ABSTRACT
Osteosarcoma is a malignant orthopedic tumor that is frequently diagnosed in the pediatric population. Several studies have summarized the functions of circular RNAs (circRNAs) in the progression of osteosarcoma. This study aimed to investigate a novel circRNA, hsa_circ_0020378 (circ_0020378), and elucidate its functions and underlying mechanisms during osteosarcoma progression. The expression levels of circ_0020378, miR-339-3p, and COL1A1 in osteosarcoma cells and tissues were determined using RT-qPCR or Western blotting. CCK8, transwell migration, colony formation, and xenograft experiments were performed to assess the malignancy of osteosarcoma cells. Luciferase and RNA immunoprecipitation (RIP) experiments were employed to validate the interactions of miR-339-3p with circ_0020378 and COL1A1 3′UTR. Osteosarcoma cells and tissues showed significant upregulation of circ_0020378 and COL1A1 and downregulation of miR-339-3p. Silencing circ_0020378 in osteosarcoma cells inhibited their proliferation, colony formation, and migration. The inhibitive influence of circ_0020378 silencing during osteosarcoma tumorigenesis in vitro was verified in vivo. Circ_0020378 sponged miR-339-3p which targeted COL1A1 3′UTR. Circ _0020378 silencing disrupted the tumor-promoting effect of the miR-339-3p inhibitor in osteosarcoma cells. Furthermore, miR-339-3p inhibitor attenuated the suppressive effect of COL1A1 downregulation on malignant osteosarcoma cell phenotypes. Circ_0020378 stimulates osteosarcoma progression by downregulating miR-339-3p/COL1A1 expression. These findings provide a theoretical basis for the discovery of novel osteosarcoma targets.
KEYWORDS:
Introduction
Osteosarcoma is a malignant bone disorder that largely originates from the metaphysis of long bone.Citation1 It is frequently diagnosed in adolescents and children and has an incidence of ~ 6–8 cases per 1,000,000.Citation1 In addition to the increasing incidence of osteosarcoma, its highly metastatic properties have resulted in an increasing morbidity burden.Citation2 Owing to obscure symptoms, distant metastasis is frequently observed at initial diagnosis in approximately 85% of patients, with a 5-year survival rate below 70%.Citation3 Therefore, it is critical to understand the mechanisms underlying the progression of osteosarcoma.
Circular RNAs (circRNAs) are a heterogeneous subgroup of non-coding RNAs with closed loop forms.Citation4 Similar to other non-coding RNAs, circRNAs have emerged as critical modulators of gene expression networks via transcriptional, epigenetic, and post-transcriptional mechanisms.Citation5 CircRNAs are involved in various biological processes associated with tumorigenesis and progression, including osteosarcoma.Citation6 For instance, CircECE1 facilitates metabolic remodeling and proliferation of osteosarcoma.Citation7 Elevated circRNA_0004674 expression impairs the sensitivity of osteosarcoma cells to chemotherapeutic drugs.Citation8 CircMYO10 serves as a chromatin remodeling factor that positively influences the β-catenin/TCF-dependent transcription which is an important promoter in the progression of various cancers, including osteosarcoma.Citation9 Therefore, it is necessary to adequately address the function of circRNAs in osteosarcoma. In this study, we identified hsa_circ_0020378 (circ_0020378) as a novel aberrantly expressed circRNA. However, its function in osteosarcoma has yet to be reported. Hence, this study aimed to elucidate the mechanisms and functions of osteosarcoma.
MicroRNAs (miRNAs) are transcribed RNAs approximately 20 nucleotides long. Their active engagement in the acquisition of hallmark capabilities that allow tumorigenesis and progression has been described in different cancers.Citation10 For example, a report showed that miRNA-206 targets Notch3 and impedes osteosarcoma outgrowth in vitro.Citation11 Zhu et al. reported the involvement of miR-200b-3p and miR-744-3p in acquired resistance to drugs against osteosarcoma.Citation12 MiR-339-3p has been well documented as a tumor suppressor in various cancers, such as osteosarcoma.Citation13–15 However, the regulatory mechanisms of circ_0020378 are not well understood.
The Collagen Type I Alpha 1 chain (COL1A1) gene consists of 51 exons and is located on chromosome 17q21.33. This gene encodes the pro-alpha 1 chains of type I collagen, which contains a triple helix composed of two alpha 1 chains and one alpha 2 chain. Its polymorphisms are associated with a predisposition to bone disorders, such as idiopathic osteoporosis, Caffey’s disease, and osteosarcoma.Citation16 Furthermore, increasing evidence has shown that COL1A1 is a hub gene that promotes tumor growth, as COL1A1 amplification is positively associated with immune infiltration and remodeling of the immune tumor microenvironment, enabling mesothelioma progression.Citation17 Elevated COL1A1 levels promote the survival of cisplatin-resistant non-small cell lung cancer (NSCLC) cells resistant to cisplatin.Citation18 However, the biofunction and mechanism of action of COL1A1 in osteosarcoma are yet to be elucidated.
In our preliminary assays, we discovered robust circ_0020378 expression in osteosarcoma tissues. Considering that it is predominantly situated within the cytoplasm of osteosarcoma cells, we further revealed that it constitutes a regulatory network of competing endogenous RNA (ceRNA). These results indicate a possible interaction between circ_0020378 and miR-339-3p to regulate COL1A1 expression during osteosarcoma progression. Our findings may have important implications for combating osteosarcoma.
Methods
Subjects and tissue preparation
From January 2020 to January 2022, we recruited a total of 18 patients diagnosed with osteosarcoma for the first time. At least three pathologists histopathologically confirmed the obtained osteosarcoma specimens as well as the matched normal bone tissues. The clinical specimens cryopreserved at −80°C. Ethical approval was obtained from the Huangshi Central Hospital and Affiliated Hospital of Hubei Polytechnic University (ethical approval number: zx2022017). Clinical tissue specimens were processed in strict compliance with the ethical standards of the Declaration of Helsinki. All the patients signed an informed consent form. shows patient characteristics.
Table 1. The characteristics of patients with osteosarcoma.
Cell culture and transfection
Normal osteoblasts (hFOB1.19) and osteosarcoma cells (HOS, Saos-2, and SW1353) were obtained from ProCell (China). SW1353 (Leibovitz’s L-15 Medium; Procell), Saos-2 (McCoy’s 5A; Procell), HOS (MEM; Procell), and hFOB1.19 (DMEM/F12; Procell) were maintained in media containing 1% penicillin-streptomycin (Procell) and 10% fetal bovine serum (FBS, Procell). Culture was performed under a 5% CO2 atmosphere at 37°C.
Small interfering RNAs (siRNAs) targeting circ_0020378 (si-circ_0020378), COL1A1 (si-COL1A1), miR-339-3p inhibitor and mimic, and their respective negative controls (si-NC, inhibitor NC, and mimic NC) were purchased from GenePharma, China. Recombinant lentiviruses (multiplicity of infection [MOI] = 50) carrying short hairpin RNAs (shRNAs) targeting either circ_0020378 (sh-circ) or sh-NC were purchased from GenePharma. Lipo3000 was used to introduce synthetic nucleotides (siRNAs, mimics, and inhibitors) into Saos-2 and HOS cells. After a 48-h transfection period, the efficacy of transfection was verified by RT-qPCR. For shRNA lentivirus infection, approximately 80% confluent Saos-2 cells were incubated with the virus particles (MOI = 50). After 14 days, the survivors were screened by adding 0.5 μg/mL puromycin. Positive monoclonal cells were expanded and verified using RT-qPCR.
RT-qPCR
Following the included protocol, total RNA was extracted with the aid of the TRIzol reagent (Invitrogen, USA). MLV reverse transcriptase (Thermo Fisher Scientific, USA) or the miRNA 1st Strand cDNA Synthesis Kit (Vazyme, China) were used to synthesize cDNA from the extracted RNA. The SYBR GreenER qPCR SuperMix Universal kit (Thermo Fisher Scientific, Inc.) was used for RT-qPCR, which was performed using an ABI Gene Amp PCR System 9700 (ABI, USA). The data were analyzed by means of the 2−∆∆Cq method, with U6 (miR-339-3p) and GAPDH (circ_0020378 and COL1A1) for normalization. The primers used are listed in .
Table 2. Real-time PCR primer sequences.
Nuclear and cytoplasmic separation and RNase R treatment assays
Using the Cytoplasmic & Nuclear RNA Purification Kit (Beyotime, China) and RT-qPCR, we determined the levels of nuclear and cytoplasmic circ_0020378 in HOS and Saos-2 cells. Furthermore, RT-qPCR was performed to assess circ_0020378 expression in Saos-2 and HOS cells after subjecting them to a 10-h RNase R treatment.
Assessment of cell proliferation
CCK8 kit (Beyotime, China) was used to monitor osteosarcoma cell proliferation. In 96-well culture plates, 3000 Saos-2 and HOS cells were seeded in each well. At 0 h, 24 h, 48 h, and 72 h post-cultivation, the cell media were supplied with 10 μL CCK-8 solution before being subjected to two more hours of incubation. The optical density (OD) was measured at 450 nm using a microplate reader.
Assessment of cell migration
To assess cell migration, 300 μL medium without FBS containing 5 × 10.Citation4 Saos-2 and HOS cells was transferred into Transwell insert, and then placed onto a new 12-well containing 500 μL medium with 10% FBS. This arrangement allowed cells to migrate to the underside of the inserts. After 48 h, migratory cells below the inserts were treated for 10 min with 4% paraformaldehyde. Subsequently, fixed cells were stained with 1% crystal violet for 20 min. Cells on the bottom of the filter were counted under a microscope (Nikon, Japan).
Colony formation assay
The 6-well plates were seeded with 1 × 10.Citation3 HOS or Saos-2 cells. After a 14-day incubation period, the cells were fixed with methanol for 30 min and subsequently treated with 0.2 crystal violet.
Xenograft assay
Six 5-week-old male BALB/c nude mice (~20 g each) were sourced from the Center for Animal Experiments of Wuhan University (China) and randomized into two groups (sh-circ or sh-NC). All mice were subjected to a 12:12 h light:dark photoperiod. Water and food were provided ad libitum. After one week of acclimatization, each mouse received a subcutaneous injection of 4 × 10.Citation6 Saos-2 cells carrying either sh-NC or sh-circ. During the 5-weeks of feeding, tumor size was monitored weekly. Finally, the mice were exposed to CO2 for euthanasia. Tumor xenografts were removed and weighed. The Animal Care and Use Committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University approved these animal experiments which were executed in strict compliance with the Declaration of Helsinki (ethical approval number: (动)202210023).
Luciferase assay
The wild-type (WT) circ_0020378 and COL1A1 3′UTR sequences that contained binding sites for miR-339-3p were inserted into reporter vectors to produce the circ_0020378-WT and COL1A1 3′UTR-WT luciferase reporter vectors, respectively. Circ_0020378-MUT and COL1A1 3′UTR-MUT, and their corresponding mutant (MUT) vectors were also constructed. The WT and MUT luciferase vectors were transfected into HOS and Saos-2 cells along with either an miR-339-3p mimic or a mimic NC. Forty-eight hours later, luciferase activity was measured using the Luciferase Dual Assay Kit (Promega, Madison Kit, Millipore, USA).
RNA immunoprecipitation (RIP) assay
The EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA) was used to detect the expression of circ_0020378 and miR-339-3p in the immunoprecipitated complexes. HOS and Saos-2 cells were exposed to the RIP lysis buffer. Magnetic beads with coatings of Ago2 or IgG antibodies were incubated with the resultant lysate at 4°C. Finally, circ_0020378 and miR-339-3p expression levels were assessed by RT-qPCR.
Western blot
A RIPA lysis buffer (Beyotime) was used to extract proteins from Saos-2 and HOS cells. The proteins were quantitatively assessed using the BCA kit (Beyotime). Proteins were loaded onto a 12% SDS-PAGE gel and ran at 150 V until the dye front reached the bottom of the gel. A PVDF membrane was used for SDS-PAGE membrane transfer and then blocked at room temperature for 30 min using 5% nonfat milk. After probing with anti-COL1A1 antibody (Cat#: ab216791, 1:1000, Abcam, USA) and anti-GAPDH antibody (Cat#: ab8245, 1:1000, Abcam, USA) overnight at 4°C, the membranes were soaked in an anti-mouse HRP secondary antibody (Cat#: 31430, 1:1000, Invitrogen, USA) for an hour at room temperature. Pierce ECL Western Blot Substrate (Pierce, USA) was used for staining before ImageJ analysis.
Statistical analysis
Data gathered from experiments, which were performed in triplicates, are expressed as mean ± SD (n = 3). Student’s t-test was used to compare data of two groups. Outcomes from multiple groups were analyzed using a one-way ANOVA with Tukey’s post-hoc test. Pearson’s correlation coefficient was used to analyze the correlation between miR-339-3p, COL1A1, and circ_0020378. Statistical significance was set at P < .05.
Results
High circ_0020378 expression in osteosarcoma
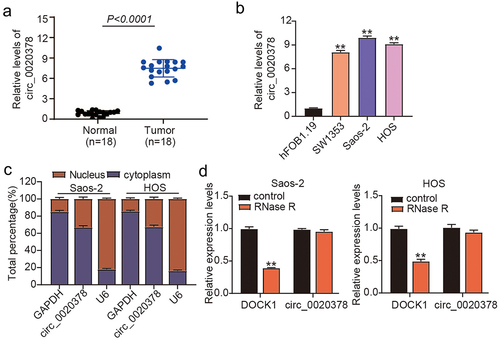
To decode the role of circ_0020378 in osteosarcoma progression, 10 osteosarcoma patients were recruited. RT-qPCR analysis was performed to determine its differential expression in osteosarcoma. Results revealed that cancer tissues had higher circ_0020378 levels than tumor-free tissues (). Consistently, osteosarcoma cells (SW1353, Saos-2, and HOS) exhibited robust expression of circ_0020378 relative to hFOB1.19 cells (). About a 9-fold increment in circ_0020378 was detected in HOS and Saos-2 cells; hence, these cells were selected for subsequent experiments. To delineate the characteristics of circ_0020378 in osteosarcoma cells, we determined its subcellular localization and circRNA structure. As shown in , cytoplasmic enrichment of circ_0020378 was detected in both HOS and Saos-2 cells. Furthermore, circ_0020378 was not sensitive to RNase R digestion (). Circ_0020378’s differential expression and specific trait interested us in uncovering its role in osteosarcoma.
Figure 1. High expression level of circ_0020378 in osteosarcoma.
Circ_0020378 silence elicits anti-tumor activity against osteosarcoma in vivo and in vitro
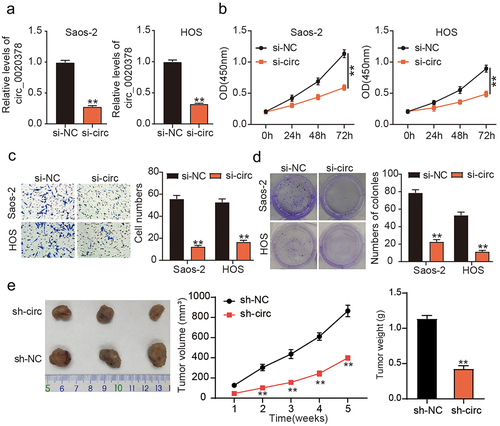
Considering the upregulation of circ_0020378 in osteosarcoma, circ_0020378 was silenced in HOS and Saos-2 cells. Forty-eight hours post-transfection, RT-qPCR analysis detected a significant decline in circ_0020378 expression in osteosarcoma cells transfected with si-circ (). CCK8 assay revealed that circ_0020378 silencing reduced the proliferative potential of HOS and Saos-2 cells (). Moreover, transwell migration assays confirmed the inhibitory effect of si-circ_0020378 on the migration of both osteosarcoma cell lines (). A reduced colony formation rate induced by circ_0020378 silencing was also observed in osteosarcoma cells (). To substantiate the function of circ_0020378 in osteosarcoma in vitro, Saos-2 cells harboring either sh-NC or sh-circ were injected subcutaneously into each mouse. After 35 days, the tumors were surgically dissected and weighed. As shown in , the volume and weight of the xenografts from the sh-circ group were noticeably reduced. These findings suggest that circ_0020378 performs a pro-tumor function during osteosarcoma tumorigenesis, both in vivo and in vitro.
Figure 2. circ_0020378 silence elicits anti-tumor activity to osteosarcoma in vitro and in vivo.
Circ_0020378 targets miR-339-3p
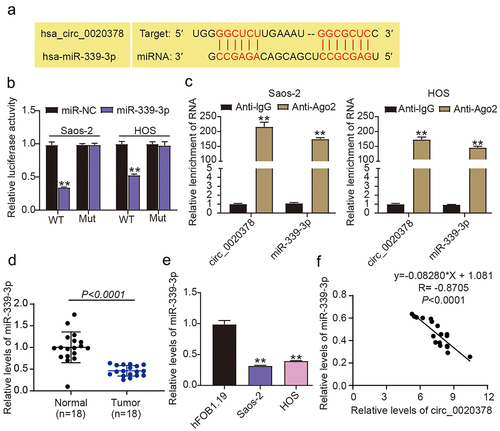
To delineate the machinery underlying the above-mentioned role of circ_0020378, we utilized circInteractome, and the prediction results revealed base pairings between miR-339-3p and circ_0020378 (). Subsequently, the luciferase reporter experiments validated that the miR-339-3p mimic inhibited circ_0020378-WT-driven luciferase activity, but failed to affect the luciferase activity driven by circ_0020378-MUT (). Through the anti-Ago2 assay, we discovered simultaneous immunoprecipitation of miR-339-3p and circ_0020378 in osteosarcoma cells, while no immunoprecipitation was observed in the anti-IgG RIP assay (). These results suggest that circ_0020378 preferentially binds to miR-339-3p in osteosarcoma cells. Since circ_0020378 targets miR-339-3p, we further analyzed miR-339-3p expression in osteosarcoma cells and tissues. RT-qPCR results revealed that osteosarcoma specimens exhibited poorer miR-339-3p expression than tumor-free tissues (). As anticipated, a constitutive miR-339-3p downregulation was also found in the osteosarcoma cells relative to hFOB1.19 cells (). Furthermore, the Pearson correlation coefficient showed that in osteosarcoma tissues, miR-339-3p was inversely correlated with circ_0020378 (). Therefore, circ_0020378 interacted directly with miR-339-3p and interfered with its availability.
Figure 3. Circ_0020378 targets miR-339-3p.
Circ_0020378 silencing disrupts the tumor-promoting influence of the miR-339-3p inhibitor on osteosarcoma cells
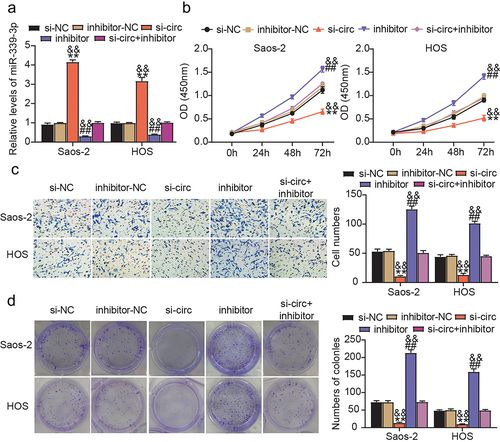
To define miR-339-3p’s biofunction in circ_0020378-mediated tumor promotion in osteosarcoma in vitro, we simultaneously introduced si-circ and the miR-339-3p inhibitor into Saos-2 and HOS cells. Forty-eight hours later, RT-qPCR was performed to assess the miR-339-3p levels. The data revealed that circ_0020378 silencing attenuated miR-339-3p inhibition in osteosarcoma cells resulting from miR-339-3p inhibitor transfection (). Functionally, treatment with the miR-339-3p inhibitor alone stimulated the proliferation of Saos-2 and HOS cells; however, this effect was offset by circ_0020378 silencing (). The increase in migration and rate of colony formation was also exhibited by the osteosarcoma cells after treatment with the miR-339-3p inhibitor (). Nevertheless, this effect was nearly nullified when si-circ and miR-339-3p inhibitors were co-introduced. Collectively, circ_0020378 is capable of diminishing miR-339-3p expression via competitive binding and can abrogate miR-339-3p-induced suppression of osteosarcoma cell proliferation and migration.
Figure 4. Circ_0020378 silence disrupts the tumor-promoting effect of miR-339-3p inhibitor in osteosarcoma cells.
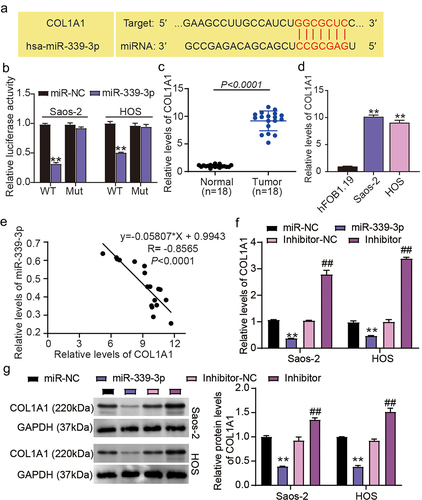
MiR-339-3p targets COL1A1
Generally, miRNA could recognize various mRNA 3′UTR and execute its function. To decipher miR-339-3p’s activity in osteosarcoma, TargetScan was used to predict the target mRNA. The COL1A1 mRNA was targeted by miR-339-3p (). To understand how miR-339-3p-mediated editing events impact COL1A1 expression, we constructed COL1A1 3′UTR MUT and WT luciferase vectors and simultaneously introduced them into HOS and Saos-2 cells. The assessment of luciferase intensity revealed that miR-339-3p repressed the COL1A1 3′UTR WT-driven transcriptional activity but exhibited no impact on the COL1A1 3′UTR MUT-driven one (). Interestingly, compared with normal tissues and osteoblast cells (hFOB1.19), osteosarcoma cells and tissues showed elevated levels of COL1A1 mRNA (). A negative correlation between COL1A1 and miR-339-3p was also detected in osteosarcoma specimens (). Moreover, RT-qPCR and western blot assays showed that upregulation of miR-339-3p led to a decrease in COL1A1 expression, whereas downregulation of miR-339-3p induced an increase in COL1A1 expression (). These indicate that miR-339-3p binds to COL1A1 3′UTR and negatively reshapes COL1A1 transcription.
Figure 5. MiR-339-3p targets COL1A1.
MiR-339-3p attenuates the malignant osteosarcoma cell phenotypes via the downregulation of COL1A1
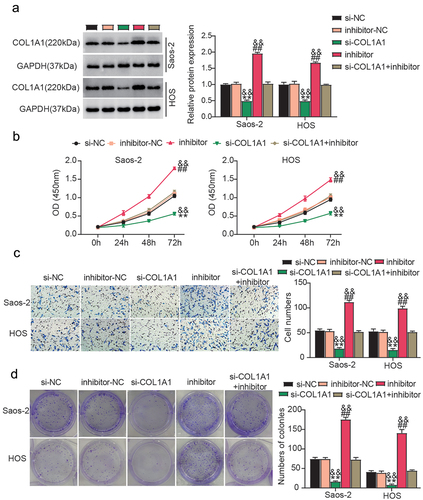
Having determined the interaction between miR-339-3p and COL1A1, we further investigated whether COL1A1 downregulation is critical for miR-339-3p activity in osteosarcoma. Hence, we simultaneously delivered miR-339-3p inhibitor and si-COL1A1 into Saos-2 and HOS cells. Western blotting analysis revealed that the miR-339-3p inhibitor restored COL1A1 knockdown in the osteosarcoma cells (). Functionally, the loss of COL1A1 resulted in delayed osteosarcoma cell proliferation, which was recovered by the addition of the miR-339-3p inhibitor (). Moreover, COL1A1 silencing in HOS and Saos-2 cells resulted in a decline in the migration and colony formation rates, which were offset by the co-silencing of miR-339-3p and COL1A1 (). Collectively, miR-339-3p suppressed osteosarcoma cell proliferation and migration by blocking COL1A1 expression.
Figure 6. miR-339-3p attenuates the malignant phenotypes of osteosarcoma cells by downregulating COL1A1.
Discussion
Herein, we revealed that circ_0020378 was frequently upregulated in osteosarcoma, and its silencing remarkably attenuated osteosarcoma cell migration and proliferation and curbed cancer growth in vivo. The miR-339-3p inhibitor abrogated tumor repression induced by circ_0020378 silencing. Furthermore, COL1A1 silencing offset the pro-tumor effect of the miR-339-3p inhibitor in osteosarcoma. Therefore, the oncogenic role of circ_0020378 in osteosarcoma was characterized based on its ceRNA activity against miR-339-3p/COL1A1. Targeting circ_0020378/miR-339-3p/COL1A1 may be a novel method for treating osteosarcoma.
Several reports have demonstrated that circRNAs participate in tumorigenesis by regulating gene expression at different levels.Citation19 An increasing number of circRNAs have been identified during the multistep process of human tumor pathogenesis.Citation20 For example, circRNA_100876 maintains the chronic proliferation of osteosarcoma cells and deregulates the homeostasis of normal tissue architecture and function, thereby enabling cancer onset.Citation21 CircATRNL1 elicits reprogramming of energy metabolism in osteosarcoma cells, favoring cancer progression.Citation22 Our data is the first to reveal that the novel circRNA circ_0020378 is robustly expressed in osteosarcoma cells and tissues. Loss-of-function experiments further demonstrated that circ_0020378 silencing hampered the migration and proliferation of osteosarcoma cells. To the best of our knowledge, this is the first study to determine the function of circ_0020378 in osteosarcoma.
Considering the typical role of cytoplasmic circRNAs as ceRNAs, our data confirmed that circ_0020378 targets and sequesters miR-339-3p. Analysis of miRNA expression profiles in the sera of patients with NSCLC suggested that miR-339-3p potently enhances the risk of tumor development.Citation23 MiR-339-3p is downregulated in colorectal cancer, and reportedly suppresses metastasis and tumor growth in vitro.Citation15 Additionally, miR-339-3p has been reported to be a tumor suppressor in melanoma because it inhibits invasive aggressiveness.Citation24 In osteosarcoma, miR-339-3p is sponged by CircDOCK1 and blunts the sensitivity of osteosarcoma cells to cisplatin.Citation13 Therefore, miR-339-3p may inhibit cancer tumors. Consistently, we found that the miR-339-3p inhibitor stimulated the migration and proliferation of osteosarcoma cells. However, miRNAs have been shown to have multiple targets. This implied that miR-339-3p may be recognized by different circRNAs. Indeed, our data showed that its promotive effect on the malignant osteosarcoma cell phenotypes was abrogated by circ_0020378. The negative correlation between circ _0020378 and miR-339-3p further reinforced its ceRNA activity with miR-339-3p.
Accumulating evidence has substantiated the role of COL1A1 as a tumor driver in different malignancies. A recent analysis of Gene Expression Omnibus (GEO) datasets showed that elevated COL1A1 expression is associated with unfavorable prognosis and marked immune cell infiltration in lung cancer patients.Citation25 COL1A1 enhances gastric cell migration and metastasis in vitro and in vivo.Citation26 In osteosarcoma, COL1A1 polymorphisms increase cancer susceptibility and mortality. Our data are the first to show that COL1A1 silencing hampers the migration and proliferation of osteosarcoma cells. COL1A1 is also a tumor promoter in osteosarcoma. However, the mechanism underlying COL1A1-driven osteosarcoma has not yet been elucidated. Our data show that miR-339-3p targets COL1A1 and downregulates COL1A1 expression. Furthermore, the miR-339-3p inhibitor offset the reduced proliferation and migration of osteosarcoma cells when COL1A1 was silenced. Collectively, these results indicate that miR-339-3p represses the tumor-promoting effect of COL1A1 by downregulating its expression.
This study had several limitations. First, the sample size was small. Hence, in the future, we need to explore the association of circ_0020378 with clinical characteristics by collecting more clinical samples. Furthermore, some studies have confirmed that COL1A1 regulates the WNT/PCP.Citation27 and PI3K/AKT.Citation28 pathways to participate in cancer progression. Hence, the COL1A1-driven signaling pathway in osteosarcoma requires further investigation. More importantly, circ_0020378 may have target multiplicity owing to base pairing. Therefore, the detailed mechanism of circ_0020378 in osteosarcoma requires further exploration.
Conclusion
In summary, this study confirmed that a novel circRNA, circ_0020378, is upregulated in osteosarcoma. Further in vivo and in vitro experiments showed that circ_0020378 promoted osteosarcoma cell malignancy by modulating the miR-339-3p/COL1A1 axis. The findings of this study may contribute to the development of rational strategies for treating osteosarcoma.
Authors contributions
LL: conceptualization, methodology, supervision, curation and analysis of data, original draft preparation, reviewing and editing of the manuscript; WZH: visualization, validation, data analysis, writing-reviewing and editing.
Consent for publication
Consent for publication was acquired from all participants.
Consent to participate
Written informed consents were provided by all patients.
Ethics declarations
The Ethics Committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University authorized this research. Clinical tissue specimen processing was accomplished in strict observance of the ethical standards of the Declaration of Helsinki. Written consents were acquired form the patients.
The Ethics Committee of Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University granted approval to this animal study which was executed according to the ARRIVE guidelines.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
All data generated or analyzed during this study are included in this published article.
Additional information
Funding
Notes on contributors
Liang Liu
Liang Liu, received Master degree from Shantou University in 2013. Now, he is a surgeon in the Huangshi Central Hospital. His research focus on gene detection of bone tumor.
Wenze Huang
Wenze Huang, received Master degree from Wuhan University in 2013. Now, he is a surgeon in the Huangshi Central Hospital. His research focus on molecular therapy of bone tumor.
References
- Gill J, Gorlick R. Advancing therapy for osteosarcoma. Nat Rev Clin Oncol. 2021;18(10):609–11. doi:10.1038/s41571-021-00519-8.
- Belayneh R, Fourman MS, Bhogal S, Weiss KR. Update on osteosarcoma. Curr Oncol Rep. 2021;23(6):71. doi:10.1007/s11912-021-01053-7.
- Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18(1):39–50. doi:10.1080/14737140.2018.1413939.
- Liu CX, Chen LL. Circular RNAs: characterization, cellular roles, and applications. Cell. 2022;185(12):2016–2034. doi:10.1016/j.cell.2022.04.021.
- Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular rnas. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7.
- Li Z, Li X, Xu D, Chen X, Li S, Zhang L, Chan MTV, Wu WKK. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2021;54(1):e12936. doi:10.1111/cpr.12936.
- Shen S, Yao T, Xu Y, Zhang D, Fan S, Ma J. Circece1 activates energy metabolism in osteosarcoma by stabilizing c-myc. Mol Cancer. 2020;19(1):151. doi:10.1186/s12943-020-01269-4.
- Ma XL, Zhan TC, Hu JP, Zhang CL, Zhu KP. Doxorubicin-induced novel circrna_0004674 facilitates osteosarcoma progression and chemoresistance by upregulating mcl1 through mir-142-5p. Cell Death Discov. 2021;7(1):309. doi:10.1038/s41420-021-00694-8.
- Chen J, Liu G, Wu Y, Ma J, Wu H, Xie Z, Chen S, Yang Y, Wang S, Shen P, et al. Circmyo10 promotes osteosarcoma progression by regulating mir-370-3p/ruvbl1 axis to enhance the transcriptional activity of β-catenin/lef1 complex via effects on chromatin remodeling. Mol Cancer. 2019;18(1):150. doi:10.1186/s12943-019-1076-1.
- Correia de Sousa M, Gjorgjieva M, Dolicka D, Sobolewski C, Foti M. Deciphering miRNAs’ action through miRNA editing. Int J Molecular Sci. 2019;20(24):6249. doi:10.3390/ijms20246249.
- Cai WT, Guan P, Lin MX, Fu B, Wu B, Wu J. miRNA-206 suppresses the metastasis of osteosarcoma via targeting notch3. J Biol Regul Homeost Agents. 2020;34(3):775–783. doi:10.23812/20-72-A-26.
- Zhu KP, Zhang CL, Ma XL, Hu JP, Cai T, Zhang L. Analyzing the interactions of mRNAs and ncrnas to predict competing endogenous RNA networks in osteosarcoma chemo-resistance. Mol Therapy: J Amer Soc Gene Therap. 2019;27(3):518–530. doi:10.1016/j.ymthe.2019.01.001.
- Wang S, Su TT, Tong H, Shi W, Ma F, Quan Z. Circpvt1 promotes gallbladder cancer growth by sponging mir-339-3p and regulates mcl-1 expression. Cell Death Discov. 2021;7(1):191. doi:10.1038/s41420-021-00577-y.
- Wang S, Su TT, Tong H, Shi W, Ma F, Quan Z. Correction: Circpvt1 promotes gallbladder cancer growth by sponging mir-339-3p and regulates mcl-1 expression. Cell Death Discov. 2021;7(1):396. doi:10.1038/s41420-021-00669-9.
- Zhou C, Lu Y, Li X. Mir-339-3p inhibits proliferation and metastasis of colorectal cancer. Oncol Lett. 2015;10(5):2842–2848. doi:10.3892/ol.2015.3661.
- He M, Wang Z, Zhao J, Chen Y, Wu Y. Col1a1 polymorphism is associated with risks of osteosarcoma susceptibility and death. Tumor Biol. 2014;35(2):1297–1305. doi:10.1007/s13277-013-1172-6.
- Zhang C, Liu S, Wang X, Liu H, Zhou X, Liu H, Ilie L. Col1a1 is a potential prognostic biomarker and correlated with immune infiltration in mesothelioma. Biomed Res Int. 2021;2021:1–13. doi:10.1155/2021/5320941.
- Jia R, Wang C. MiR-29b-3p reverses cisplatin resistance by targeting COL1A1 in non-small-cell lung cancer A549/DDP cells. Cancer Manag Res. 2020;12:2559–2566. doi:10.2147/CMAR.S246625.
- Yu T, Wang Y, Fan Y, Fang N, Wang T, Xu T, Shu Y. Circrnas in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90. doi:10.1186/s13045-019-0776-8.
- Xi Y, Fowdur M, Liu Y, Wu H, He M, Zhao J. Differential expression and bioinformatics analysis of circrna in osteosarcoma. Biosci Rep. 2019;39(5). doi:10.1042/BSR20181514.
- Jin J, Chen A, Qiu W, Chen Y, Li Q, Zhou X, Jin D. Dysregulated circrna_100876 suppresses proliferation of osteosarcoma cancer cells by targeting microRNA-136. J Cellular Biochem. 2019;120(9):15678–15687. doi:10.1002/jcb.28837.
- Zhang Q, Wang L, Cao L, Wei T. Novel circular RNA circatrnl1 accelerates the osteosarcoma aerobic glycolysis through targeting mir-409-3p/ldha. Bioengineered. 2021;12(2):9965–9975. doi:10.1080/21655979.2021.1985343.
- Trakunram K, Chaniad P, Geater SL, Keeratichananont W, Chittithavorn V, Uttayamakul S, Buya S, Raungrut P, Thongsuksai P. Serum mir-339-3p as a potential diagnostic marker for non-small cell lung cancer. Cancer Biol Med. 2020;17(3):652–663. doi:10.20892/j.issn.2095-3941.2020.0063.
- Weber CE, Luo C, Hotz-Wagenblatt A, Gardyan A, Kordaß T, Holland-Letz T, Osen W, Eichmüller SB. Mir-339-3p is a tumor suppressor in melanoma. Cancer Res. 2016;76(12):3562–3571. doi:10.1158/0008-5472.CAN-15-2932.
- Geng Q, Shen Z, Li L, Zhao J. Col1a1 is a prognostic biomarker and correlated with immune infiltrates in lung cancer. PeerJ. 2021;9:e11145. doi:10.7717/peerj.11145.
- Li Y, Sun R, Zhao X, Sun B. Runx2 promotes malignant progression in gastric cancer by regulating col1a1. Cancer biomarkers: section a of disease markers. Cancer Biomark. 2021;31(3):227–238. doi:10.3233/CBM-200472.
- Zhang Z, Wang Y, Zhang J, Zhong J, Yang R. Col1a1 promotes metastasis in colorectal cancer by regulating the wnt/pcp pathway. Mol Med Report. 2018;17(4):5037–5042. doi:10.3892/mmr.2018.8533.
- Huang P, Li M, Tang Q, Jiang K, Luo Y. Circ_0000523 regulates mir-1184/col1a1/pi3k/akt pathway to promote nasopharyngeal carcinoma progression. Apoptosis 2022;27(9–10):751–761. doi:10.1007/s10495-022-01743-y.
