ABSTRACT
Colorectal cancer (CRC) is a malignancy with high incidence and poor prognosis. It is urgent to identify valuable biomarkers for early diagnosis and potent therapeutic targets. It has been reported that SATB1 is associated with the malignant progression in CRC. To explore the role of SATB1 in CRC progression and the underlying mechanism, we evaluated the expression of SATB1 in the paired CRC tissues with immunohistochemistry. The results showed that the expression of SATB1 in lymph node metastasis was higher than that in primary lesion, and that in distant organ metastasis was higher than that in primary lesion. The retrospective analysis showed that patients with high expression of SATB1 had a significantly worse prognosis than those with negative and moderate expression. In vitro experiments that employing SATB1 over-expressing and depleted CRC cell lines confirmed that SATB1 contributes to cell proliferation and colonization, while inhibiting cell motility. Furthermore, the tissue immunofluorescence assay, Co-IP and Western blot were conducted to reveal that SATB1 induced translocation of β-catenin and formed a protein complex with it in the nuclei. In conclusion, SATB1 mediated tumor colonization and β-catenin nuclear localization are associated with the malignant progression and poor prognosis of CRC.
Introduction
Colorectal cancer (CRC) is the most common malignancy and the second leading cause of cancer-associated deaths all over the world.Citation1–3 The low survival rate of CRC is largely due to a variety of complications caused by its metastasis. Although extensive studies have been conducted, the pathogenesis of CRC remains unclear. Due to the high morbidity and mortality, it is urgently necessary to identify more valuable biomarkers for early diagnosis and more potent therapeutic targets in CRC.
The nuclear matrix binding protein SATB1 has emerged to be a key regulator in chromatin remodeling.Citation4 SATB1 provides a regulatory network for orchestrating the transcription of a set of genes whose dysregulation directly affects the cellular phenotype. Modification of SATB1 was previously considered to be a dynamic molecular switch which can regulate the expression of a variety of genes in the development and differentiation of T cells.Citation5 Studies also demonstrated strong association between SATB1 and a large number of malignant tumors, including CRC.Citation6,Citation7 Our previous work with the clinical specimens showed that the expression level of SATB1 was significantly higher in the CRC primary lesions and lymph node metastases than in the intestinal mucosa, and the high expression of SATB1 was significantly correlated with poor differentiation of tumor tissues and the aberrant expression of β-catenin in CRC.Citation7
The Wnt/β-catenin signaling pathway plays a critical role in the formation and progression of CRC.Citation8,Citation9 At present, it has been widely used to explain the phenomena of epithelial-mesenchymal transition, metastasis and chemotherapy resistance of CRC. As one of the most evolutionary conserved pathways, Wnt signaling pathway is involved in multiple cellular events, such as cell proliferation and differentiation, largely by modulating gene transcription.Citation10
In the present study, we evaluated the expression level of SATB1 protein in paired human CRC tissue specimens and found that the abnormal high expression of SATB1 is associated with malignant progression of CRC. Then, with in vitro experiments, we confirmed that SATB1 promotes CRC cell proliferation and colonization. Furthermore, we observed that the overexpressed SATB1 induced nuclear accumulation of β-catenin both in vitro and in vivo, which indicated that SATB1 is involved in modulating β-catenin signaling in CRC.
Materials and methods
Patients
Two-hundred CRC specimens were identified retrospectively from Suzhou Hospital affiliated to Nanjing Medical University. These patients had undergone surgery with curative intent between 2011 and 2012. The cohort consisted of 200 cases of primary lesions of CRC, 80 cases of lymph node metastasis, 5 cases of distant organ metastasis, and 50 cases of normal para-carcinoma mucosa. The entire cohort consisted of 93 female and 107 male patients, with a mean age of 65 years, and a range of 35–90 years. All 200 cases of CRC in this cohort were adenocarcinoma, including 42 cases of well-differentiated adenocarcinoma, 127 cases of moderately differentiated adenocarcinoma, and 31 cases of poorly differentiated adenocarcinoma. All tumors were prepared into 4 μm slides, stained with hematoxylin and eosin, then applied with histopathological evaluation. Clinical data such as gender, age, degree of tumor differentiation, metastasis was obtained from the medical record. The study was approved by our Institutional Review Board.
Immunohistochemistry and Co-immunolocalization
Immunohistochemical analysis was performed on 3 μm tissue sections using Ventana BenchMark auto immunostainer. Antibodies against SATB1(1:50, ab92307, Abcam), β-catenin (1:1, β-catenin-1, DAKO) were used. Immunohistochemistry results of SATB1 were evaluated as follow. The nuclear intensity of SATB1 stain was graded as strong (3+), moderate (2+), weak (1+), or negative (0). The percentage of tumor cells stained with STAB1 was denoted as 4 (>75% tumor cells), 3 (51–75% tumor cells), 2 (26–50% tumor cells), 1 (2–25% tumor cells), or 0 (<1% tumor cells). The final score defined as intensity score × percentage score was calculated. High expression was defined as a score > 9, intermediate expression was defined as a score between 3 and 9, negative was defined as a score < 3. FITC and Rhodamine labeled secondary antibodies were used for immunofluorescence, and DAPI was used to stain the nucleus. The immunofluorescence sections were observed and photographed with Olympus FV1200 confocal laser microscope, and the images were analyzed with FV10-ASW 4.1 Viewer software.
Cell culture and Lenti-virus package
SW480, HCT116, SW620 and HEK293T cell lines were grown in DMEM (HyClone, Logan, UT) supplemented with 10% FBS (VACCA, Murphysboro, IL),100 U/ml penicillin, and .1 mg/ml streptomycin at 37°C in a humidified atmosphere of 5% CO2. HCT15 and Jurkat T cells were grown in RPMI 1640 (HyClone, Logan, UT).
The sequences encoding SATB1 cDNA and shRNA were subcloned into the pLVX lentiviral vector respectively from our previous plasmids.Citation11 The lentiviral vector, packaging plasmid and envelope plasmid were co-transfected into HEK293T cells using PEI. The supernatant containing virus was collected 72 hr after transfection. SW480 cells were then transduced with the lentivirus at a 1:1 ratio. Overexpression and knockdown of SATB1 were detected by RT-qPCR and immunoblotting analysis.
Western blot
Cells were cultured and treated as described. Cell pellets were lysed using RIPA buffer and the protein concentration were determined by the BCA protein assay kit. The cell extracts were separated in an 8% SDS-PAGE gel and then transferred to PVDF membranes. The blots were incubated with different antibodies against SATB1 (1:2000, ab92307, Abcam), β-Actin (1:2500, A1978, Sigma) at 4°C for 16 hr. After washing with TST, the membranes were incubated with anti-mouse antibodies (7076, Cell Signaling) for 1 hr at room temperature. Signals were detected by chemiluminescence using ECL kit (Amersham).
Soft agar assays
Cells (1 × 103 were collected and resuspend in 750 μl 2×DMEM. Mixed the agarose stock with the culture medium containing cells at a 1:1 ratio, and then overlaid on a .2% agarose base in 6-well plates. The colonies were stained with 5% MTT on the 21st day. The number of colonies was counted with ImageJ software.
Transwell assays
Cells (5 × 104 were collected and resuspend in 200 μl DMEM with 2% FBS, and added to the top chamber with 8-μm pore (Corning, USA). Complete medium with 10% FBS was added to the lower chamber. After 24 hr the cells were fixed and stained with .5% crystal violet for 30 min. The randomly selected fields were observed with a microscope and photographed. The number of cells was evaluated with software.
Cell counting kit-8 viability assays
1 × 103 cells were seeded in 96-well plates with 200 μl medium. Add 10% CCK-8 (Vazyme, China) in each well and incubate at 37°C for 30 min. The absorbance at 450 nm was measured every day. Cell proliferation was calculated as the ratio of the absorbance (OD) value at day 1 to the OD value of the sample.
Preparation of nuclear extracts
Nuclear protein was extracted using NE-PERTM kit (Thermo Fisher) according to the manufacturer’s instructions. Cells were harvested and washed with cold PBS. Add ice-cold CER I to resuspend the cell pellets and then incubated on ice for 10 min. Add ice-cold CER II to the tube, and incubate on ice for 1 min. After that samples were centrifuged at 12,000 g for 5 min. The insoluble fraction produced were treated with ice-cold NER, and continue vortexing for 15s every 10 min, for a total of 40 min. After centrifugation at 12,000 g for 10 min, the remaining supernatant was the nuclear extract. The protein concentration was determined by the BCA protein assay kit.
Co-immunoprecipitation
1 mg total protein of cells were diluted with 1 ml RIPA buffer that containing protease inhibitors. Each sample was incubated with 10 μg antibody (anti-SATB1 or anti-β-catenin) at 4°C overnight. Control samples were incubated with nonimmune, species-specific IgG. The protein complexes were enriched by co-incubating with Protein A/G agarose beads (Santa Cruz) at 4°C for 4 hr. Then the beads were centrifuged at 2,000 g for 1 min. Pellets were collected and washed with RIPA buffer for 5 times and then resuspended in 40 µl of 1×Electrophoresis Sample Buffer. After boiling for 5 min, the samples were applied to Western blot.
Statistical analysis
Analyses were performed using GraphPad Prism 7.0 and SPSS statistics 26. Differences between groups were reported as mean ± SD, and a two-tailed T-test was used to assess differences. To estimate the statistical differences between more than two groups, one-way analysis of variance (ANOVA) was used. Kaplan-Meier method and Log-Rank test were used for survival analysis. A p value < .05 was considered statistically significant.
Results
SATB1 is highly expressed in the CRC tissues and is associated with poor prognosis
To evaluate the SATB1 protein expression level in Chinese CRC cohort, we inspected SATB1 protein level in 200 specimens of CRC lesions and 50 specimens of para cancer colorectal mucosa using immunohistochemistry. The IHC staining showed that SATB1 was positively expressed in a few normal tissues and the most of tumor tissues, localized in the nucleus (). As summarized in and , the high and moderate expression rates of SATB1 in tumor tissue were obviously higher than in normal mucosa. In follow-up and survival analysis of these 200 patients, we found that patients with high expression of SATB1 (12 of 37 cases survived) had worse prognosis, while patients with negative and moderate expression of SATB1 had no significant difference in prognosis (40/67, 55/96 survivors, respectively) ().
Figure 1. Expression of SATB1 in the CRC tissues and its association with prognosis. (a) SATB1 staining in normal mucosal glandular epithelium and CRC tissues. In normal intestinal mucosa, SATB1 showed no staining in epithelial cells, whereas it showed strong positive staining in interstitial lymphocytes. In the tumor epithelial cells, SATB1 showed variable staining, including negative, moderate and strong staining. (b) The number of cases with different degree of SATB1 expression. (c) Kaplan-Meier survival curve of CRC patients with different degree of SATB1 expression. *p < .05 (log-rank test).
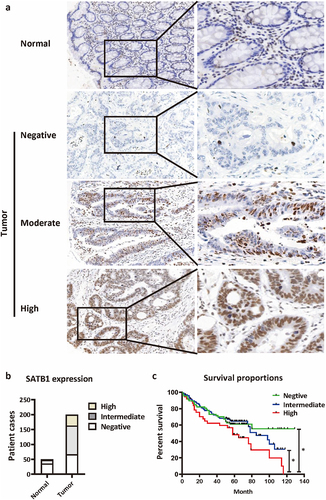
Table 1. SATB1 expression in primary CRC and lymph node metastasis.
SATB1 expression in the metastatic lesions is higher than that in primary lesions of CRC
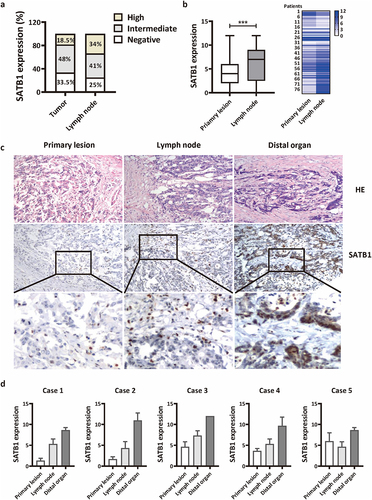
To further investigate the correlation of SATB1 protein level with metastatic potential of CRC, we further examined SATB1 protein level in 80 corresponding specimens of lymph node metastasis and 5 specimens of distant organ metastasis. The results showed that the metastatic lymph nodes with high expression of SATB1 account for about 34% of the total. This rate is nearly double that of the primary lesion (18.5%) ( and ). Comparing 80 cases of the paired primary lesion and lymph node metastases, we found that the expression of SATB1 was significantly higher in the latter than in the former (). Moreover, in CRC patients with distant metastasis, SATB1 expression level increased gradually from the primary lesion to the lymph node and then to the distal organ (). The distant metastatic organs of these 5 patients were left ovary, right ovary, peritoneum, right kidney and posterior vaginal wall. The expression of SATB1 was low in the primary tumor tissues of case 1 and case 2, and moderate expression was detected in the corresponding lymph node metastatic lesions. In the distant metastatic lesions, the expression was further enhanced, and almost all the cancer cells showed brown nuclear staining, indicating strong positive expression. In case 3 and case 4, SATB1 protein was moderately expressed in some areas of the primary tumor. SATB1 was also moderately expressed in the lymph node metastasis, but strongly expressed in the distant metastasis. In case 5, the expression of SATB1 protein was moderate in the primary tumor, slightly decreased in the lymph node metastasis, but further increased in the distant metastasis ().
Figure 2. SATB1 expression in the metastatic lesions is higher than that in primary lesions of CRC. (a) The percentage of negative, intermediate and high SATB1 expression in 200 primary cancer tissue and 80 lymph node metastases. (b) The expression of SATB1 in 80 paired primary cancer tissue and lymph node metastases. ***p < .001 (paired t-test). (c) SATB1 staining in primary cancer tissue, lymph node metastasis and distant organ metastasis from one patient. (d) Semi-quantitative analysis of SATB1 expression in five cases with paired primary tumors, lymph node metastases, and distant metastases.
SATB1 promotes CRC cell proliferation and colony formation
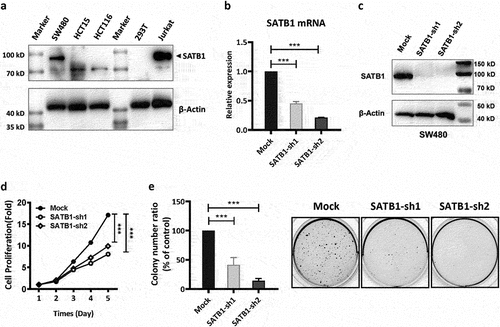
To explore the role of SATB1 in the progression of colorectal cancer, we first detected expression of SATB1 in three CRC cell lines, including SW480, HCT15 and HCT116. Western blot showed a certain amount of SATB1 expression was detected in SW480 (). Then knockdown using two different shRNA for SATB1 in SW480 was confirmed by Immunoblot analysis and Q-PCR (). The CCK-8 assay showed SATB1-silenced SW480 cells grew at a significantly lower rate than mock-transfected cells (). The soft agar assay also indicated knocking down SATB1 expression significantly suppressed the anchorage-independent growth ability of SW480 cells (). Interestingly, although an accelerated cell proliferation rate was observed in the SATB1-overexpressing SW480 cells, the enhanced expression of SATB1 did not exhibit dramatic effect on colony formation (Figure S1).
Figure 3. SATB1 promote CRC proliferation and colony formation. (a)The expression of SATB1 protein in SW480, HCT15, HCT116 and 293T cells was detected by western blot. (b), (c) RT-PCR and western blot were used to verify the interference results of SATB1 protein expression in SW480 cells. (d) CCK-8 assay showed that the proliferation ability of SATB1-knockdown cell line was decreased. (e) Clone formation experiment showed that SATB1-knockdown cell lines had reduced clone formation. The results represent at least three independent experiments. The data are represented as the mean ± SD ***p < .001 (two-tailed student’s t-test).
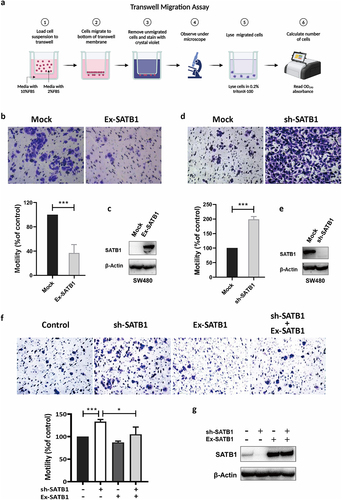
SATB1 inhibits CRC cell migration
To further investigate whether SATB1 promotes tumor metastasis through regulating cell motility, we performed the transwell assay with SATB1-overexpressing SW480 cells. The results showed that cells with highly expressed SATB1 exhibited less aggressive migratory potential (). In contrast, depletion of SATB1 significantly enhanced the migration of CRC cells (). We then overexpressed SATB1 in the SATB1 knockdown cells with lentivirus (). The transwell assay showed that rescuing SATB1 knockdown reduced cell motility (). We propose that SATB1 mediated inhibitory effect on cell migration may contribute to the colonization of metastatic cells.
Figure 4. SATB1 inhibit CRC migration. (a) Schematic diagram illustrating transwell migration assay (created in BioRender.com). (b), (c) Western blot assay verified the SATB1-overexpression cell line, transwell assay showed that the migration ability of the overexpression cell line was weakened. (d), (e) the SATB1-knockdown cell line was verified by western blot, transwell assay showed that the migration ability of the knockout cell line was enhanced. (f) Transwell assay showed that rescuing SATB1 knockdown reduced cell migration ability. (g) Western blot verified SATB1 expression in each cell line. The results represent at least three independent experiments. The data are represented as the mean ± SD ***p < .001 (two-tailed Student’s t-test).
High expression of SATB1 is associated with nuclear translocation of β-catenin in CRC tissues
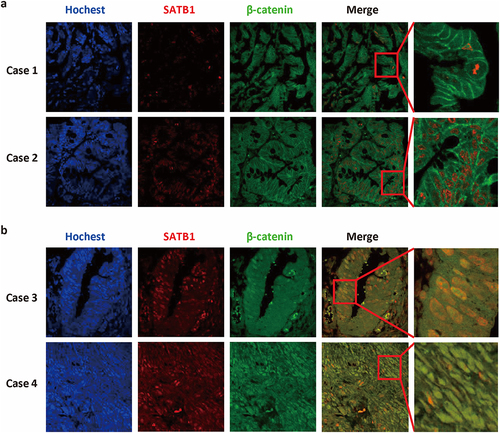
The canonical Wnt pathway that involves the nuclear translocation of β-catenin mainly controls cell proliferation. A large number of studies have shown that activating mutations of the canonical Wnt/β-catenin signaling can be detected in more than 90% of CRC tissues. Our previous work has shown that high level of SATB1 protein was positively correlated with a reduced level of membrane-bound β-catenin in CRC cells. Therefore, we further examined the subcellular localization of β-catenin and SATB1 in colorectal cancer tissues by immunofluorescence. The result of double staining showed that SATB1, marked red by Rhodamine, is located in the nucleus, while β-catenin, green by FITC, is normally located in the cell membrane in the cases with no or low expression of SATB1 (). In the cases with high expression level of SATB1, the staining intensity of β-catenin was stronger, and the co-localization of the two proteins, SATB1 and β-catenin, was observed in nuclei ().
SATB1 facilitated nuclear localization of β-catenin
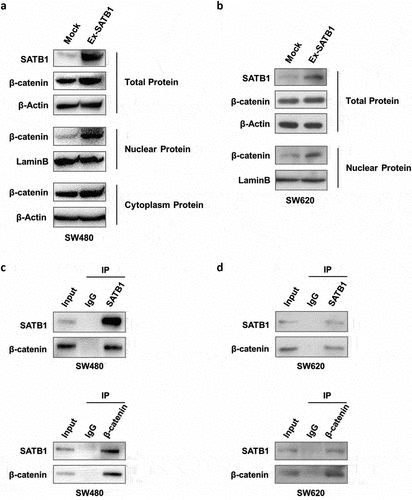
To explore the relationship between SATB1 and β-catenin, we conducted Western blot with whole cell lysate, nuclear and cytoplasmic protein respectively to analyze the expression of β-catenin in SATB1-overexpressing SW480 cells. The results revealed that SATB1 induced β-catenin expression, and facilitated nuclear accumulation of β-catenin. We then overexpressed SATB1 transiently in SW620, another SATB1 positive CRC cell line (Figure S2), and also observed nuclear accumulation of β-catenin (). The co-immunoprecipitation assay further confirmed that the translocated β-catenin interacted with SATB1 to form a protein complex in both SW480 cells and SW620 cells, which indicated that SATB1 is involved in modulating β-catenin signaling ().
Figure 6. SATB1 regulated β-catenin at multiple levels. (a) Western blot detected the expression of SATB1 and β-catenin with the whole cell lysate, nuclear and cytoplasmic extract of SW480 cells. (b) Western blot detected the expression of SATB1 and β-catenin with the whole cell lysate and nuclear extract of SW620 cells. (c), (d) Co-immunoprecipitation detect SATB1/β-catenin protein interaction in SW480 and SW620 cells.
Discussion
In this study we reported a SATB1 expression pattern in CRC. Our retrospective study revealed that the protein level of SATB1 is associated with metastasis potential and the poor prognosis of CRC. With in vitro experiments we further confirmed that SATB1 induced β-catenin translocated into the nuclear and promotes colorectal cancer tumorigenesis.
The mechanisms of SATB1 in regulating T cell development and differentiation has been well established.Citation12,Citation13 and the role of SATB1 in cancer development has attracted more and more attention in recent years. Current reports have shown that increased expression of SATB1 is associated with a variety of cancers, including hepatocellular carcinoma,Citation14 gastric cancer,Citation15 ovarian cancer,Citation16 and laryngeal squamous cell carcinoma.Citation17 However, the correlation between SATB1 expression and tumor biological behavior is controversial. Most studies have suggested that overexpression of SATB1 is closely related to low differentiation, high stage, and poor prognosis of tumors. The study by Selinger et al. revealed that SATB1 protein loss predicted a poorer prognosis in lung squamous cell carcinoma.Citation18 Al-Sohaily also reported that loss of SATB1 protein nuclear expression in colorectal cancer implies poor prognosis and insensitivity to adjuvant chemotherapy.Citation19 Other studies suggest that SATB1 is not an independent prognostic factor and is only associated with prognosis in SATB2-negative tumors.Citation20 In our study, we found that more abundant expression of SATB1 can be detected in the metastatic foci. However, the overexpressed SATB1 in CRC cells exhibits dual function, promoting single-cell colonization and inhibiting cell motility. Different from cell lines in vitro, the tumor tissue is a heterogeneous population of cells. Early observations showed that tumors contain subclones that differ in respect to karyotype and sensitivity to chemotherapy.Citation21 More recent profiling endeavors, using Single-Cell Sequencing of various tumors, revealed multiple clones with distinct gene expression pattern within a single tumor.Citation22 We proposed that tumor metastasis begins with a mass incident involving multiple subclones. Different clones have different roles in the organization. Some are responsible for degrading ECM, some are responsible for damaging barriers. Finally, only a few tumor cells reach the specific distant organs, and colonize to form metastatic tumor. Although SATB1 may not be beneficial for tumor cells to break through the endothelial barrier, the SATB1-positive subclone would have advantages to survive the harsh environment and colonized. Therefore, identification of biomarkers for predicting tumor metastasis and prognosis still needs to analyze the characteristics of multiple cell populations in the tumor tissues.
The Wnt signaling pathway plays an important role in tumor progression and metastasis by regulating a series of cellular events.Citation23–25 The Wnt signaling pathways include canonical and noncanonical pathways.Citation26 In the canonical Wnt pathway, signaling triggers the nuclear translocation of β-catenin, and activates the transcription of the downstream genes via TCF/LEF complex. So, it is also known as Wnt/β-catenin pathway, and is regarded as a manipulator of cell proliferation. It has been reported that SATB1 is also involved in the Wnt/β-catenin regulatory network. SATB1 is one of the target genes of Wnt/β-catenin pathway, the translocated β-catenin induces SATB1 expression through TCF7L2. Then the up-regulated SATB1 interacts with β-catenin, and subsequently binds to Wnt responsive genes to induce their transcription.Citation27 In our study, we also observed the co-localization of SATB1 and β-catenin in the CRC tissues. Then we demonstrated that SATB1 contributes to the nuclear accumulation of β-catenin and form the SATB1/β-catenin complex. Therefore, we proposed that the two proteins form a positive feedback network of mutual regulation. SATB1 depletion resulted reduction of proliferation and tumorigenic potential of CRC cell line may be caused by modulating of Wnt/β-catenin pathway. However, the mechanism by which SATB1, a nuclear matrix binding protein, regulates nuclear translocation of β-catenin remains to be further investigation.
In summary, this study described that SATB1 mediated tumorigenesis is associated with malignant progression and poor prognosis of CRC. And the results of in vitro and in vivo experiments suggest SATB1 is involved in modulating β-catenin signaling. Further elucidation of SATB1/β-catenin regulatory network will provide a promising target for the treatment of CRC.
Authors’ contributions
L.S. and J.H.L. designed the study and analyzed the data. F.W. contributed to histology review. X.F.W. and F.Y.Z. performed the in vitro experiment. S.J.M. collected the data. All authors read and approved the final manuscript.
Ethical approval and consent to participate
This publication is approved by the Ethics Committee of the Affiliated Suzhou Hospital of Nanjing Medical University (K-2022-018-K01). The participants in the study gave verbal regarding.
Supplementary Data_240131.docx
Download MS Word (283 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The datasets used and analyzed in the present study are available from the corresponding author upon reasonable request.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2024.2320307
Additional information
Funding
Notes on contributors
Luan Sun
Luan Sun received her B.S. and Ph.D. degree from Nanjing Medical University in 2006, and 2011. Currently, she is an associate professor in the Department of Cell Biology at Nanjing Medical University College of Basic Medical Sciences. Her research focuses on T-cell biology and CAR-T cell therapy.
Feng Wang
Feng Wang received her B.S. and M.S. degree in School of Medicine from Soochow University in 2004 and 2014. Now she is an associate chief physician in the Department of Pathology of Suzhou Municipal Hospital. Her research focuses on pathological diagnosis of digestive tract tumors.
Xufei Wang
Xufei Wang studies a Master’s degree in Cell Biology at Nanjing Medical University. His research focuses on the mechanisms of CAR-T cell exhaustion.
Feiying Zhang
Feiying Zhang studies a Bachelor degree in Clinical Medicine Science at Nanjing Medical University.
Sujuan Ma
Sujuan Ma received her B.S. degree from Liaocheng University in 2008 and M.S. degree from Xiamen University. She is currently working in the Department of Cell Biology at Nanjing Medical University College of Basic Medical Sciences.
Jinghuan Lv
Jinghuan Lv received her B.S. and M.S. degree from Nanjing Medical University in 2003 and 2009. She received her Ph.D. degree in Pathology from Nanjing University in 2017. Now she is an associate professor of Nanjing Medical University, and director of the Department of Pathology at Suzhou Municipal Hospital. Her research focuses on the diagnosis and pathogenesis of gastrointestinal tumors.
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA-Cancer J Clin. 2020;70(1):7–11. doi:10.3322/caac.21590.
- Siegel RL, Miller KD, Sauer AG, Fedewa SA, Butterly LF, Anderson JC, Cercek A, Smith RA, Jemal A. Colorectal cancer statistics, 2020. CA-Cancer J Clin. 2020;70(3):145–164. doi:10.3322/caac.21601.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LAJemal A, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA-Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492.
- Naik R R, Galande S. SATB family chromatin organizers as master regulators of tumor progression. Oncogene. 2019;38(12):1989–2004. doi:10.1038/s41388-018-0541-4.
- Pavan Kumar P, Purbey PK, Sinha CK, Notani D, Limaye A, Jayani RS, Galande S. Phosphorylation of SATB1, a global gene regulator, acts as a molecular switch regulating its transcriptional activity in vivo. Mol Cell. 2006;22(2):231–43. doi:10.1016/j.molcel.2006.03.010.
- Mir R, Pradhan SJ, Patil P, Mulherkar R, Galande S. Wnt/β-catenin signaling regulated SATB1 promotes colorectal cancer tumorigenesis and progression. Oncogene. 2016;35(13):1679–1691. doi:10.1038/onc.2015.232.
- Lv JH, Wang F, Shen MH, Wang X, Zhou X-J. SATB1 expression is correlated with β-catenin associated epithelial–mesenchymal transition in colorectal cancer. Cancer Biol Ther. 2016;17(3):254–261. doi:10.1080/15384047.2016.1139239.
- Zhang XL, Li CC, Wu Y, Cui P. The research progress of Wnt/β-catenin signaling pathway in colorectal cancer. Clin Res Hepatol Gastroenterol. 2023;47(3). doi:10.1016/j.clinre.2023.102086.
- Disoma C, Zhou YZ, Li SN, Peng J, Xia Z. Wnt/β-catenin signaling in colorectal cancer: is therapeutic targeting even possible? Biochimie. 2022. 195:39–53. doi:10.1016/j.biochi.2022.01.009.
- Liu JQ, Xiao Q, Xiao JN, Niu CX, Li YY, Zhang XJ, Zhou ZW, Shu G, Yin G. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7(1). doi:10.1038/s41392-021-00762-6.
- Gong F, Sun L, Wang Z, Shi J, Li W, Wang S, Han X, Sun Y. The BCL2 gene is regulated by a special AT-rich sequence binding protein 1-mediated long range chromosomal interaction between the promoter and the distal element located within the 3′-UTR. Nucleic Acids Res. 2011;39(11):4640–4652. doi:10.1093/nar/gkr023.
- Burute M, Gottimukkala K Galande K, Galande S. Chromatin organizer SATB1 is an important determinant of T-cell differentiation. Immunol Cell Biol. 2012;90(9):852–9. doi:10.1038/icb.2012.28.
- Kitagawa Y, Ohkura N, Kidani Y, Vandenbon A, Hirota K, Kawakami R, Yasuda K, Motooka D, Nakamura S, Kondo M, et al. Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat Immunol. 2017;18(2):173–183. doi:10.1038/ni.3646.
- Tu W, Luo M, Wang Z, Yan W, Xia Y, Deng H, He J, Han PTian P, Tian D. Upregulation of SATB1 promotes tumor growth and metastasis in liver cancer. Liver Int. 2012;32(7):1064–78. doi:10.1111/j.1478-3231.2012.02815.x.
- Yuan CL, Li L, Zhou X, Han L. Expression of SATB1 and HER2 in gastric cancer and its clinical significance. Eur Rev Med Pharmacol Sci. 2016;20(11):2256–64.
- Xiang J, Zhou L, Li S, Xi X, Zhang J, Wang Y, Yang Y, Liu X, Wan X. AT-rich sequence binding protein 1: contribution to tumor progression and metastasis of human ovarian carcinoma. Oncol Lett. 2012;3(4):865–870. doi:10.3892/ol.2012.571.
- Li YC, Bu LL, Mao L, Ma SR, Liu JF, Yu GT, Deng WW, WF Z, ZJ S. SATB1 promotes tumor metastasis and invasiveness in oral squamous cell carcinoma. Oral Dis. 2017;23(2):247–254. doi:10.1111/odi.12602.
- Selinger CI, Cooper WA, Al-Sohaily S, Mladenova DN, Pangon L, Kennedy CW, McCaughan BC, Stirzaker C, MR K-C. Loss of special AT-rich binding protein 1 expression is a marker of poor survival in lung cancer. J Thorac Oncol. 2011;6(7):1179–1189. doi:10.1097/JTO.0b013e31821b4ce0.
- Al-Sohaily S, Henderson C, Selinger C, Pangon L, Segelov E, MR K-C, Warusavitarne J. Loss of special AT-rich sequence-binding protein 1 (SATB1) predicts poor survival in patients with colorectal cancer. Histopathology. 2014;65(2):155–63. doi:10.1111/his.12295.
- Nodin B, Johannesson H, Wangefjord S, O’Connor DP, Lindquist KE, Uhlen M, Jirstrom K Eberhard K. Molecular correlates and prognostic significance of SATB1 expression in colorectal cancer. Diagn Pathol. 2012;7(1):115. doi:10.1186/1746-1596-7-115.
- Shapiro JR, WK Y, WR S. Isolation, karyotype, and clonal growth of heterogeneous subpopulations of human malignant gliomas. Cancer Res. 1981;41(6):2349–2359.
- Lawson DA, Kessenbrock K, Davis RT, Pervolarakis N Werb N, Werb Z. Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol. 2018;20(12):1349–1360. doi:10.1038/s41556-018-0236-7.
- Kormish JD, Sinner D, AM Z. Interactions between SOX factors and Wnt/beta-catenin signaling in development and disease. Dev Dynam. 2010;239(1):56–68. doi:10.1002/dvdy.22046.
- Birchmeier W. The Wnt/beta-catenin signaling pathway in development and disease. Pathol Res Pract. 2007;203(5):341–341.
- Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127(3):469–480. doi:10.1016/j.cell.2006.10.018.
- Veeman MT, Axelrod JD, Moon RT. Axelrod JDMoon RT. A second canon: functions and mechanisms of beta-catenin-independent wnt signaling. Dev Cell. 2003;5(3):367–377. doi:10.1016/S1534-5807(03)00266-1.
- Mir R, Pradhan SJ, Patil P, Mulherkar R, Galande S. Wnt/beta-catenin signaling regulated SATB1 promotes colorectal cancer tumorigenesis and progression. Oncogene. 2016;35(13):1679–1691. doi:10.1038/onc.2015.232.