ABSTRACT
Breast cancer (BC), characterized by its diverse molecular profiles and clinical outcomes, presents a significant challenge in the development of effective therapeutic strategies. Metabolic reprogramming, a defining characteristic of cancer, has emerged as a promising target for novel therapies. SLC7A11, an amino acid transporter that facilitates cysteine uptake in exchange for glutamate, plays a crucial role in sustaining the altered metabolism of cancer cells. This study delves into the comprehensive analysis of SLC7A11 at the genomic, transcriptomic, and protein levels in extensive BC datasets to elucidate its potential role in different BC subtypes. SLC7A11 gene copy number and mRNA expression were evaluated using the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort (n = 1,980) and Breast Cancer Gene Expression Miner (n = 4,712). SLC7A11 protein was assessed using immunohistochemistry in a large BC cohort (n = 1,981). Additionally, The Cancer Genome Atlas (TCGA) dataset was used to explore SLC7A11 DNA methylation patterns using MethSurv (n = 782) and association of SLC7A11 mRNA expression with immune infiltrates using TIMER (n = 1,100). High SLC7A11 mRNA and SLC7A11 protein expression were significantly associated with high tumor grade (p ≤ .02), indicating a potential role in cancer progression. Interestingly, SLC7A11 copy number gain was observed in HER2+ tumors (p = .01), suggesting a subtype-specific association. In contrast, SLC7A11 mRNA expression was higher in the basal-like/triple-negative (TN; p < .001) and luminal B tumors (p = .02), highlighting its differential expression across BC subtypes. Notably, high SLC7A11 protein expression was predominantly observed in Estrogen Receptor (ER)-negative and Triple Negative (TN) BC, suggesting a role in these aggressive subtypes. Further analysis revealed that SLC7A11 was positively correlated with other amino acid transporters and enzymes associated with glutamine metabolism, implying a coordinated role in metabolic regulation. Additionally, SLC7A11 gene expression was positively associated with neutrophil and macrophage infiltration, suggesting a potential link between SLC7A11 and tumor immunity. Our findings suggest that SLC7A11 plays a significant role in BC metabolism, demonstrating differential expression across subtypes and associations with poor patient outcomes. Further functional studies are warranted to elucidate the precise mechanisms by which SLC7A11 contributes to BC progression and to explore its potential as a therapeutic target.
Introduction
Breast cancer (BC), the top malignancy in women,Citation1 is a heterogeneous disease with various biological subtypes that differ in morphology, biological subtypes, response to therapy and clinical behavior.Citation2 This variability further underscores the notion of varying cancer hallmarks and drivers within the different subtypes of breast cancer.
Metabolic reprogramming ensures tumors have a sufficient supply of nutrients to support their rapid growth and proliferation, playing a central role in energy production and biosynthesis.Citation3 This altered metabolism likely contributes to tumor progression and therapeutic failure, which further reinforces the importance of novel therapeutic strategies targeting metabolic reprogramming. Glutamine metabolism is a particular focus of attention as cancer cells can be heavily dependent on this most abundant, non-essential amino acid for nutritional uptake fueling cancer cell unremitted growth.Citation4 Glutamine metabolism and associated metabolic networks including expression of related amino acid transporters are essential for cancer cell survival.Citation5 Rapidly proliferating cancer cells take up glutamine via these amino acid transporters and subsequently transform it into glutamate by glutaminase, a process called glutaminolysis.Citation6 This intracellular glutamate is exchanged with extracellular cystine, the oxidized dimer form of cysteine, at a ratio of 1:1 through Solute Carrier Family 7 Member 11 (SLC7A11) which is later converted into cysteine. SLC7A11 is a sodium independent, chloride-dependent anionic L-cystine/L-glutamate antiporter on the cell surface.Citation7 Intracellular cysteine is generally produced de novo or re-utilized through protein degradation.Citation8–10 However, in oxidative stress, de novo biosynthesis or a catabolic supply of cysteine is not sufficient to meet the high requirement for antioxidant synthesis by cancer cells. Hence, most rely on amino acid transporters, such as SLC7A11, that can import extracellular cystineCitation8 and thus metabolic reprogramming orchestrates the energy supply reflecting the altered metabolic needs of tumors.Citation11
SLC7A11 mediated cystine uptake plays a positive but rate limiting role in glutathione synthesis, contributing to the pathogenesis of cancer where glutathione activates the antioxidant defense machinery that protects cancer cells from apoptosis, oxidative stress and ferroptosis.Citation7,Citation9,Citation12,Citation13 SLC7A11 is expressed at a low level in normal cells but remains a major transporter for cancer cells that are largely reliant on extracellular cystine for survival,Citation10 including liver, glioma, and lung cancer directing disease survival.Citation8,Citation12,Citation14–23 The opposing expression levels between normal and cancer cell suggest the SLC7A11 could act as a potential target for cancer treatment. In BC, SLC7A11 is suggested to play a crucial role in cancer stem cells impacting metastasis.Citation24 In TNBC, the most aggressive biological subtype with poor survival, it also plays an important role in disease progression and proliferation.Citation25,Citation26
Given the diverse origins and divergent disease progression of breast cancer (BC) subtypes, these subtypes are likely to possess distinct genomic, transcriptomic, and proteomic alterations that influence clinical outcomes and treatment responses. Metabolic heterogeneity is also evident among BC biological subtypes, with differential expression of glutamine metabolism-related proteins, a phenomenon that may be driven by ER expression.Citation27–29 Therefore, it is hypothesized that SLC7A11 expression also exhibits heterogeneity in BC, leading to varying outcomes across different subtypes. This prompted a comprehensive investigation to assess the impact of SLC7A11 in BC, evaluating its transcriptomic and proteomic expression in BC subtypes as a potential prognostic marker.
Results
SLC7A11 expression in breast cancer
High SLC7A11 mRNA expression (log2 intensity > 5.5) was observed in 239/1,980 (12%) of the cases. Most tumors showed normal SLCA11 gene copy number (CN) with only 23/1,980 (1.2%) cases showing SLC7A11 CN gain whereas 66/1,980 (3.3%) cases had gene CN loss. No significant association was seen between SLC7A11 gene CN variation (CNV) and mRNA expression (p = .180, data not shown).
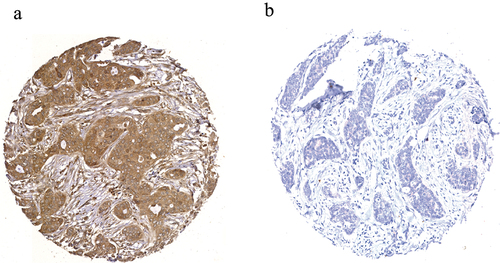
Staining of SLC7A11 in full-face BC tissue sections demonstrated a homogenous pattern of immunoreactivity. SLC7A11 protein expression was mainly observed in the cytoplasm of the invasive breast tumor cells, with intensity levels varying from absent to high (). The distribution of SLC7A11 expression was unimodal and left-skewed and X-tile software was used to identify the optimal cutoff point in predicting breast cancer specific survival (BCSS). High SLC7A11 protein (>100 H-score) was observed in 784/1,981 (40%) of cases.
SLC7A11 DNA methylation in breast cancer
Several CpG islands showed high SLC17A11 DNA methylation within the genomic body of the gene including cg21877274 and cg24869834 (Supplementary Figure S2). The first exon in the 5’ untranslated region showed little methylation.
Association of SLC7A11 expression with clinicopathological characteristics
Within the METABRIC cohort, SLC7A11 CN gain was significantly associated with invasive breast carcinoma of no special type (NST) (p = .0002; ). High SLC7A11 mRNA expression was associated with high tumor grade (p = .01, ) but not with tumor size or nodal stage (). Within the Breast Cancer Gene-Expression Miner dataset, SLC7A11 mRNA expression was significantly associated with higher tumor grade, lymph node negative tumors, and high NPI (all p ≤ .0005; Supplementary Figure S3a-c).
Figure 2. SLC7A11 mRNA expression and its association with clinicopathological parameters and molecular subtypes in the METABRIC cohort: a) SLC7A11 and tumor size, b) SLC7A11 and tumor grade, c) SLC7A11 and lymph node stage, d) SLC7A11 and METABRIC Integrative clusters, e) SLC7A11 and PAM50 subtypes, f) SLC7A11 and ER, g) SLC7A11 and PR, h) SLC7A11 and HER2.
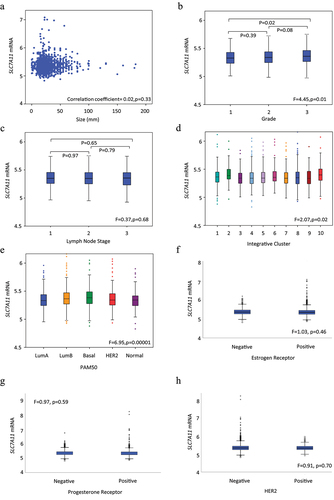
Table 1. SLC7A11 copy number variation in the METABRIC breast cancer cohort and their association with clinicopathological parameters and breast cancer subtypes.
At the protein level, high SLC7A11 was significantly associated with both high and low tumor grade (p = .02, ) which was reflected in its separate components where it was associated with high mitotic count (p = .02, ) and high pleomorphism (p = .002, ) but in contrast also showed association with tubule formation (p = .007; ). High SLC7A1l was further significantly associated with special and tubular histological types (p = .005; ). There was no association between SLC7A11 with tumor size, nodal stage, or NPI.
Table 2. Clinicopathological associations of SLC7A11 protein expression in Nottingham BC cohort.
Association of SLC7A11 expression with BC subtypes
While SLC7A11 CN gain was mainly associated with HER2+ cases (p = .01, ), high levels of SLC7A11 mRNA expression were observed in basal-like tumors (p < .001, ) and METABRIC Integrative Clusters 2 (Luminal B) and 10 which embraces TNBC (p = .02, ). There was no association between SLC7A11 mRNA with ER, PR, or HER2. Within the Breast Cancer Gene-Expression Miner dataset, high SLC7A11 was significantly associated with ER-, PR-, and HER2- BC (all p < .01; Supplementary Figure S3f-h). Both basal-like and luminal B tumors displayed high SLC7A11 mRNA expression (p < .0001; Supplementary Figure S3d-e). High SLC7A11 protein was significantly associated with ER- tumors (p = .0006, ). There was a higher expression of SLC7A11 protein in TNBC (p ≤ .04, ).
Table 3. Association of SLC7A11 protein expression with breast cancer subtypes in the Nottingham BC cohort.
Association of SLC7A11 expression with immune infiltration
Using the TIMER dataset, SLC7A11 gene expression was positively associated, albeit weakly, in breast tumors with neutrophil (p = 2.0×10−24) and macrophage (p = 4.5×10−6) infiltration (). HER2+, luminal A and luminal B tumors also showed a weak to moderate correlation between SLC7A11 mRNA with neutrophil and macrophage infiltration (p ≤ .009, ). A weak negative correlation between SLC7A11 mRNA and CD4+ immune cells was observed in HER2+, luminal A and luminal B tumors (p ≤ .037, ). SLC7A11 CN gain or loss were not associated with immune cell infiltrates in BC or any subtype (data not shown).
Table 4. SLC7A11 gene expression and association with immune infiltrates using timer.
SLC7A11 expression and other associated genes
The METABRIC dataset was used to investigate the correlation between SLC7A11 mRNA and the expression of other associated genes (). The genes were selected based on other publications, being either regulatory genes or others that share SLC7A11 biological function.Citation30–33 There was a positive correlation between SLC7A11 mRNA expression and the expression of the regulatory gene, ATF4 (p = .004, ). SLC7A11 was also positively correlated with the ancillary genes, SLC3A2 and CD44 (all p ≤ .01, ). While the former was associated within all molecular subtypes (all p ≤ .03, ) except HER2+, correlation with CD44 was only observed within luminal B tumors (p = .03, ). Whilst there was a positive correlation observed between SLC7A11 with the high glutamine affinity solute carriers SLC1A5 and SLC7A5 (all p ≤ .001, ). There were no significant associations with other glutamine affinity solute carriers SLC7A8 or SLC38A2. Likewise, there was no association between SLC7A11 and the glutamate transporters SLC1A3, SLC1A6 and SLC1A7 or the cystine/glutamate antiporter SLC1A1. There was, however, a positive association between SLC7A11 and SLC1A2 (p = .04, ) which lost its significance when the different BC subtypes were examined separately. The association of SLC7A11 protein expression with other associated proteins was also examined (). At the protein level, SLC7A11 was significantly expressed with breast tumors that expressed high MYC, Ki67, and Glutaminase (GLS; all p ≤ .01, ). There was no significant association between SLC7A11 and Glutamate Dehydrogenase (GLUD1) (p > .05, ), the enzyme which degrades glutamate to α-ketoglutarate. Nonetheless, a positive association was observed with SLC7A11 and SLC1A5, SLC7A5, SLC7A8, SLC3A2 and SLC38A2 (all p ≤ .04, ). Moreover, SLC7A11 was highly expressed in BCs which express high p53 protein (p = .01, ).
Table 5. Correlation of SLC7A11 mRNA expression with the expression of other related genes in the METABRIC cohort.
Table 6. Association of SLC7A11 protein expression and other associated proteins in the Nottingham BC cohort.
Association of SLC7A11 and patient outcome
Survival analysis of high SLC7A11 mRNA was significantly associated with shorter BCSS (p = .02, ). Multivariate analysis showed that SLC7A11 mRNA was a predictive of poor survival independent of tumor grade, tumor size, and lymph node stage (p = .008, ) and the result remained significant within the luminal A tumors (p = .02, ). The relationships between SLC7A11 mRNA expression and patient outcome were verified using Breast Cancer Gene-Expression Miner (Supplementary Figure S4). High SLC7A11 mRNA expression showed association with shorter overall survival (OS) in all cases (p = .0004, Supplementary Figure S4a) and in HER2+ cases (p = .02, Supplementary Figure S4e). In contrast, SLC7A11 protein was not associated with patient outcome, in terms of BCSS and DMFS either in the whole cohort or when the different BC subtypes were examined (). High SLC7A11 mRNA expression showed association with shorter OS (p = 3.6×10–5, Supplementary Figure S5a) in the KM plotter dataset whereas no significance was found in terms of SLC7A11 protein expression (Supplementary Figure S5b). DNA methylation of both cg21877274 and cg24869834 CpG sites on SLC7A11 correlated with poorer patient survival rates (p = .0014 and p = .0074 respectively; Supplementary Figure S2b,c).
Figure 3. SLC7A11 mRNA expression and its association in breast cancer biological subtypes with patient breast cancer specific survival in the METABRIC cohort: a) SLC7A11 in all cases, b) SLC7A11 in luminal A, c) SLC7A11 in luminal B, d) SLC7A11 vs triple Negative, and e) SLC7A11 in HER2+ breast cancer.
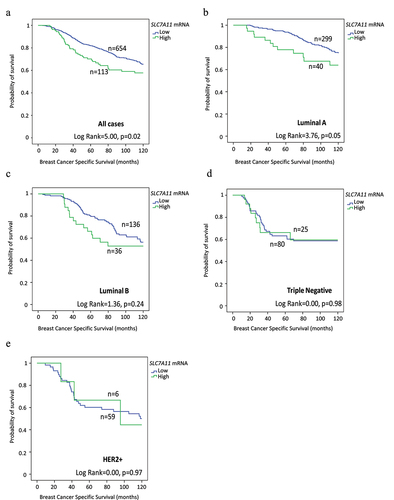
Figure 4. SLC7A11 protein expression and its association with patient breast cancer specific survival (BCSS) in the Nottingham cohort: a) SLC7A11 in all cases, b) SLC7A11 in ER+ low proliferative, c) SLC7A11 in ER+ high proliferative, d) SLC7A11 in triple Negative, and e) SLC7A11 in HER2+ breast cancer.
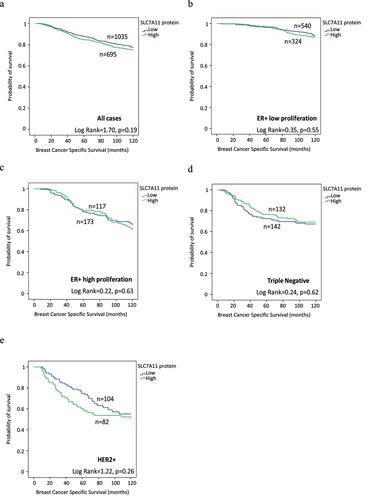
Table 7. SLC7A11 mRNA expression and patient outcome in BC molecular subtypes using the METABRIC cohort.
Discussion
BC is a biologically heterogeneous diseaseCitation34 with various molecular subtypes that influences its metabolic programming and nutritional needs for its growth and proliferation, which can ultimately affect patient survival. Thus, understanding the metabolic profiling along with their biological characteristics could help to identify potential therapeutic targets and prognostic markers. The present study explored SLC7A11 expression in BC by assessing its genomic, transcriptomic, and proteomic levels for the first time in several large breast cancer cohorts. It revealed that SLC7A11 could potentially play a significant role in BC subtypes influencing the patient outcome.
In this study, we show SLC7A11 CN loss was more common than CN gain which was generally associated with a small number of HER2+ cases. However, there was no association between SLC7A11 CNV and mRNA expression, suggesting that CN gain does not contribute to SLC7A11 transcription or SLC7A11 translation. SLC7A11 mRNA and SLC7A11 protein expression were primarily associated with poor clinicopathological parameters and SLC7A11 DNA methylated predicted poor patient survival. Further analysis revealed that SLC7A11 was positively correlated with other amino acid transporters and enzymes associated with glutamine metabolism, implying a coordinated role in metabolic regulation. Additionally, SLC7A11 gene expression was positively associated with neutrophil and macrophage infiltration, suggesting a potential link between SLC7A11 and tumor immunity.
While focusing on BC subtypes, the poor prognostic ER negative and TNBC showed high SLC7A11 mRNA and SLC7A11 protein expression. This relation was supported by a previous study where Timmerman et alCitation26 reported that SLC7A11 is significantly expressed in TNBC. TNBC tend to act as a glutamine auxotroph, which correlates SCL7A11 and cystine consumption. This potentially indicates that TNBC with high SLC7A11 expression can show an aggressive BC advancement. TNBC currently as only chemotherapy as its treatment option due to the absence of growth in response to hormones and SLC7A11 is found to modulate chemotherapy or vice versa.Citation35,Citation36 Cancer cells are rendered more resistant to chemotherapy if SLC7A11 is overexpressed.Citation37 This supports the fact that SLC7A11 can represent itself as a potential target to observe it as a treatment response factor in TNBC and such suggestions were backed by earlier studies.Citation25,Citation35,Citation38
Regarding exploring the additional genes associated with SLC7A11, transcription factor ATF4 was significantly associated with SLC7A11 mRNA. This transcription factor is thought to foster SLC7A11 expression and upregulating SLC7A11 transcription.Citation39–41 AFT4 regulates redox homeostasis and amino acid metabolism.Citation38 As a part of redox homeostasis in the cancer cell, SLC7A11 presence contributes to increasing glutathione, which later promotes metabolic reprogramming. This supports the notion that under stress condition; proteasome inhibition or glucose starvation, SLC7A11 requires ATF4 for its expression.Citation40 This relation can be targeted for SLC7A11 reduction supported by previous studies showed AFT4 inhibition reduces SLC7A11 expression.Citation39,Citation41–43 Hereby, SLC7A11 is also influenced by CD44, the adhesion molecule, which stabilizes SLC7A11 which subsequently promotes the uptake of cystine for glutathione synthesis,Citation33 which is further supported by the fact that CD44 deficiency can result into intracellular glutathione depletionCitation43 resulting in ROS induction, inhibiting tumor formation and inducing ferroptosis. From our study, SLC7A11 mRNA shows a strong relation with CD44, distinctively within luminal B tumors, and this reflects previous evidence.Citation44–46
This study further investigated the association of SLC7A11 expression with other glutamine transporters. SLC7A11 was strongly associated with SLC3A2, SLC1A5, SLC7A5, and SLC38A2, which reinforces that amino acid transporter activity is altered to meet the altered environment in BC. SLC3A2 is required for SLC7A11 for its optimum activation, where SLC7A11 heterodimerises with SLC3A2 via a disulfide bridge. Efficient exchange of glutamate requires both subunits where the transport activity which is primarily maintained by SLC7A11, highly specific for cystine and glutamate, whereas SLC3A2 which mainly acts as a chaperone protein to regulate the trafficking function of SLC7A11.Citation47–49 SLC3A2 absence also brings a substantial decrease of SLC7A11 activity indicating a crucial role for SLC7A11 stability.Citation50,Citation51
Our study also showed SLC7A11 protein was strongly associated with GLS expression, the key enzyme for glutamine catabolism in line with an earlier study.Citation52 GLS activity is paused by the presence of sufficient intracellular glutamate but the high expression of SLC7A11, which decreases the cellular glutamate by exporting, potentially reactivate GLS activity through negative feedback contributing to cell nutrient supply, especially in TNBC.Citation53 Thus, the combination of GLS and SLC7A11 inhibitors can offer a potential treatment approach for TNBC.
In BC, MYC and TP53 genes are altered and have direct role on BC prognosis and treatment options.Citation54 c-Myc has a direct role in glutamate metabolismCitation55 and SLC7A11 is directly induced by c-Myc, or vice versa, and our results support this association. In addition, p53 protein (encoded by TP53) is strongly associated with SLC7A11Citation18 and p53 mutations alter SLC7A11 expression.Citation51,Citation56 p53 also acts upon glutathione synthesis, which is mediated by SLC7A11, when tumor cells are serine starved, thus preserving cellular anti-oxidant capacity.Citation57 p53 also negatively regulates SLC7A11 and as p53 is considered a pro-ferroptotic factor, it can increase the sensitivity to ferroptosis.Citation31 Furthermore, high SLC7A11 expression was significantly associated with Ki67 reinforcing previous studies.Citation18,Citation58 Since Ki67 is a poor prognostic biomarker in invasive breast cancer,Citation59 its positive association with SLC7A11 depicts the fact that subtype with overexpression of SLC7A11 can demonstrate aggressive disease progression.
SLC7A11 mRNA was correlated with the infiltration levels of neutrophils and macrophages irrespective of biological subtype. Neutrophils, believed to promote metastasis in BC,Citation60 can affect intracellular GSH levels and thus can influence SLC7A11 activity.Citation61 Macrophages are thought to participate in ferroptosisCitation62 and SLC7A11 could act as a connection between the two. High presence of these immune cells can potentially offer a synergistic impact for BC treatment focusing on SLC7A11.
Whilst high SLC7A11 mRNA was an independent predictor of poor OS all cases and in luminal A tumors this was not translated into protein expression. This discrepancy can be attributed to transcriptional factors, miRNA, linc-RNA, DNA methylation, and other translational and post-translational modifications as well as protein stability, other co-existing metabolic pathway, cellular condition and coupling with other protein could change the protein activity so as of SLC7A11.Citation63–67 Transcriptional process and mRNA decaying process can lead discordance between protein and mRNA expression. SLC7A11 mRNA has a relation with SLC3A2 and CD44 which can also affect its protein stability, so these regulatory mechanisms require further investigation.
In conclusion, this study revealed a robust characterization of the glutamate/cysteine transporter SLC7A11 as an independent prognostic factor in BC. Several SLC and other factors associated with the glutamate transport axis were related to SLC7A11 and their combined functionality gives opportunities to explore them as potential therapeutic targets. Further functionals studies on SLC7A11 in BC are therefore warranted.
Material and methods
SLC7A11 genomic and transcriptomic expression
SLC7A11 CNV and mRNA expression were evaluated in a cohort of 1,980 BC tumors in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC).Citation68 The METABRIC study provides genomic and transcriptional profiling data on BC using the Affymetrix SNP 6.0 and Illumina HT-12v3 platforms, respectively. In this cohort, patients enrolled as ER- and lymph node (LN) positive received adjuvant treatment while patients with ER+ and/or LN negative did not receive adjuvant chemotherapy. Dichotomization of SLC7A11 mRNA expression was conducted using X-tile (version 3.6.1, Yale University, USA), based on the prediction of Breast Cancer Specific Survival (BCSS). Association of CNV including loss and gain and mRNA expression along with clinicopathological parameters and molecular subtypes were investigated. mRNA expression with other genes related to glutamine metabolism and patient’s survival were also investigated. To validate SLC7A11 mRNA expression, Breast Cancer Gene Expression Miner v4.5 (bcgenex.centregauducheau.fr) incorporating The Cancer Genome Atlas (TCGA) and SCAN-B RNA sequencing data (n = 4,712) and Kaplan–Meier Plotter database (RNA-seq data for breast cancer) (n = 2976) were used as external validation datasets.
SLC7A11 CNV (n = 1080) and mRNA (n = 1100) expression were also investigated in BC using TIMER (http://timer.cistrome.org/) to estimate the abundance of SLC7A11 in six subsets (B cells, CD4+ T cells, CD8+ T cells, macrophages, neutrophils, and dendritic cells).Citation69 MethSurv (biit.cs.ut.ee/methsurv) was used to explore the DNA methylation of SLC7A11 in the TCGA data according to patient survival.Citation70 DNA methylation status was dichotomized using the ‘best’ option.
Patients’ cohort for SLC7A11 protein expression
Immunohistochemistry (IHC) was conducted on a well-characterized BC series from patients at Nottingham City Hospital during 1987–2006 (n = 1,981). The patients were diagnosed with early-stage primary operable invasive BC and aged ≤70 years. Patient management was uniform and based on tumor characteristics including the Nottingham Prognostic Index (NPI) and hormone receptor status. Patients with NPI > 3.4 received tamoxifen if ER+ (± goserelin, in premenopausal patients). Conversely, classical cyclophosphamide, methotrexate and 5-fluorouracil (CMF) were used in ER-patients who were fit enough to receive chemotherapy. No patients received neoadjuvant therapy. Clinical history, tumor characteristics, information on therapy and outcomes were prospectively maintained. Outcome data including development and time to distant metastasis (DM) and BCSS. DM-free survival (DMFS) was defined as the time (in months) from the date of primary surgery to the appearance of DM. The BCSS was defined as the time (in months) from the date of primary surgery to the date of BC-related death. SLC7A11 protein expression was correlated with clinicopathological parameters, BC subtypes, other genes related to glutamine metabolism and patient’s survival. The Kaplan–Meier Plotter database (protein data for breast cancer) (n = 1064) was used as an external validation dataset.
SLC7A11 antibody validation
SLC7A11 primary antibody specificity (Rabbit monoclonal, Ab37185, Abcam, UK) was determined using Western blotting in cell line lysates: HCC1500, ZR-751, MDA-MB-436, MCF7 and T47D (American Type Culture Collection; Rockville, MD, USA at a dilution of 1:2000. Donkey anti-rabbit (1:15,000, IRDye680 CW, 926–32213, LI-COR Bioscience) was used as a fluorescent secondary antibody. Mouse monoclonal anti-β-actin primary antibody (1:5,000, A5441, Sigma-Aldrich) with donkey anti-mouse fluorescent secondary (1:15,000, IRDye 800CW, 926–68072, LI-COR Bioscience) was used as a control. The Odyssey Fc machine (LI-COR Bioscience) was used to visualize blots showing specific bands at the predicted size of approximately 55 KDa (Supplementary Figure S1).
Tissue microarrays and IHC
Tissue Microarrays (TMAs) were produced following standard techniques as described.Citation71 Polymer-based IHC was performed on 4 µm thick section using the Novolink Polymer Detection System (Leica Biosystems, RE7150-K) according to the manufacturer’s instructions. Heat mediated antigen retrieval was carried out using citrate buffer at pH 6.0. SLC7A11 antibody was used at a 1:50 dilution for 1 hour at room temperature. Beta2-microglobulin at a concentration of 1:2,000 was used as a positive control.
Assessment of SLC7A11 protein expression
Stained TMA sections were assessed using high resolution digital images (NanoZoomer, Hamamatsu Photonics, Welwyn Garden City, UK) at 20× magnification viewed using Philips Xplore software (Philips Digital Pathology Solutions, Amsterdam, Netherlands). Assessment of protein staining was based on a semi-quantitative H-Score which includes an assessment of both intensity of staining and percentage of stained cells.Citation72 Adequate tumor burden was assessed ensuring that the TMA core contained more than 15% of invasive tumor (excluding DCIS). Intensity of the staining ranged from a scale of 0 to 3 (0 = no staining, 1= weak, 2 = moderate, 3 = strong) and the percentage of positively stained invasive BC cells was estimated subjectively. A final H-score of 0–300 was generated based on % of cells stained at each intensity. A second observer blindly scored 10% cores to assess scoring ability and concordance. There was high inter-observer concordance between the scorers (Kappa score = 0.64). Dichotomization of SLC7A11 protein expression was determined based on the prediction of BCSS using X-tile software.
Immunohistochemical staining and dichotomization of the other biomarkers included in this study were as per previous publications.Citation73–77 ER and progesterone receptor (PR) positivity was defined as ≥ 1% staining. Immunoreactivity of HER2 in TMA cores was scored using standard HercepTest guidelines (Dako). Chromogenic in situ hybridization (CISH) was used to quantify HER2 gene amplification in borderline cases using the HER2 FISH pharmDx™ plus HER2 CISH pharmDx™ kit (Dako) and was assessed according to the American Society of Clinical Oncology guidelines. BC molecular subtypes were defined based on tumor IHC profile and the Elston-EllisCitation78 mitotic score as: ER+/HER2- low proliferation (mitotic score 1), ER+/HER2- high proliferation (mitotic score 2 and 3); HER2+ class: HER2+ regardless of ER status; triple negative: ER-, PR- and HER2-.Citation79 Basal-like phenotype was defined as tumors expressing cytokeratin (Ck) 5/6, and/or Ck14 and/or Ck17.
Statistical analysis
Statistical analysis was performed using SPSS 22 statistical software (SPSS Inc., Chicago, IL, USA). Chi-square test was performed to analyze categorical variables. For continuous variables, Pearson correlation coefficient was used to test correlation between two continuous normalized data, whereas the differences between three or more groups were assessed using one-way analysis of variance (ANOVA) with the post-hoc Tukey multiple comparison test. Survival analysis was done using Kaplan-Meier with Log Rank test for outcome. Cox’s proportional hazard method was implied to identify independent prognostic factors. P-values were adjusted using Bonferroni correction for multiple testing. Variables with p-values <0.05 were considered statistically significant.
Ethical approval
This study was approved by the Nottingham Research Ethics Committee 2 under the title ‘Development of a molecular genetic classification of breast cancer’ and the North West – Greater Manchester Central Research Ethics Committee under the title ‘Nottingham Health Science Biobank (NHSB)’ reference number 15/NW/0685.
Author contributions
P.N., L.H.A., R.E.: writing, review, editing, methodology, formal analysis and interpretation. B.M., B.E., A.F.: writing – original draft, reviewing and editing. I.O.E., E.A.R. supervision, writing – review and editing A.G: conceptualization, formal analysis and interpretation, supervision, writing – review and editing. All authors have read and approved the final submitted manuscript.
Supplemental Material
Download MS Word (1.1 MB)Acknowledgments
We thank the Nottingham Health Science Biobank and Breast Cancer Now Tissue Bank for the provision of tissue samples. The authors are part of the PathLAKE digital pathology consortium. These new Centres are supported by a £50 m investment from the Data to Early Diagnosis and Precision Medicine strand of the government’s Industrial Strategy Challenge Fund, managed and delivered by UK Research and Innovation (UKRI).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2023.2291855
Additional information
Funding
Notes on contributors
Preyanka Nath
Preyanka Nath holds master’s degrees from the University of Nottingham, UK, and Brac University, Bangladesh, and a bachelor’s degree from the Shahjalal University of Science & Technology in Bangladesh. They are a lecturer at the North South University in Bangladesh.
Lutfi H. Alfarsi
Lutfi Alfarsi holds a PhD from the University of Nottingham, UK, a bachelor’s degree from Manchester Metropolitan University, UK, and a master’s degree from Nottingham Trent University, UK. They are a Director of a healthcare provider in Saudi Arabia.
Rokaya El-Ansari
Rokaya El-Ansari holds a PhD from the University of Nottingham, UK, a medical degree from Tripoli University, Libya, and a master’s degree from the University of Leicester, UK. They are a histopathologist in the UK.
Brendah K. Masisi
Brendah Masisi holds a PhD from the University of Nottingham, UK, a master’s degree from the University of the West of England, UK, and a bachelor’s degree from the University of Botswana, Botswana. They are a lecturer at the University of Botswana.
Busra Erkan
Busra Erkan is a PhD student at the University of Nottingham, UK. They hold a bachelor’s degree from Istanbul Halic University, Turkey, and a master’s degree from the University of Leicester, UK.
Ali Fakroun
Ali Fakroun is a PhD student at the University of Nottingham, UK. They hold a bachelor’s degree from Cardiff University, Wales, and a master’s degree from the University of Nottingham, UK.
Ian O. Ellis
Ian Ellis is a Professor of Cancer Pathology at the University of Nottingham, UK, and Honorary Consultant Pathologist at Nottingham University Hospitals NHS Trust, UK. He is a Past President of the Pathological Society of Great Britain and Ireland. He has been Specialty Advisor to The Royal College of Pathologists, and is past Chairman of the UK National Co-ordinating Committee for Breast Pathology. He has acted as an advisor to the DoH, UICC, WHO, and IARC.
Emad A. Rakha
Emad Rakha is a Professor of Breast Pathology at the University of Nottingham and Honorary Consultant Pathologist at Nottingham University Hospitals NHS Trust, UK. They hold a PhD from the University of Nottingham. They are co-chair of the UK National Breast Pathology Interpretive Quality Assurance Scheme and General Secretary of the International Society of Breast Pathology.
Andrew R. Green
Andrew R. Green is an Associate Professor at the University of Nottingham, UK. They hold a PhD from the University of Hull, UK, and a bachelor’s degree from the University of East London, UK. They are Deputy Director of the Nottingham Breast Cancer Research Centre and group leader of the breast cancer metabolism group.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–14. doi:10.3322/caac.21660.
- Dawson SJ, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. EMBO J. 2013;32(5):617–628. doi:10.1038/emboj.2013.19.
- Phan LM, Yeung SC, Lee MH. Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol Med. 2014;11(1):1–19. doi:10.7497/j.issn.2095-3941.2014.01.001.
- Wang Z, Liu F, Fan N, Zhou C, Li D, Macvicar T, Dong Q, Bruns CJ, Zhao Y. 2020. Targeting glutaminolysis: new perspectives to understand cancer development and novel strategies for potential target therapies. Front Oncol. 10:589508. doi:10.3389/fonc.2020.589508.
- Matés JM, Di Paola FJ, Campos-Sandoval JA, Mazurek S, Márquez J. Therapeutic targeting of glutaminolysis as an essential strategy to combat cancer. Semin Cell Dev Biol. 2020;98:34–43. doi:10.1016/j.semcdb.2019.05.012.
- Choi Y-K, Park K-G. Targeting glutamine metabolism for cancer treatment. Biomol & Therapeutics. 2018;26(1):19. doi:10.4062/biomolther.2017.178.
- Lewerenz J, Hewett SJ, Huang Y, Lambros M, Gout PW, Kalivas PW, Massie A, Smolders I, Methner A, Pergande M. The cystine/glutamate antiporter system xc− in health and disease: from molecular mechanisms to novel therapeutic opportunities. Antioxid Redox Signal. 2013;18(5):522–555. doi:10.1089/ars.2011.4391.
- Chio IIC, Tuveson DA. Ros in cancer: the burning question. Trends Mol Med. 2017;23(5):411–429. doi:10.1016/j.molmed.2017.03.004.
- Koppula P, Zhuang L, Gan B. Cystine transporter slc7a11/xct in cancer: ferroptosis, nutrient dependency, and cancer therapy. Protein Cell. 2021;12(8):599–620. doi:10.1007/s13238-020-00789-5.
- Zhu J, Berisa M, Schwörer S, Qin W, Cross JR, Thompson CB. Transsulfuration activity can support cell growth upon extracellular cysteine limitation. Cell Metab. 2019;30(5):865–876. e865. doi:10.1016/j.cmet.2019.09.009.
- Wei Z, Liu X, Cheng C, Yu W, Yi P. 2021. Metabolism of amino acids in cancer. Front Cell Dev Biol. 8:603837. doi:10.3389/fcell.2020.603837.
- Koppula P, Zhang Y, Zhuang L, Gan B. Amino acid transporter slc7a11/xct at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. 2018;38(1):1–13. doi:10.1186/s40880-018-0288-x.
- Lu SC. Glutathione synthesis. Biochimica et Biophysica Acta (BBA)-general subjects. Biochim et Biophys Acta (BBA) - Gen Subj. 2013;1830(5):3143–3153. doi:10.1016/j.bbagen.2012.09.008.
- Combs JA, DeNicola GM. The non-essential amino acid cysteine becomes essential for tumor proliferation and survival. Cancers. 2019;11(5):678. doi:10.3390/cancers11050678.
- Hu K, Li K, Lv J, Feng J, Chen J, Wu H, Cheng F, Jiang W, Wang J, Pei H. Suppression of the slc7a11/glutathione axis causes synthetic lethality in kras-mutant lung adenocarcinoma. J Clin Invest. 2020;130(4):1752–1766. doi:10.1172/JCI124049.
- Ji X, Qian J, Rahman SJ, Siska PJ, Zou Y, Harris BK, Hoeksema MD, Trenary IA, Heidi C, Eisenberg R. Xct (slc7a11)-mediated metabolic reprogramming promotes non-small cell lung cancer progression. Oncogene. 2018;37(36):5007–5019. doi:10.1038/s41388-018-0307-z.
- Lin W, Wang C, Liu G, Bi C, Wang X, Zhou Q, Jin H. 2020. Slc7a11/xct in cancer: biological functions and therapeutic implications. Am J Cancer Res. 10:3106.
- Ma Z, Zhang H, Lian M, Yue C, Dong G, Jin Y, Li R, Wan H, Wang R, Wang Y, et al. Slc7a11, a component of cysteine/glutamate transporter, is a novel biomarker for the diagnosis and prognosis in laryngeal squamous cell carcinoma. Oncol Rep. 2017;38(5):3019–3029. doi:10.3892/or.2017.5976.
- Robert SM, Buckingham SC, Campbell SL, Robel S, Holt KT, Ogunrinu-Babarinde T, Warren PP, White DM, Reid MA, Eschbacher JM. Slc7a11 expression is associated with seizures and predicts poor survival in patients with malignant glioma. Sci Transl Med. 2015;7(289):289ra286–289ra286. doi:10.1126/scitranslmed.aaa8103.
- Savaskan NE, Heckel A, Hahnen E, Engelhorn T, Doerfler A, Ganslandt O, Nimsky C, Buchfelder M, Eyüpoglu IY. Small interfering rna–mediated xct silencing in gliomas inhibits neurodegeneration and alleviates brain edema. Nat Med. 2008;14(6):629–632. doi:10.1038/nm1772.
- Sharbeen G, McCarroll JA, Akerman A, Kopecky C, Youkhana J, Kokkinos J, Holst J, Boyer C, Erkan M, Goldstein D. Cancer-associated fibroblasts in pancreatic ductal adenocarcinoma determine response to slc7a11 inhibition. Cancer Res. 2021;81(13):3461–3479. doi:10.1158/0008-5472.CAN-20-2496.
- Zhang L, Huang Y, Ling J, Zhuo W, Yu Z, Luo Y, Zhu Y. Overexpression of slc7a11: A novel oncogene and an indicator of unfavorable prognosis for liver carcinoma. Future Oncol. 2018;14(10):927–936. doi:10.2217/fon-2017-0540.
- Zhu JH, De Mello RA, Yan QL, Wang JW, Chen Y, Ye QH, Wang ZJ, Tang HJ, Huang T. Mir-139-5p/slc7a11 inhibits the proliferation, invasion and metastasis of pancreatic carcinoma via pi3k/akt signaling pathway. Biochim Biophys Acta Mol Basis Dis. 2020;1866(6):165747. doi:10.1016/j.bbadis.2020.165747.
- Lanzardo S, Conti L, Rooke R, Ruiu R, Accart N, Bolli E, Arigoni M, Macagno M, Barrera G, Pizzimenti S. Immunotargeting of antigen xct attenuates stem-like cell behavior and metastatic progression in breast cancer. Cancer Res. 2016;76(1):62–72. doi:10.1158/0008-5472.CAN-15-1208.
- Hasegawa M, Takahashi H, Rajabi H, Alam M, Suzuki Y, Yin L, Tagde A, Maeda T, Hiraki M, Sukhatme VP. Functional interactions of the cystine/glutamate antiporter, cd44v and muc1-c oncoprotein in triple-negative breast cancer cells. Oncotarget. 2016;7(11):11756. doi:10.18632/oncotarget.7598.
- Timmerman LA, Holton T, Yuneva M, Louie RJ, Padró M, Daemen A, Hu M, Chan DA, Ethier SP, Van‘t Veer LJ. Glutamine sensitivity analysis identifies the xct antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell. 2013;24(4):450–465. doi:10.1016/j.ccr.2013.08.020.
- Kim S, Kim DH, Jung W-H, Koo JS. Expression of glutamine metabolism-related proteins according to molecular subtype of breast cancer. Endocr Relat Cancer. 2013;20(3):339–348. doi:10.1530/ERC-12-0398.
- Kulkoyluoglu-Cotul E, Arca A, Madak-Erdogan Z. Crosstalk between estrogen signaling and breast cancer metabolism. Trends In Endocrinol Metabolism. 2019;30(1):25–38. doi:10.1016/j.tem.2018.10.006.
- Gong Y, Ji P, Yang Y-S, Xie S, Yu T-J, Xiao Y, Jin M-L, Ma D, Guo L-W, Pei Y-C. Metabolic-pathway-based subtyping of triple-negative breast cancer reveals potential therapeutic targets. Cell Metab. 2021;33(1):51–64. e59. doi:10.1016/j.cmet.2020.10.012.
- Verschoor ML, Singh G. Ets-1 regulates intracellular glutathione levels: key target for resistant ovarian cancer. Mol Cancer. 2013;12(1):138. doi:10.1186/1476-4598-12-138.
- Jiang L, Kon N, Li T, Wang SJ, Su T, Hibshoosh H, Baer R, Gu W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature. 2015;520(7545):57–62. doi:10.1038/nature14344.
- Bhutia YD, Ganapathy V. Glutamine transporters in mammalian cells and their functions in physiology and cancer. Biochim Biophys Acta. 2016;1863(10):2531–2539. doi:10.1016/j.bbamcr.2015.12.017.
- Nagano O, Okazaki S, Saya H. Redox regulation in stem-like cancer cells by cd44 variant isoforms. Oncogene. 2013;32(44):5191–5198. doi:10.1038/onc.2012.638.
- Kimbung S, Johansson I, Danielsson A, Veerla S, Egyhazi Brage S, Frostvik Stolt M, Skoog L, Carlsson L, Einbeigi Z, Lidbrink E. Transcriptional profiling of breast cancer metastases identifies liver metastasis–selective genes associated with adverse outcome in luminal a primary breast cancer. Clin Cancer Res. 2016;22(1):146–157. doi:10.1158/1078-0432.CCR-15-0487.
- Ge C, Cao B, Feng D, Zhou F, Zhang J, Yang N, Feng S, Wang G, Aa J. The down-regulation of slc7a11 enhances ros induced p-gp over-expression and drug resistance in mcf-7 breast cancer cells. Sci Rep. 2017;7(1):3791. doi:10.1038/s41598-017-03881-9.
- Lu H, Samanta D, Xiang L, Zhang H, Hu H, Chen I, Bullen JW, Semenza GL. Chemotherapy triggers HIF-1–dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc Natl Acad Sci U S A. 2015;112(33):E4600–4609. doi:10.1073/pnas.1513433112.
- Polewski MD, Reveron-Thornton RF, Cherryholmes GA, Marinov GK, Cassady K, Aboody KS. Increased expression of system xc− in glioblastoma confers an altered metabolic state and temozolomide resistance. Mol Cancer Res. 2016;14(12):1229–1242. doi:10.1158/1541-7786.MCR-16-0028.
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM. The integrated stress response. EMBO Rep. 2016;17(10):1374–1395. doi:10.15252/embr.201642195.
- Wolf IM, Fan Z, Rauh M, Seufert S, Hore N, Buchfelder M, Savaskan NE, Eyüpoglu IY. Histone deacetylases inhibition by saha/vorinostat normalizes the glioma microenvironment via xct equilibration. Sci Rep. 2014;4(1):6226. doi:10.1038/srep06226.
- Ye P, Mimura J, Okada T, Sato H, Liu T, Maruyama A, Ohyama C, Itoh K. Nrf2- and atf4-dependent upregulation of xct modulates the sensitivity of t24 bladder carcinoma cells to proteasome inhibition. Mol Cell Biol. 2014;34(18):3421–3434. doi:10.1128/mcb.00221-14.
- Chen D, Fan Z, Rauh M, Buchfelder M, Eyupoglu IY, Savaskan N. Atf4 promotes angiogenesis and neuronal cell death and confers ferroptosis in a xct-dependent manner. Oncogene. 2017;36(40):5593–5608. doi:10.1038/onc.2017.146.
- Chen D, Rauh M, Buchfelder M, Eyupoglu IY, Savaskan N. The oxido-metabolic driver atf4 enhances temozolamide chemo-resistance in human gliomas. Oncotarget. 2017;8(31):51164–51176. doi:10.18632/oncotarget.17737.
- Liu T, Jiang L, Tavana O, Gu W. The deubiquitylase otub1 mediates ferroptosis via stabilization of slc7a11. Cancer Res. 2019;79(8):1913–1924. doi:10.1158/0008-5472.can-18-3037.
- Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, Oshima M, Ikeda T, Asaba R, Yagi H. Cd44 variant regulates redox status in cancer cells by stabilizing the xct subunit of system xc− and thereby promotes tumor growth. Cancer Cell. 2011;19(3):387–400. doi:10.1016/j.ccr.2011.01.038.
- Wada F, Koga H, Akiba J, Niizeki T, Iwamoto H, Ikezono Y, Nakamura T, Abe M, Masuda A, Sakaue T. High expression of cd 44v9 and xct in chemoresistant hepatocellular carcinoma: potential targets by sulfasalazine. Cancer Sci. 2018;109(9):2801–2810. doi:10.1111/cas.13728.
- Ju H-Q, Lu Y-X, Chen D-L, Tian T, Mo H-Y, Wei X-L, Liao J-W, Wang F, Zeng Z-L, Pelicano H. Redox regulation of stem-like cells though the cd44v-xct axis in colorectal cancer: mechanisms and therapeutic implications. Theranostics. 2016;6(8):1160. doi:10.7150/thno.14848.
- Fairweather SJ, Shah N, Brӧer S.Heteromeric solute carriers: function, structure, pathology and pharmacology.Protein Reviews. 21. 2021:13–127. doi:10.1007/5584_2020_584.
- Fotiadis D, Kanai Y, Palacín M. The slc3 and slc7 families of amino acid transporters. Mol Aspects Med. 2013;34(2–3):139–158. doi:10.1016/j.mam.2012.10.007.
- Kandasamy P, Gyimesi G, Kanai Y, Hediger MA. Amino acid transporters revisited: new views in health and disease. Trends Biochem Sci. 2018;43(10):752–789. doi:10.1016/j.tibs.2018.05.003.
- Shin C-S, Mishra P, Watrous JD, Carelli V, D’Aurelio M, Jain M, Chan DC. The glutamate/cystine xct antiporter antagonizes glutamine metabolism and reduces nutrient flexibility. Nat Commun. 2017;8(1):1–11. doi:10.1038/ncomms15074.
- Liu DS, Duong CP, Haupt S, Montgomery KG, House CM, Azar WJ, Pearson HB, Fisher OM, Read M, Guerra GR, et al. Inhibiting the system xC−/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat Commun. 2017;8(1):14844. doi:10.1038/ncomms14844.
- Muir A, Danai LV, Gui DY, Waingarten CY, Lewis CA, Vander Heiden MG. 2017. Environmental cystine drives glutamine anaplerosis and sensitizes cancer cells to glutaminase inhibition. eLife. 6. doi:10.7554/eLife.27713
- Lampa M, Arlt H, He T, Ospina B, Reeves J, Zhang B, Murtie J, Deng G, Barberis C, Hoffmann D, et al. Glutaminase is essential for the growth of triple-negative breast cancer cells with a deregulated glutamine metabolism pathway and its suppression synergizes with mtor inhibition. PLoS ONE. 2017;12(9):e0185092. doi:10.1371/journal.pone.0185092.
- Fumagalli C, Ranghiero A, Gandini S, Corso F, Taormina S, De Camilli E, Rappa A, Vacirca D, Viale G, Guerini-Rocco E. Inter-tumor genomic heterogeneity of breast cancers: comprehensive genomic profile of primary early breast cancers and relapses. Breast Cancer Res. 2020;22(1):1–11. doi:10.1186/s13058-020-01345-z.
- Shroff EH, Eberlin LS, Dang VM, Gouw AM, Gabay M, Adam SJ, Bellovin DI, Tran PT, Philbrick WM, Garcia-Ocana A, et al. Myc oncogene overexpression drives renal cell carcinoma in a mouse model through glutamine metabolism. Proc Natl Acad Sci USA. 2015;112(21):6539–6544. doi:10.1073/pnas.1507228112.
- Li Y, Li N, Shi J, Ahmed T, Liu H, Guo J, Tang W, Guo Y, Zhang Q. 2019. Involvement of glutathione depletion in selective cytotoxicity of oridonin to p53-mutant esophageal squamous carcinoma cells. Front Oncol. 9:1525. doi:10.3389/fonc.2019.01525.
- Maddocks OD, Berkers CR, Mason SM, Zheng L, Blyth K, Gottlieb E, Vousden KH. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature. 2013;493(7433):542–546. doi:10.1038/nature11743.
- Toyoda M, Kaira K, Ohshima Y, Ishioka NS, Shino M, Sakakura K, Takayasu Y, Takahashi K, Tominaga H, Oriuchi N, et al. Prognostic significance of amino-acid transporter expression (lat1, asct2, and xct) in surgically resected tongue cancer. Br J Cancer. 2014;110(10):2506–2513. doi:10.1038/bjc.2014.178.
- Davey MG, Hynes SO, Kerin MJ, Miller N, Lowery AJ. Ki-67 as a prognostic biomarker in invasive breast cancer. Cancers Basel. 2021;13(17):4455. doi:10.3390/cancers13174455.
- Uribe-Querol E, Rosales C Neutrophils in cancer: two sides of the same coin. J Immunol res. 2015.
- Nagasaki T, Schuyler AJ, Zhao J, Samovich SN, Yamada K, Deng Y, Ginebaugh SP, Christenson SA, Woodruff PG, Fahy JV. 15lo1 dictates glutathione redox changes in asthmatic airway epithelium to worsen type 2 inflammation. J Clin Invest. 2022;132(1):132. doi:10.1172/JCI151685.
- Yang Y, Wang Y, Guo L, Gao W, Tang T-L, Yan M. Interaction between macrophages and ferroptosis. Cell Death Disease. 2022;13(4):355. doi:10.1038/s41419-022-04775-z.
- Goodall GJ, Wickramasinghe VO. Rna in cancer. Nat Rev Cancer. 2021;21(1):22–36. doi:10.1038/s41568-020-00306-0.
- Itai Y, Rappoport N, Shamir R. Integration of gene expression and DNA methylation data across different experiments. Nucleic Acids Res. 2023;51(15):7762–7776. doi:10.1093/nar/gkad566.
- Lu H, Zhou Q, He J, Jiang Z, Peng C, Tong R, Shi J. Recent advances in the development of protein–protein interactions modulators: mechanisms and clinical trials. Signal Transduct Target Ther. 2020;5(1):213. doi:10.1038/s41392-020-00315-3.
- Statello L, Guo C-J, Chen L-L, Huarte M. Gene regulation by long non-coding rnas and its biological functions. Nat Rev Mol Cell Biol. 2021;22(2):96–118. doi:10.1038/s41580-020-00315-9.
- Alla AS. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi:10.1126/science.aad3346.
- Curtis C, Shah SP, Chin S-F, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346. doi:10.1038/nature10983.
- Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, XS L. Timer: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307.
- Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. 2018. Methsurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 10:277–288. doi:10.2217/epi-2017-0118.
- Green A, Powe D, Rakha E, Soria D, Lemetre C, Nolan C, Barros F, Macmillan R, Garibaldi J, Ball G. Identification of key clinical phenotypes of breast cancer using a reduced panel of protein biomarkers. Br J Cancer. 2013;109(7):1886–1894. doi:10.1038/bjc.2013.528.
- Meyerholz DK, Beck AP. Principles and approaches for reproducible scoring of tissue stains in research. Lab Invest. 2018;98(7):844–855. doi:10.1038/s41374-018-0057-0.
- Green AR, Aleskandarany MA, Agarwal D, Elsheikh S, Nolan CC, Diez-Rodriguez M, Macmillan RD, Ball GR, Caldas C, Madhusudan S, et al. Myc functions are specific in biological subtypes of breast cancer and confers resistance to endocrine therapy in luminal tumours. Br J Cancer. 2016;114(8):917–928. doi:10.1038/bjc.2016.46.
- Craze ML, El-Ansari R, Aleskandarany MA, Cheng KW, Alfarsi L, Masisi B, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, et al. Glutamate dehydrogenase (glud1) expression in breast cancer. Breast Cancer Res Treat. 2019;174(1):79–91. doi:10.1007/s10549-018-5060-z.
- El Ansari R, Alfarsi L, Craze ML, Masisi BK, Ellis IO, Rakha EA, Green AR. The solute carrier slc7a8 is a marker of favourable prognosis in er-positive low proliferative invasive breast cancer. Breast Cancer Res Treat. 2020;181(1):1–12. doi:10.1007/s10549-020-05586-6.
- El Ansari R, Craze ML, Miligy I, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA, Green AR. The amino acid transporter slc7a5 confers a poor prognosis in the highly proliferative breast cancer subtypes and is a key therapeutic target in luminal b tumours. Breast Cancer Res. 2018;20(1):21. doi:10.1186/s13058-018-0946-6.
- Morotti M, Zois CE, El-Ansari R, Craze ML, Rakha EA, Fan SJ, Valli A, Haider S, Goberdhan DCI, Green AR, et al. Increased expression of glutamine transporter snat2/slc38a2 promotes glutamine dependence and oxidative stress resistance, and is associated with worse prognosis in triple-negative breast cancer. Br J Cancer. 2020;124(2):10.1038/s41416-020-01113–y. doi:10.1038/s41416-020-01113-y.
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19(5):403–410. 2002;41: 151-152, discussion 152-153. doi:10.1111/j.1365-2559.1991.tb00229.x.
- Senkus E, Kyriakides S, Ohno S, Penault-Llorca F, Poortmans P, Rutgers E, Zackrisson S, Cardoso F. Primary breast cancer: Esmo clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2015;26(5):v8–30. doi:10.1093/annonc/mdv298.