00839 is up-regulated in the bladder cancer, and the high expression of LINC00839 is associated with advanced TNM stages.
Figure 1. LINC00839 is deregulated in the bladder cancer.
LINC00839 promotes migration, invasion and Gemcitabine resistance of bladder cancer cells
Since we had identified that LINC00839 is deregulated in bladder cancer and previous studies have shown that LINC00839 induces metastasis of different cancers, here we asked whether LINC00839 promotes the migration and invasion in bladder cancer. Ectopic LINC00839 significantly induced the expression of Vimentin (VIM), whereas the expression of CDH1 was repressed by LINC00839 in T24 and J82 cells (). Silencing LINC00839 displayed an opposite effect on VIM and CDH1 compared to ectopic LINC00839 (). Therefore, LINC00839 promotes EMT in bladder cancer cells. Furthermore, ectopic LINC00839 promoted the migration and invasion capability of T24 cells (), whereas silencing LINC00839 restrained the migration and invasion capability of T24 cells (). The above results suggest that LINC00839 is a promoter of EMT, and thereby induces the migration and invasion of bladder cancer cells. In addition, ectopic LINC00839 induced the proliferation of T24 cells (). Conversely, silencing LINC00839 inhibited T24 cell proliferation ().
Figure 2. LINC00839 promotes the migration, invasion and EMT of bladder cancer cells.
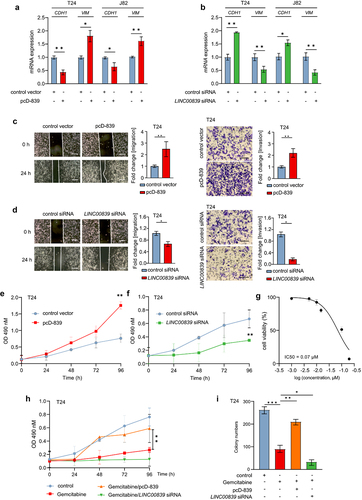
Next, we evaluated the effect of LINC00839 on the resistance of Gemcitabine, a frequently used chemotherapy drug in the treatment of bladder cancer patients. The half maximal inhibitory concentration (IC50) of Gemcitabine in T24 cells is 0.07 µM (), therefore we applied the Gemcitabine concentration of 0.1 µM in the following experiments. Ectopic LINC00839 significantly induced the proliferation rate of T24 cells treated with Gemcitabine. Conversely, silencing of LINC00839 repressed the proliferation rate and enhanced the effect of Gemcitabine on the proliferation inhibition (). Furthermore, ectopic LINC00839 induced the colony formation capability of T24 cells treated with Gemcitabine (). Therefore, LINC00839 functions as an oncogenic lncRNA, which promotes migration, invasion and Gemcitabine resistance in bladder cancer cells.
LINC00839 is a direct target of EGR1
Previous studies have shown that the deregulated lncRNAs are controlled by a myriad of transcription factors in cancer. Here we defined the upstream transcription factor of LINC00839. Interestingly, previous study indicated that EGR1 is a promising biomarker of bladder cancer. The high expression of EGR1 is associated with the progression of bladder cancer.Citation23 EGR1 also functions as a transcription factor and promotes the metastasis via directly inducing MMP1, SNAI1 and SNAI2.Citation19,Citation20 However, in certain cases, EGR1 displays tumor suppressive role by directly inducing the expression of tumor suppressive circRNA in bladder cancer.Citation24 Therefore, it is intriguing to determine the exact role of EGR1 in bladder cancer. Since we had shown that LINC00839 induced the migration and invasion of bladder cancer cells, we demonstrated the possible associations between EGR1 and LINC00839. By analyzing the ChIP-seq data obtained from Cistrome DB, EGR1 appeared to be a transcription factor of LINC00839. EGR1-occupancy was observed in the promoter region of LINC00839 (). The frequency of binding motif base information is shown in . The target between EGR1 and LINC00839 was validated by q-ChIP analysis (). Ectopic EGR1 repressed the expression of LINC00839 () in T24 and J82 cells. Therefore, EGR1 is a transcription factor that directly suppresses LINC00839 in bladder cancer cells.
Figure 3. LINC00839 is a direct target of EGR1.
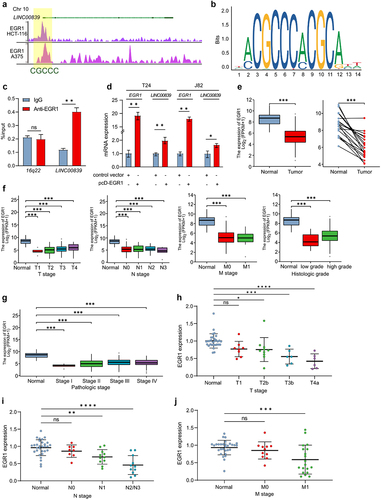
Since we had identified that the deregulation of LINC00839 is positively associated with the advanced stages of bladder cancer and EGR1 directly represses LINC00839, next we determined the function of EGR1 in bladder cancer. The analysis on TCGA-BLCA dataset showed that bladder cancer tissues tend to display a lower expression of EGR1 (). In addition, tissues of the advanced TNM stages of bladder cancer showed a suppressed EGR1 level compared to normal tissue (). The expression of EGR1 was negatively associated with the advanced pathologic stages () and high histologic score (). Indeed, qPCR analysis validated that the expression of EGR1 was generally lower in tissues of T1-T4a, N0-N3 and M0-M1 stages compared to the adjacent normal tissue derived from 30 bladder cancer patients (). In summary, our results suggest that EGR1 functions as tumor suppressor by inhibiting LINC00839 in bladder cancer.
EGR1 represses migration and invasion of bladder cancer cells
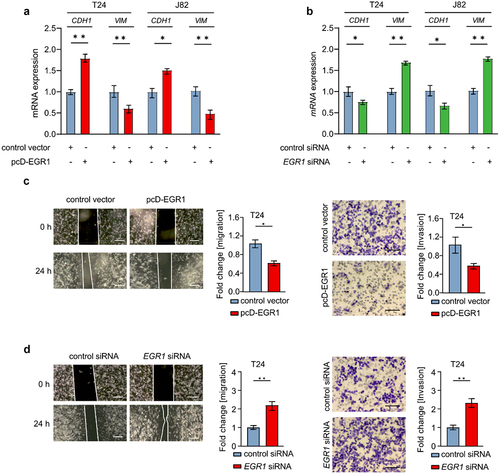
Since EGR1 is a tumor suppressor of bladder cancer, next we demonstrated the effect of EGR1 on EMT and migration/invasion in bladder cancer cells. Indeed, ectopic EGR1 repressed the expression of VIM and SNAI1 in T24 and J82 cells. Conversely, CDH1 was up-regulated following EGR1 overexpression (). Silencing EGR1 induced the VIM and SNAI1 expression, whereas the CDH1 expression was repressed in T24 and J82 cells (). Therefore, EGR1 inhibits EMT of bladder cancer cells. Furthermore, ectopic EGR1 repressed the migration and invasion ability of T24 cells (). The migration and invasion of T24 cells were promoted following EGR1 repression (). In summary, EGR1 suppresses EMT and migration/invasion of bladder cancer cells.
Figure 4. EGR1 represses migration and invasion of bladder cancer cells.
LINC00839 interacts with miR-142
Numerous studies have shown that lncRNAs interact with different miRNAs, which modulate downstream target genes. Therefore, we asked whether LINC00839 could modulate downstream genes via targeting specific miRNAs. Certain miRNAs can function biologically as oncogenes and tumor suppressor genes,Citation25 and miR-142 is frequently mentioned and analyzed in previous studies. Repressed miR-142 expression was observed in the serum of bladder cancer patients.Citation26 Interestingly, by screening in TargetScan database, LINC00839 appeared to be a target of miR-142. A seed match sequence of miR-142 was observed in the 3’-UTR of LINC00839 (). The target was validated by luciferase assay. Ectopic miR-142 decreased the luciferase activity of the reporter vector containing wild-type LINC00839 3’-UTR, whereas the reporter vector with mutation of miR-142 seed match sequence in the 3’-UTR of LINC00839 was refractory to miR-142 overexpression (). In addition, the expression of LINC00839 was repressed following miR-142 overexpression in T24 and J82 cells (). Interestingly, ectopic LINC00839 repressed the expression miR-142 in T24 and J82 cells (), indicating that LINC00839 interacts with miR-142 via the seed matching sequence in the 3’-UTR of LINC00839.
Figure 5. LINC00839 interacts with miR-142.
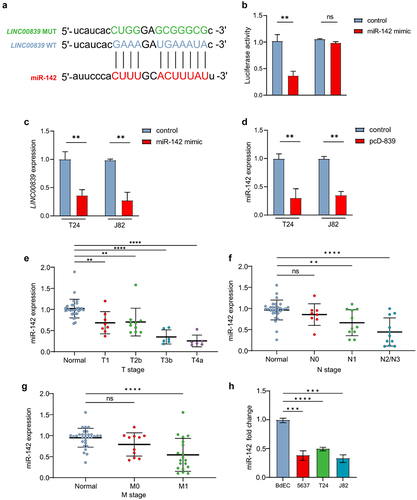
miR-142 has been characterized as tumor suppressive miRNA in cancers including bladder cancer. In line with previous studies, the expression of miR-142 was significantly lower in the cancer tissues derived from T2-T4a, N1-N3 and M0-M1 stages compared to the adjacent normal tissue derived from 30 bladder cancer patients compared to the normal adjacent tissues (). miR-142 was also down-regulated in three indicated bladder cancer cell lines compared to the epithelial cell of bladder (). Taken together, miR-142 is a tumor suppressive miRNA and interacts with LINC00839 through the seed match sequence in bladder cancer.
LINC00839/miR-142 axis modulates the expression of SOX5
Interestingly, previous studies have identified that the oncogene SOX5 is a miR-142 target.Citation27 Indeed, the target between miR-142 and SOX5 was validated by luciferase assay in bladder cancer cell lines (). In addition, ectopic miR-142 repressed the expression of SOX5 at mRNA and protein level (). However, overexpression of SOX5 failed to repress miR-142 in T24 cells (), indicating that SOX5 is located downstream of miR-142. Since we had identified that LINC00839 interacts with miR-142, here we evaluated the effect of LINC00839 on SOX5 expression in bladder cancer cells. Ectopic LINC00839 induced the expression of SOX5 in mRNA and protein level in T24 cells (). Interestingly, overexpression of miR-142 largely abrogated the effect of LINC00839 on SOX5 in T24 cells (). Therefore, LINC00839 induces the expression of SOX5 via repressing miR-142.
Figure 6. LINC00839/miR-142 axis modulates the expression of SOX5.
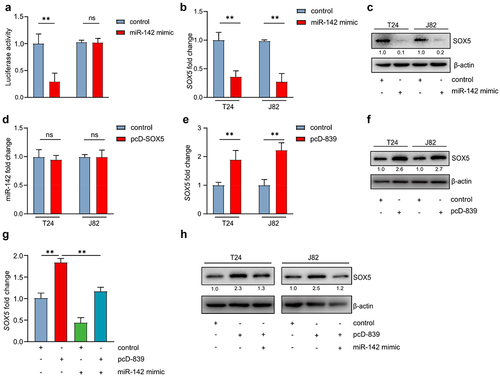
EGR1 suppresses SOX5 via a coherent feed-forward loop
Since EGR1 had been identified as a suppressive transcription factor of LINC00839, we deducted that EGR1 may repress the expression SOX5 via inhibiting LINC00839. Indeed, ectopic EGR1 significantly reduced the SOX5 mRNA and protein level in T24 and J82 cells (). Intriguingly, ectopic EGR1 kept SOX5 in a relatively low level in the condition of LINC00839 overexpression in T24 cells compared to the cells only with ectopic LINC00839 (). However, silencing LINC00839 only partly abrogated the effect of silencing EGR1 on SOX5 expression (), which indicates that an alternative route may also mediate the regulation between EGR1 and SOX5 independent of LINC00839. Since EGR1 serves as a transcription factor in bladder cancer cells, we analyzed the ChIP-seq data from Cistrome DB and found EGR1-occupancy in the SOX5 gene (). Therefore, SOX5 is a possible direct target of EGR1. Q-ChIP analysis validated the target between SOX5 and EGR1 (). The above results suggest that EGR1, LINC00839 and SOX5 form a coherent feed-forward loop, which confers a robust gene regulation.
Figure 7. EGR1 suppresses SOX5 via a coherent feed-forward loop.
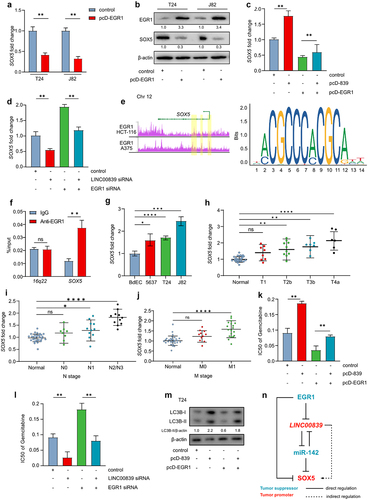
Furthermore, the expression of SOX5 was up-regulated in the bladder cancer cells () and in the bladder cancer tissues from the advanced TNM stages (). In addition, ectopic EGR1 sensitized T24 cells to the Gemcitabine treatment, which was represented by a lower IC50 of Gemcitabine. The effect of ectopic EGR1 was reversed by ectopic LINC00839 (). Silencing of EGR1 induced the Gemcitabine resistance in T24 cells, whereas silencing of LINC00839 largely abrogated the effect (). Previous studies have indicated that EGR1 can sensitize chemotherapy by preventing cyto-protective autophagy in ovarian cancer.Citation28 Here we evaluated the level of LC3B-II, an indicator of autophagy, in T24 cells. Indeed, ectopic LINC00839 induced autophagy evidenced by an enhanced turnover of endogenous LC3B-II. Overexpression of EGR1 prevented autophagy indicated by a repressed LC3B-II turnover. LINC00839 largely abrogated the effect of EGR1 on repressing autophagy (). Thereby LINC00839 may promote the Gemcitabine resistance at least via inducing autophagy.
Discussion
Here, we characterized the long non-coding RNA LINC00839 is an oncogenic lncRNA that promotes EMT, migration/invasion and Gemcitabine resistance of bladder cancer cells. In addition, LINC00839 is up-regulated in the bladder cancer tissues derived from bladder cancer patients. The expression of LINC00839 is positively associated with the advanced TNM stages and the high histologic scores of bladder cancer. Furthermore, we identified that the transcription factor EGR1, as a tumor suppressor, directly represses the expression of LINC00839 in bladder cancer cells. Interestingly, we screened that miR-142 commonly targets LINC00839 and a well-studied oncogene SOX5. Of note, the expression of miR-142 can only be repressed by the ectopic LINC00839, whereas SOX5 failed to repress miR-142, indicating that SOX5 is located downstream of LINC00839/miR-142 axis. Furthermore, EGR1 directly suppresses the expression of SOX5, and thereby EGR1 regulates SOX5 directly or indirectly. EGR1, LINC00839 and SOX5 form a coherent feed-forward loop in bladder cancer (summarized in ).
Previous studies have shown that LINC00839 promotes proliferation, migration, and invasion of different cancers.Citation3,Citation6,Citation29 In line with these results, ectopic LINC00839 induces migration and invasion of bladder cancer cells. By modulating the expression of EMT markers, LINC00839 is capable of controlling EMT in bladder cancer. Interestingly, LINC00839 activates PI3K/AKT pathway, and thereby contributes to the chemoresistance in breast cancer.Citation30 In line with this result, ectopic expression of LINC00839 contributed to the Gemcitabine resistance in bladder cancer cells in our study indicating that LINC00839 may serve as a promising biomarker for screening the patients with chemotherapy resistance. EGR1, as a tumor suppressor, tilts PI3K/AKT pathway and MAPK pathway.Citation31 Because EGR1 directly represses the expression LINC00839, EGR1 may suppress PI3K/AKT pathway via inhibiting LINC00839, but this hypothesis needs further experimental validations. In addition, LINC00839 is a direct target of c-MYC and mediates the oncogenic function of c-MYC in breast cancer.Citation30 EGR1 is a direct non-canonical c-MYC target, which requires ARF to be involved.Citation32 Therefore, c-MYC may either directly induce LINC00839 or indirectly repress LINC00839 via inducing EGR1.
The function of miR-142 remains unclear due to the contradictory effects in different cancers. miR-142 inhibits proliferation and promotes apoptosis of bladder cancer via targeting ZEB2 and TUG1.Citation33 However, miR-142 functions as an oncogene in prostate cancer by targeting FOXO1Citation34 and lncRNA MAGI2-AS3.Citation35 Here, we confirmed that miR-142 is a tumor suppressive miRNA in bladder cancer via targeting LINC00839 and the oncogene SOX5. The low-expression miR-142 in bladder cancer tissues supports the conclusion. Previous studies have indicated that EGR1 can sensitize chemotherapy by preventing cyto-protective autophagy in ovarian cancer.Citation28 miR-142 also enhances chemosensitivity of breast cancer cells by inhibiting autophagy.Citation16 In line with previous studies, EGR1 repressed autophagy via inhibiting LINC00839 in bladder cancer cells. Since EGR1 directly represses LINC00839, miR-142 may serve as a down-stream regulator of EGR1 in autophagy, and thereby mediates the function of EGR1/LINC00839 axis in the chemotherapy resistance in bladder cancer. The function of EGR1/LINC00839/miR-142 axis in autophagy-associated chemotherapy resistance needs further experiments to validate.
Furthermore, we validated that the transcription factor EGR1 directly represses the expression of SOX5, indicating that EGR1 can regulate SOX5 via at least two approaches: 1. EGR1 represses SOX5 via repressing LINC00839, which results in an elevated level of miR-142; 2. EGR1 directly represses SOX5. The two different ways confer a robust regulation between EGR1 and SOX5 in bladder cancer. Taken together, SOX5 is located very down stream of EGR1/LINC00839/miR-142 axis and mediates the function of this axis in bladder cancer.
Gemcitabine is widely used in non-muscle-invasive bladder cancer via intravesical therapy.Citation36 EGR1 was observed up-regulated in bone marrow after receiving biotherapy in bladder cancer mice model.Citation37 Therefore, the EGR1/LINC00839/miR-142 axis may serve as a therapeutic target to reduce Gemcitabine resistance in intravesical therapy. For bladder cancer of advance stages, the combined intravenous injection of Gemcitabine with other chemotherapy drugs is frequently used.Citation38 However, we are still unclear about the differences of the resistant mechanisms underlying the two ways of Gemcitabine administration. Further in vivo studies, therefore, are warranted. Since we identified LINC00839 and miR-142 are differentially expressed in bladder cancer, both of them may serve as biomarkers in screening bladder cancer and indicate the Gemcitabine resistance.
Conclusions
LINC00839 is an oncogenic lncRNA promoting migration, invasion and Gemcitabine resistance of bladder cancer. The tumor suppressive miR-142 commonly targets LINC00839 and SOX5. In addition, the transcription factor EGR1 directly represses LINC00839 and SOX5. Therefore, EGR1, LINC00839 and SOX5 form a coherent feed-forward loop which modulates the migration, invasion and Gemcitabine resistance of bladder cancer.
Methods
Cell cultures
Human bladder cancer cell lines 5637, T24 and J82 were obtained from the American Type Culture Collection (ATCC) and maintained in RPMI-1640 supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin, and 1% L-glutamine. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2.
Patient samples
To determine the gene expressions in tissues of bladder cancer patients, we collected bladder cancer tissues of different TNM stages from total 30 patients. The detailed patient information of TNM stages is listed in Table S1. These involved tissue sections were collected from Feb. 2019 to Nov. 2021 in The First Affiliated Hospital of Jiamusi University. The Ethics Committee of Jiamusi University approved the experiments performed with human samples (No. 202326).
Plasmid
The ectopic expression of EGR1 and LINC00839 was achieved by transfected cells with overexpression vector containing CDS region of EGR1 and LINC00839 mRNA. The CDS of EGR1 and LINC00839 was obtained by PCR with the cDNA template collected from the epithelial cell of bladder. Subsequently, the EGR1 CDS, SOX5 CDS and LINC00839 were inserted into pcDNA3.1 vector, thereafter named as pcD-EGR1, pcD-SOX5 and pcD-839, respectively. The inserted sequences were confirmed by Sanger-sequencing. The construction of ectopic vectors was performed by OBiO (Shanghai, China).
Transfection
Cells were transfected with siRNA or vectors using Lipofectamine 3000 (Invitrogen, USA) according to the manufacturer’s instructions. Briefly, 25 nM of siRNA or 2 µg of vector was mixed with 10 µl of Lipofectamine 3000 in 300 µl of Opti-MEM and added to the cells in a 6-well plate. After 6 hours of incubation, the transfection medium was replaced with fresh medium, and cells were subsequently incubated for 48 hours before following experiments. To repress the expression of LINC00839 and EGR1, LINC00839 or EGR1 siRNA pools containing four siRNAs (5 nmol, Qiagen, Germany) were used.
RNA isolation and quantitative PCR
Total RNA was extracted from the cells seeded in 6-well plate by using 1 ml of TRIzol reagent per well. Cellular RNA isolation was performed as previously described.Citation39 For tissues, RNA was isolated by using RNeasy Mini Kit (Qiagen, Germany) following the handbook. Thirty-milligram tissues were supplied for disruption and homogenization. cDNA was synthesized from 1 μg of RNA using the High-Capacity cDNA Reverse Transcription Kit (Roche). qPCR was performed using SYBR Green PCR Master Mix (Invitrogen, USA) on a StepOnePlus Real-Time PCR System (Invitrogen, USA). The relative expression of genes was calculated by using 2(-ΔΔCT) method and normalized to the expression of GAPDH. The primers used for qPCR are listed in Table S2.
Western blot analysis
Cells were lysed in RIPA buffer supplemented with protease and phosphatase inhibitors. Protein concentration was determined using the BCA protein assay kit. Equal amounts of protein were separated on SDS-PAGE and transferred onto nitrocellulose membranes. Membranes were incubated with primary antibodies EGR1 (Cell Signaling Technology, 4153, diluted in 5% skim milk at 1:1000), SOX5 (Novus, AF5268, diluted in 5% skim milk at 1:1000), LC3B (Abcam, ab192890, diluted in 5% skim milk at 1:1000) and β-actin (Invitrogen, MA1–140, diluted in 5% skim milk at 1:10000) overnight at 4°C, followed by incubation with secondary antibodies conjugated to horseradish peroxidase. Proteins were visualized using enhanced chemo-luminescence. (Thermo Fisher, USA). The relative protein expression was quantified by using Quantity One software, and the result was normalized to the control group and β-actin.
Wound-healing assay
Cells were seeded in 6-well plates and allowed to grow to the confluence over 95%. Before scratching, 10 µg/ml of mitomycin C was added to plates and incubated for 1 hour. A scratch was generated by using a 10 μl pipette tip, and the cell debris was washed away with pre-warmed PBS twice. Images of the scratch were taken at 0 and 24 hours using an inverted microscope.
Transwell assay
After different treatments, 2 × 10Citation5 cells were seeded in the upper chamber of a transwell plate with an 8-μm pore size membrane pre-coated with matrigel (Corning, USA). The lower chamber was filled with medium containing 10% FBS. After 24 hours of incubation, cells that had migrated to the lower chamber were fixed with 4% paraformaldehyde for 30 minutes and stained with 0.1% of crystal violet for 20 minutes. Images were taken using an inverted microscope. Cell numbers were counted by Image J software.
Q-ChIP analysis
Cells were crosslinked with formaldehyde, and chromatin was extracted and sheared by sonication. Immunoprecipitation was performed using EGR1 antibody against the protein. DNA was purified and analyzed by qPCR using primers specific for the promoter region of target genes. The primers used in q-ChIP analysis are listed in Table S3.
Luciferase assay
Cells were transfected with miR-142 mimic, pGL3 reporter plasmid containing 3’-UTR or mutant 3’-UTR and internal control vector by using Lipofectamine 3000. After 48 hours of incubation, cells were lysed in reporter lysis buffer (Promega, USA) and luciferase activity was measured using the Luciferase Assay System.
Bioinformatics analysis
The differential expression of LINC00839 and EGR1 was analyzed from the TCGA-BLCA patient datasets downloaded from https://www.cancer.gov/ccg/research/genome-sequencing/tcga. The ChIP-seq data were analyzed from the Cistrome DB (http://cistrome.org/db/#/). The target between miR-142 and downstream genes was screened by using ENCORI and TargetScan database.
Statistics
All of the data were analyzed by GraphPad Prism v.9.0. Differences between the indicated groups were calculated using GraphPad Prism Student’s t-test (two groups) and ANOVA test (more than two groups). For multiple group comparisons in patient cohorts, we performed Welch’s t-test. p < .05 was considered statistically significant.
List of abbreviations
| EMT | = | epithelial-mesenchymal transition |
| UTR | = | Untranslational region |
| PCR | = | Polymerase chain reaction |
| ATCC | = | American Type Culture Collection |
| VIM | = | Vimentin |
| BLCA | = | Urothelial Bladder Carcinoma |
| TCGA | = | The Cancer Genome Atlas |
| q-ChIP | = | Quantitative-chromosomal immunoprecipitation |
Authors’ contributions
Data curation, ZW and BW; formal analysis, ZW; project administration and supervision, SM; validation, ZW; writing – original draft, ZW and SM; writing – review & editing, ZW and SM. All authors read and approved the final manuscript.
Availability of data and materials
The authors confirm that the data supporting the findings of this study are available within the article and its supplementary materials.
Ethics approval and consent to participate
The Ethics Committee of Jiamusi University approved the experiments performed with human tissue samples (No. 202326).
Supplemental Material
Download MS Word (18.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2023.2270106
Additional information
Funding
Notes on contributors
Zunxian Wang
Zunxian Wang obtained a Bachelor degree in Clinical Medicine from Jiamusi University, and obtained her master degree in oncology from Harbin Medical University. She is now a doctoral student in Jiamusi University. She is currently working in the Department of Oncology of the First Affiliated Hospital of Jiamusi University.
Bo Wei
Bo Wei obtained Bachelor degree in Clinical Medicine from Jiamusi University, master degree from Harbin Medical University, and Doctoral degree from Jiamusi University. He is currently working in the Urology Department of the First Affiliated Hospital of Jiamusi University.
Shuxia Ma
Shuxia Ma obtained Bachelor degree in Medicine (in Russian) from Harbin Medical University, Master degree in Immunology from Jiamusi University Medical College, and Doctoral degree in Pathogeny Microbiology from Harbin Medical University. She is currently working at the Basic Medical School of Jiamusi University in China.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–13. doi:10.3322/caac.21492.
- Lee YC, Lam HM, Rosser C, Theodorescu D, Parks WC, Chan KS. The dynamic roles of the bladder tumour microenvironment. Nat Rev Urol. 2022;19(9):515–533. doi:10.1038/s41585-022-00608-y.
- Liu X, Chen J, Zhang S, Liu X, Long X, Lan J, Zhou M, Zheng L, Zhou J. LINC00839 promotes colorectal cancer progression by recruiting RUVBL1/Tip60 complexes to activate NRF1. EMBO Rep. 2022;23(9):e54128. doi:10.15252/embr.202154128.
- Vishnubalaji R, Shaath H, Elkord E, Alajez NM. Long non-coding RNA (lncRNA) transcriptional landscape in breast cancer identifies LINC01614 as non-favorable prognostic biomarker regulated by TGFβ and focal adhesion kinase (FAK) signaling. Cell Death Discov. 2019;5(1):109. doi:10.1038/s41420-019-0190-6.
- Zhou X, Chang Y, Zhu L, Shen C, Qian J, Chang R. LINC00839/miR-144-3p/WTAP (WT1 associated protein) axis is involved in regulating hepatocellular carcinoma progression. Bioengineered. 2021;12(2):10849–10861. doi:10.1080/21655979.2021.1990578.
- Yu X, Jiang Y, Hu X, Ge X. LINC00839/miR-519d-3p/JMJD6 axis modulated cell viability, apoptosis, migration and invasiveness of lung cancer cells. Folia Histochem Cytobiol. 2021;59(4):271–281. doi:10.5603/FHC.a2021.0022.
- Zhang S, Cao H, Ye L, Wen X, Wang S, Zheng W, Zhang Y, Huang D, Gao Y, Liu H, et al. Cancer-associated methylated lncRnas in patients with bladder cancer. Am J Transl Res. 2019;11(6):3790–3800.
- Zhang Q, Wei J, Li N, Liu B. LINC00839 promotes neuroblastoma progression by sponging miR-454-3p to up-regulate NEUROD1. Neurochem Res. 2022;47(8):2278–2293. doi:10.1007/s11064-022-03613-0.
- Pahlavan Y, Mohammadi Nasr M, Dalir Abdolahinia E, Pirdel Z, Razi Soofiyani S, Siahpoush S, Nejati K. Prominent roles of microRNA-142 in cancer. Pathol Res Pract. 2020;216(11):153220. doi:10.1016/j.prp.2020.153220.
- Li Y, Chen D, Jin LU, Liu J, Li Y, Su Z, QI Z, SHI M, JIANG Z, YANG S, et al. Oncogenic microRNA-142-3p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Oncol Lett. 2016;11(2):1235–1241. doi:10.3892/ol.2015.4021.
- Lv M, Zhang X, Jia H, Li D, Zhang B, Zhang H, Hong M, Jiang T, Jiang Q, Lu J, et al. An oncogenic role of miR-142-3p in human T-cell acute lymphoblastic leukemia (T-ALL) by targeting glucocorticoid receptor-α and cAMP/PKA pathways. Leukemia. 2012;26(4):769–777. doi:10.1038/leu.2011.273.
- Xiao P, Liu WL. MiR-142-3p functions as a potential tumor suppressor directly targeting HMGB1 in non-small-cell lung carcinoma. Int J Clin Exp Pathol. 2015;8(9):10800–10807.
- Lin RJ, Xiao DW, Liao LD, Chen T, Xie ZF, Huang WZ, Wang W-S, Jiang T-F, Wu B-L, Li E-M, et al. MiR-142-3p as a potential prognostic biomarker for esophageal squamous cell carcinoma. J Surg Oncol. 2012;105(2):175–182. doi:10.1002/jso.22066.
- Islam F, Gopalan V, Vider J, Lu CT, Lam AK. MiR-142-5p act as an oncogenic microRNA in colorectal cancer: clinicopathological and functional insights. Exp Mol Pathol. 2018;104(1):98–107. doi:10.1016/j.yexmp.2018.01.006.
- Shi Y, Wang J, Tao S, Zhang S, Mao L, Shi X, Wang W, Cheng C, Shi Y, Yang Q, et al. miR-142-3p improves paclitaxel sensitivity in resistant breast cancer by inhibiting autophagy through the GNB2-AKT-mTOR pathway. Cell Signal. 2023;103:110566. doi:10.1016/j.cellsig.2022.110566.
- Liang L, Fu J, Wang S, Cen H, Zhang L, Mandukhail SR, Du L, Wu Q, Zhang P, Yu X, et al. MiR-142-3p enhances chemosensitivity of breast cancer cells and inhibits autophagy by targeting HMGB1. Acta Pharm Sin B. 2020;10(6):1036–1046. doi:10.1016/j.apsb.2019.11.009.
- Jia L, Xi Q, Wang H, Zhang Z, Liu H, Cheng Y, Guo X, Zhang J, Zhang Q, Zhang L, et al. miR-142-5p regulates tumor cell PD-L1 expression and enhances anti-tumor immunity. Biochem Biophys Res Commun. 2017;488(2):425–431. doi:10.1016/j.bbrc.2017.05.074.
- Pagel JI, Deindl E. Early growth response 1--a transcription factor in the crossfire of signal transduction cascades. Indian J Biochem Biophys. 2011;48(4):226–235.
- Vetter G, Le Bechec A, Muller J, Muller A, Moes M, Yatskou M, Al Tanoury Z, Poch O, Vallar L, Friederich E, et al. Time-resolved analysis of transcriptional events during SNAI1-triggered epithelial to mesenchymal transition. Biochem Biophys Res Commun. 2009;385(4):485–491. doi:10.1016/j.bbrc.2009.05.025.
- Wang Y, Qin C, Zhao B, Li Z, Li T, Yang X, Zhao Y, Wang W. EGR1 induces EMT in pancreatic cancer via a P300/SNAI2 pathway. J Transl Med. 2023;21(1):201. doi:10.1186/s12967-023-04043-4.
- Grotegut S, von Schweinitz D, Christofori G, Lehembre F. Hepatocyte growth factor induces cell scattering through MAPK/Egr-1-mediated upregulation of Snail. EMBO J. 2006;25(15):3534–3545. doi:10.1038/sj.emboj.7601213.
- Worden B, Yang XP, Lee TL, Bagain L, Yeh NT, Cohen JG, Van Waes C, Chen Z. Hepatocyte growth factor/scatter factor differentially regulates expression of proangiogenic factors through Egr-1 in head and neck squamous cell carcinoma. Cancer Res. 2005;65(16):7071–7080. doi:10.1158/0008-5472.CAN-04-0989.
- Egerod FL, Bartels A, Fristrup N, Borre M, Orntoft TF, Oleksiewicz MB, Brünner N, Dyrskjøt L. High frequency of tumor cells with nuclear Egr-1 protein expression in human bladder cancer is associated with disease progression. BMC Cancer. 2009;9(1):385. doi:10.1186/1471-2407-9-385.
- Ren L, Jiang M, Xue D, Wang H, Lu Z, Ding L, Xie H, Wang R, Luo W, Xu L, et al. Nitroxoline suppresses metastasis in bladder cancer via EGR1/circNDRG1/miR-520h/smad7/EMT signaling pathway. Int J Biol Sci. 2022;18(13):5207–5220. doi:10.7150/ijbs.69373.
- Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y, Khanin R. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PloS One. 2010;5(9):e13067. doi:10.1371/journal.pone.0013067.
- Yu Z, Lu C, Lai Y. A serum miRnas signature for early diagnosis of bladder cancer. Ann Med. 2023;55(1):736–745. doi:10.1080/07853890.2023.2172206.
- Hu XH, Dai J, Shang HL, Zhao ZX, Hao YD. SP1-mediated upregulation of lncRNA ILF3-AS1 functions a ceRNA for miR-212 to contribute to osteosarcoma progression via modulation of SOX5. Biochem Biophys Res Commun. 2019;511(3):510–517. doi:10.1016/j.bbrc.2019.02.110.
- He J, Yu JJ, Xu Q, Wang L, Zheng JZ, Liu LZ, Jiang B-H. Downregulation of ATG14 by EGR1-MIR152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy. 2015;11(2):373–384. doi:10.1080/15548627.2015.1009781.
- An JX, Ma ZS, Yu WJ, Xie BJ, Zhu FS, Zhou YX, Cao FL. LINC00839 promotes the progression of gastric cancer by sponging miR-1236-3p. Bull Exp Biol Med. 2022;173(1):81–86. doi:10.1007/s10517-022-05498-z.
- Chen Q, Shen H, Zhu X, Liu Y, Yang H, Chen H, Xiong S, Chi H, Xu W. A nuclear lncRNA Linc00839 as a Myc target to promote breast cancer chemoresistance via PI3K/AKT signaling pathway. Cancer Sci. 2020;111(9):3279–3291. doi:10.1111/cas.14555.
- Yu X, Shen N, Zhang ML, Pan FY, Wang C, Jia WP, Liu C, Gao Q, Gao X, Xue B, et al. Egr-1 decreases adipocyte insulin sensitivity by tilting PI3K/Akt and MAPK signal balance in mice. EMBO J. 2011;30(18):3754–3765. doi:10.1038/emboj.2011.277.
- Boone DN, Qi Y, Li Z, Hann SR. Egr1 mediates p53-independent c-Myc–induced apoptosis via a noncanonical ARF-dependent transcriptional mechanism. Proc Natl Acad Sci U S A. 2011;108(2):632–637. doi:10.1073/pnas.1008848108.
- Liu Q, Liu H, Cheng H, Li Y, Li X, Zhu C. Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. Onco Targets Ther. 2017;10:2461–2471. doi:10.2147/OTT.S124595.
- Tan YF, Chen ZY, Wang L, Wang M, Liu XH. MiR-142-3p functions as an oncogene in prostate cancer by targeting FOXO1. J Cancer. 2020;11(6):1614–1624. doi:10.7150/jca.41888.
- Hu R, Wu P, Liu J. LncRNA MAGI2-AS3 inhibits prostate cancer progression by targeting the miR-142-3p. Horm Metab Res. 2022;54(11):754–759. doi:10.1055/a-1891-6864.
- Shelley MD, Jones G, Cleves A, Wilt TJ, Mason MD, Kynaston HG. Intravesical gemcitabine therapy for non-muscle invasive bladder cancer (NMIBC): a systematic review. BJU Int. 2012;109(4):496–505. doi:10.1111/j.1464-410X.2011.10880.x.
- Tan QL, Zhou CY, Cheng L, Luo M, Liu CP, Xu WX, Zhang X, Zeng X. Immunotherapy of bacillus Calmette‑Gu?rin by targeting macrophages against bladder cancer in a NOD/scid IL2Rg‑/‑ mouse model. Mol Med Rep. 2020;22:362–370. doi:10.3892/mmr.2020.11090.
- Bellmunt J, Albiol S, Suarez C, Albanell J. Optimizing therapeutic strategies in advanced bladder cancer: update on chemotherapy and the role of targeted agents. Crit Rev Oncol Hematol. 2009;69(3):211–222. doi:10.1016/j.critrevonc.2008.06.002.
- Mannhalter C, Koizar D, Mitterbauer G. Evaluation of RNA isolation methods and reference genes for RT-PCR analyses of rare target RNA. Clin Chem Lab Med. 2000;38(2):171–177. doi:10.1515/CCLM.2000.026.
