ABSTRACT
Circular RNA Ribonuclease P RNA Component H1 (circ_RPPH1) and microRNA (miRNA) miR-1296-5p play a crucial role in breast cancer (BC), but the molecular mechanism is vague. Evidence showed that miR-1296-5p can activate tripartite motif-containing 14 (TRIM14). Clinical indications of eighty BC patients were collected and the circ_RPPH1 expression was detected using real-time quantitative PCR. MCF-7 and MDA-MB-231 cells were transfected with overexpression or knockdown of circ_RPPH1, miR-1296-5p, or TRIM14. Cell counting kit-8, cell cloning formation, wound healing, Transwell, and flow cytometry assays were performed to investigate the malignant phenotype of BC. The dual-luciferase reporter gene analyses were applied to reveal the interaction between these target genes. Subcutaneous tumorigenic model mice were established with circ_RPPH1 overexpression MDA-MB-231 cells in vivo; the tumor weight and volume, levels of miR-1296-5 and TRIM14 mRNA were measured. Western blot and immunohistochemistry were used to detect TRIM14 in cells and mice. Circ_RPPH1 levels were notably higher in BC patients and have been found to promote cell proliferation, invasion, and migration of BC cells. Circ_RPPH1 altered cell cycle and hindered apoptosis. Circ_RPPH1 knockdown or miR-1296-5p overexpression inhibited the malignant phenotype of BC. Furthermore, miR-1296-5p knockdown reversed circ_RPPH1’s promotion effects on BC. Interestingly, TRIM14 overexpression counteracts the inhibitory effects of miR-1296-5p overexpression and circ_RPPH1 silencing on BC. Moreover, in BC tumor-bearing mice, circ_RPPH1 overexpression led to increased TRIM14 expression and facilitated tumor growth. Circ_RPPH1 enhanced BC progression through miR-1296-5p/TRIM14 axis, indicating its potential as a biomarker and therapeutic target in BC.
1. Introduction
Breast cancer (BC) is a malignant tumor in women worldwide,Citation1 and has become a major threat to the life health of women. Due to its high morbidity and mortality rates, 2.26 million women have been affected with BC in 2020Citation2 and there are 1.7 million new incidences every year.Citation3 BC is diagnosed by breast self-examination and clinical breast examination,Citation4 and mammography or ultrasonography is used for BC screening.Citation5,Citation6 The Tumor-Node-Metastasis (TNM)-based staging of BC represents different severity of BC, the larger number is on behalf of the more serious BC.Citation7 In recent years, surgery, endocrine drugs, or targeted drug therapyCitation8 and radiotherapyCitation9 have been performed in BC treatment. In addition, because of triple-negative BC, endocrine therapy brings little effect.Citation10 So, the mechanism of BC remains to be further studied and elucidated. Therefore, we need to pay more attention to studying the biomarkers and the potential mechanisms in BC.
Circular RNA (circRNA) Ribonuclease P RNA Component H1 (circ_RPPH1) is a highly expressed circRNA in BC. Evidence shown that circ_RPPH1 has a higher expression level in BC tissues compared to adjacent tissues.Citation11,Citation12 Studies have shown that circ_RPPH1 knockout delays tumor deterioration in BC.Citation13 Evidence has shown that circRNA plays a crucial role in malignant tumors through microRNAs (miRNAs) as a sponge.Citation14 Circ_RPPH1 sponges miR-296-5p and miR-146b-3p to promote BCCitation15,Citation16 suggesting that circ_RPPH1 has a crucial role in regulating BC as a miRNA sponge.
MiRNAs play crucial roles in proliferation, migration, invasion and metastasis of BC cells.Citation17,Citation18 Among them, miR-1296-5p has drawn considerable attention in recent years. MiR-1296-5p shows low expression in BC tissues and cells, and overexpression of miR-1296-5p inhibits BC malignant phenotype.Citation19 Moreover, miR-1296-5p promotes the sensitivity of BC cells to cisplatin and 5-fluorouracil.Citation20 Tripartite motif-containing (TRIM) 14 was a downstream target of miR-1296-5p, and circ_0048764 can activate the TRIM14 in BC cells by targeting miR-1296-5p.Citation21 Although several studies of circRNA-miRNA-mRNA regulatory axis in BC have been done, how circ_RPPH1 regulates the miR-1296-5p/TRIM14 axis is still unclear. Here, we find that circ_RPPH1 is elevated in BC and the clinical indications expression is related to increased circ_RPPH1 in BC patients. Additionally, we suspect that circRNAs could promote tumor progression of BC by inhibiting miR-1296-5p. We performed lentivirus or plasmid transfection on BC cells to study the effects of circ_RPPH1 on BC promotion and the mechanism of circ_RPPH1 on activating TRIM14 through inhibiting miR-1296-5p. Subcutaneous tumorigenic model mice were established with BC subcutaneous tumors to verify the mechanism of circ_RPPH1 on BC tumor Growth.
2. Results
2.1. The increased circ_RPPH1 expression in tumor tissues of BC patients
We used UALCAN database to analyze the circ_RPPH1 expression in breast invasive carcinoma (BRCA) from TGCA. The circ_RPPH1 expression is increased in primary tumor group compared to the normal group, with a statistical significance (Supplementary figure S1). The circ_RPPH1 expression in BC tissues of patients was detected using real-time quantitative PCR (RT-qPCR) (). The mRNA expression of circ_RPPH1 in BC tissues was higher than in pa-carcinoma tissues. A chi-square test was performed to examine the difference in frequency distribution. Results are as shown in . The frequency distribution differences of high and low circ_RPPH1 expression was significantly different in TNM staging. Additionally, there was no significant difference in age, tumor size, pausimenia or not, lymph node metastasis (N1) or not (N0), ER, PR, HER2.
Figure 1. Circ_RPPH1 overexpression promoted malignant phenotype of BC cells. (a) Real-time quantitative PCR (RT-qPCR) was used to detect circ_RPPH1 expression in patients with BC; circ_RPPH1 mRNA expression in BC tissues was higher than in para-carcinoma tissues (n = 80). (b-c) Cell counting kit-8 assay was used to measure the cell viability of BC cells; circ_RPPH1 overexpression [circ_RPPH1(+)] increased cell viability of MCF-7 cells and MDA-MB-231 cells (n = 6). The cell viability (%) = (OD450 of the experimental group – average OD450 of the blank group)/(average OD450 of the Control group – average OD450 of the blank group) × 100%. (d-e) Cell cloning formation assay was used to detect the proliferation of BC cells; circ_RPPH1 (+) promoted proliferation of MCF-7 cells and MDA-MB-231 cells (n = 3). (f-g) Cell wound healing assay was used for cell migration detection; circ_RPPH1 (+) promoted migration of MCF-7 cells and MDA-MB-231 cells (n = 3, scale bar = 200 μm). The cell migration rate (%) = (0 h scratch width −24 h scratch width)/0 h scratch width × 100%. (h-i) Transwell assay was used to detect the invasion of BC cells; circ_RPPH1 (+) promoted invasion of MCF-7 cells and MDA-MB-231 cells (n = 3, scale bar = 50 μm). Data were presented as the mean ± SD. **p < .01 vs. the para-carcinoma tissue; ▲▲p < .01 vs. the Control group.
![Figure 1. Circ_RPPH1 overexpression promoted malignant phenotype of BC cells. (a) Real-time quantitative PCR (RT-qPCR) was used to detect circ_RPPH1 expression in patients with BC; circ_RPPH1 mRNA expression in BC tissues was higher than in para-carcinoma tissues (n = 80). (b-c) Cell counting kit-8 assay was used to measure the cell viability of BC cells; circ_RPPH1 overexpression [circ_RPPH1(+)] increased cell viability of MCF-7 cells and MDA-MB-231 cells (n = 6). The cell viability (%) = (OD450 of the experimental group – average OD450 of the blank group)/(average OD450 of the Control group – average OD450 of the blank group) × 100%. (d-e) Cell cloning formation assay was used to detect the proliferation of BC cells; circ_RPPH1 (+) promoted proliferation of MCF-7 cells and MDA-MB-231 cells (n = 3). (f-g) Cell wound healing assay was used for cell migration detection; circ_RPPH1 (+) promoted migration of MCF-7 cells and MDA-MB-231 cells (n = 3, scale bar = 200 μm). The cell migration rate (%) = (0 h scratch width −24 h scratch width)/0 h scratch width × 100%. (h-i) Transwell assay was used to detect the invasion of BC cells; circ_RPPH1 (+) promoted invasion of MCF-7 cells and MDA-MB-231 cells (n = 3, scale bar = 50 μm). Data were presented as the mean ± SD. **p < .01 vs. the para-carcinoma tissue; ▲▲p < .01 vs. the Control group.](/cms/asset/8d106525-b4d8-44eb-b1e6-fe57c2a547f8/kcbt_a_2360768_f0001_oc.jpg)
Table 1. Correlation analysis between clinical indications and circ_RPPH1 expression in clinical BC samples (n = 80).
2.2. Circ_RPPH1 promotes cell mobility and inhibits cell apoptosis in BC cells
The circ_RPPH1 overexpression [circ_RPPH1 (+)] cells were successfully established (supplementary figure S2a). In , cell viability in circ_RPPH1 (+) group was increased. To further study the mechanisms of circ_RPPH1 on BC cell mobility, cell cloning formation, wound healing and Transwell assays were performed. showed that circ_RPPH1 increased the number of colonies in BC cells. In , the cell migration of both cells was increased by circ_RPPH1. Moreover, in , circ_RPPH1 elevated the number of invaded cells. These results indicated that circ_RPPH1 promoted the cell reproductive capacity, cell migration capacity and cell invasion capacity of BC cells.
Compared to negative control (NC) group, circ_RPPH1 (+) group had a significantly decreased percentage of G0/G1 and G2/M cells, and cells in the S phase were accordingly increased (). From the results of flow cytometry (FCM) in , circ_RPPH1 overexpressing inhibited the BC cell apoptosis rate. Additionally, Western blot was used to measure TRIM14 protein expression and showed that TRIM14 protein expression was increased in circ_RPPH1 (+) groups ().
Figure 2. Circ_RPPH1 overexpression promoted BC cell entry into S phase and inhibited cell apoptosis. (a-c) Flow cytometry was used to detect the cycle of BC cells; circ_RPPH1 overexpression [circ_RPPH1(+)] decreased cells in the G0/G1 and G2/M phase, as well as promoting cells entry into the S phase in MCF-7 cells and MDA-MB-231 cells. (d-e) Flow cytometry was also used to detect the apoptosis; circ_RPPH1 (+) inhibited apoptosis in MCF-7 cells and MDA-MB-231 cells. (f) Western blot assay was used to detect TRIM14 protein expression. Circ_RPPH1 (+) increased TRIM14 protein expression in MCF-7 and MDA-MB-231 cells. Data were presented as the mean ± SD, n = 3. ▲▲p < .01 vs. the Control group.
![Figure 2. Circ_RPPH1 overexpression promoted BC cell entry into S phase and inhibited cell apoptosis. (a-c) Flow cytometry was used to detect the cycle of BC cells; circ_RPPH1 overexpression [circ_RPPH1(+)] decreased cells in the G0/G1 and G2/M phase, as well as promoting cells entry into the S phase in MCF-7 cells and MDA-MB-231 cells. (d-e) Flow cytometry was also used to detect the apoptosis; circ_RPPH1 (+) inhibited apoptosis in MCF-7 cells and MDA-MB-231 cells. (f) Western blot assay was used to detect TRIM14 protein expression. Circ_RPPH1 (+) increased TRIM14 protein expression in MCF-7 and MDA-MB-231 cells. Data were presented as the mean ± SD, n = 3. ▲▲p < .01 vs. the Control group.](/cms/asset/fa0153de-2a90-4442-9a20-81cb422b5551/kcbt_a_2360768_f0002_oc.jpg)
2.3. Circ_RPPH1 promotes cell mobility and inhibits cell apoptosis in BC cells by inhibiting miR-1296-5p
revealed the predicted results of the binding situation of circ_RPPH1 and miR-1296-5p that the target sequence of miR-1296-5p was located at 23–29 bp of circ_RPPH1 gene 3’-UTR. In , in the circ_RPPH1 WT, compared with the NC group, the luciferase activity in the miR-1296-5p group was decreased. There was no significant difference between the NC group and the miR-1296-5p group in the circ_RPPH1 mutant (Mut). These findings confirmed that circ_RPPH1 is directly bound to miR-1296-5p. From the RT-qPCR verification results in , circ_RPPH1 and miR-1296-5p levels were reduced after transfection, indicating successful transfection.
Figure 3. Circ_RPPH1 silencing inhibited BC cell malignant phenotype by increasing miR-1296-5p. (a) The predicted results of the binding situation of circ_RPPH1 and miR-1296-5p. (b) The luciferase activities in BC cells co-transferred with wild-type (WT) or mutant (Mut) circ_RPPH1 plasmid together with miR-1296-5p mimic or mic-NC (n = 3). (c-d) Results of RT-qPCR verified the silencing of circ_RPPH1 [circ_RPPH1(-)] and miR-1296-5p [miR-1296-5 (-)] in BC cells (n = 3). (e-f) Circ_RPPH1 (-) inhibited cell viability, while miR-1296-5p (-) treatment reversed it (n = 6). (g, h) Results of cell cloning formation assay; (i, j) Results of wound healing assay; (k, l) Results of Transwell assay; they respectively revealed the inhibitory effects of circ_RPPH1 (-) on BC cell proliferation, migration, and invasion, while miR-1296-5p (-) treatment reversed it (n = 3, scale bar = 200 μm or 50 μm). Data were presented as the mean ± SD. ▲p < .05, ▲▲p < .01 vs. the Control group; ★p < .05, ★★p < .01 vs. the circ_RPPH1 silencing (-) group.
![Figure 3. Circ_RPPH1 silencing inhibited BC cell malignant phenotype by increasing miR-1296-5p. (a) The predicted results of the binding situation of circ_RPPH1 and miR-1296-5p. (b) The luciferase activities in BC cells co-transferred with wild-type (WT) or mutant (Mut) circ_RPPH1 plasmid together with miR-1296-5p mimic or mic-NC (n = 3). (c-d) Results of RT-qPCR verified the silencing of circ_RPPH1 [circ_RPPH1(-)] and miR-1296-5p [miR-1296-5 (-)] in BC cells (n = 3). (e-f) Circ_RPPH1 (-) inhibited cell viability, while miR-1296-5p (-) treatment reversed it (n = 6). (g, h) Results of cell cloning formation assay; (i, j) Results of wound healing assay; (k, l) Results of Transwell assay; they respectively revealed the inhibitory effects of circ_RPPH1 (-) on BC cell proliferation, migration, and invasion, while miR-1296-5p (-) treatment reversed it (n = 3, scale bar = 200 μm or 50 μm). Data were presented as the mean ± SD. ▲p < .05, ▲▲p < .01 vs. the Control group; ★p < .05, ★★p < .01 vs. the circ_RPPH1 silencing (-) group.](/cms/asset/aad68677-5eb3-4d97-a9cb-1239ee69666a/kcbt_a_2360768_f0003_oc.jpg)
Circ_RPPH1 (-) inhibited the cell viability of BC cells while the miR-1296-5p (-) reversed the tendency and promoted the cell viability in . Similarly, showed that circ_RPPH1 (-) reduced the number of colonies in BC cells and miR-1296-5p (-) increased it. The cell wound healing assay () and Transwell assay () also indicated similar results. In , circ_RPPH1 (-) increased the cells in G0/G1 significantly and reduced the cells in the G2/M which indicated that cells were blocked in the G0/G1, while all changes induced by circ_RPPH1 (-) in cell cycle were reversed by miR-1296-5p (-). Furthermore, circ_RPPH1 (-) promoted the cell apoptosis while miR-1296-5p (-) reversed it in . Moreover, Western blot assay showed that TRIM14 protein expression was decreased in MCF-7 and MDA-MB-231 cells with circ_RPPH1 overexpression (). This was antagonized by miR-1296-5p knockdown ().
Figure 4. Circ_RPPH1 silencing blocked the G0/G1 phase and inhibited apoptosis in BC cells by increasing miR-1296-5p. (a-c) the effects of circ_RPPH1 silencing [circ_RPPH1(-)] and miR-1296-5p silencing [circ_RPPH1(-)] of the cell cycle in BC cells. Circ_RPPH1 silencing blocked the G0/G1 phase. (d-e) Circ_RPPH1 (-) promoted apoptosis in BC cells, but miR-1296-5p (-) reversed it. (f) Western blot assay was used to detect TRIM14 protein expression. Circ_RPPH1 (-) inhibited TRIM14 protein expression in MCF-7 and MDA-MB-231 cells, while mir-1296-5p (-) antagonized this. Data were presented as the mean ± SD, n = 3. ▲p < .05, ▲▲p < .01 vs. the Control group; ★★p < .01 vs. the circ_RPPH1 (-) group.
![Figure 4. Circ_RPPH1 silencing blocked the G0/G1 phase and inhibited apoptosis in BC cells by increasing miR-1296-5p. (a-c) the effects of circ_RPPH1 silencing [circ_RPPH1(-)] and miR-1296-5p silencing [circ_RPPH1(-)] of the cell cycle in BC cells. Circ_RPPH1 silencing blocked the G0/G1 phase. (d-e) Circ_RPPH1 (-) promoted apoptosis in BC cells, but miR-1296-5p (-) reversed it. (f) Western blot assay was used to detect TRIM14 protein expression. Circ_RPPH1 (-) inhibited TRIM14 protein expression in MCF-7 and MDA-MB-231 cells, while mir-1296-5p (-) antagonized this. Data were presented as the mean ± SD, n = 3. ▲p < .05, ▲▲p < .01 vs. the Control group; ★★p < .01 vs. the circ_RPPH1 (-) group.](/cms/asset/ed904fc6-d9d9-492f-ba61-f82735a02943/kcbt_a_2360768_f0004_oc.jpg)
2.4. The interaction between miR-1296-5p and TRIM14 affects the cell mobility, cell cycles and inhibits cell apoptosis in BC cells
MiR-1296-5p overexpression [miR-1296-5p (+)] and TRIM14 overexpression [TRIM14 (+)] cells were established (supplementary figure S2b, c). revealed the binding situation of miR-1296-5p and TRIM14 that the target sequence of miR-12966-5p was located at 187–194 bp of TRIM14 gene 3’-UTR. In , in the TRIM14 WT, the luciferase detection of the miR-1296-5p group was decreased. In the TRIM14 Mut, there was no significant difference between the NC and miR-1296-5p groups.
Figure 5. MiR-1296-5p overexpression inhibited BC cell malignant phenotype via TRIM14. (a) The predicted results of the binding situation of miR-1296-5p and TRIM14. (b) The luciferase activities in BC cells co-transferred with WT or Mut TRIM14 plasmid together with miR-1296-5p mimic or mic-NC (n = 3). (c-d) the overexpressing miR-1296-5p [miR-1296-5 (+)] in MCF-7 and MDA-MB-231 cells inhibited cell viability while the overexpressing TRIM14 [TRIM14 (+)] reversed it (n = 6). (e, f) Results of cell cloning formation assay; (g, h) Results of wound healing assay; (i, j) Results of Transwell assay; they were respectively revealed the inhibitory effects of miR-1296-5 (+) on the proliferation, migration, and invasion of MCF-7 and MDA-MB-231 cells, as well as and TRIM14 (+) reversed it (n = 3, scale bar = 200 μm or 50 μm). Data were presented as the mean ± SD. ▲p < .05, ▲▲p < .01 vs. the Control group; ★★p < .01 vs. the miR-1296-5p (+) group.
![Figure 5. MiR-1296-5p overexpression inhibited BC cell malignant phenotype via TRIM14. (a) The predicted results of the binding situation of miR-1296-5p and TRIM14. (b) The luciferase activities in BC cells co-transferred with WT or Mut TRIM14 plasmid together with miR-1296-5p mimic or mic-NC (n = 3). (c-d) the overexpressing miR-1296-5p [miR-1296-5 (+)] in MCF-7 and MDA-MB-231 cells inhibited cell viability while the overexpressing TRIM14 [TRIM14 (+)] reversed it (n = 6). (e, f) Results of cell cloning formation assay; (g, h) Results of wound healing assay; (i, j) Results of Transwell assay; they were respectively revealed the inhibitory effects of miR-1296-5 (+) on the proliferation, migration, and invasion of MCF-7 and MDA-MB-231 cells, as well as and TRIM14 (+) reversed it (n = 3, scale bar = 200 μm or 50 μm). Data were presented as the mean ± SD. ▲p < .05, ▲▲p < .01 vs. the Control group; ★★p < .01 vs. the miR-1296-5p (+) group.](/cms/asset/11592dc4-c24f-4d5e-9c14-42392a5c2408/kcbt_a_2360768_f0005_oc.jpg)
MiR-1296-5p (+) inhibited the cell viability of BC cells, while the miR-1296-5p (+) cells with TRIM14 (+) treatment increased the cell viability inversely (). In addition, miR-1296-5p (+) had similar effects of circ_RPPH1 (-) on BC cell malignant phenotype () to inhibit proliferation, migration, and invasion of BC cells, while TRIM14 (+) reversed it. The FCM was used to evaluate cell cycle and apoptosis, and the results were revealed in . It was found that GO/G1 cell percentage and cell apoptosis in BC were markedly increased by miR-1296-5p (+) treatment, which were reversed by TRIM14 (+) (). Results of Western bolt () showed that miR-1296-5p (+) decreased the expression of TRIM14, while TRIM14 (+) reversed miR-1296-5p (+) effects.
Figure 6. MiR-1296-5p overexpression blocked the G0/G1 phase and promoted apoptosis in BC cells. (a-c) Flow cytometry was used to detect the cycle of BC cells; The overexpressing miR-1296-5p [miR-1296-5 (+)] increased percentage of GO/G1 cells in MCF-7 and MDA-MB-231 cells. (d-e) Flow cytometry was used to detect apoptosis of BC cells; miR-1296-5 (+) promoted cell apoptosis. Overexpressing TRIM14 [TRIM14 (+)] reversed them. Data were presented as the mean ± SD, n = 3. ▲p < .05, ▲▲p < .01 vs. the Control group; ★★p < .01 vs. the miR-1296-5p (+) group.
![Figure 6. MiR-1296-5p overexpression blocked the G0/G1 phase and promoted apoptosis in BC cells. (a-c) Flow cytometry was used to detect the cycle of BC cells; The overexpressing miR-1296-5p [miR-1296-5 (+)] increased percentage of GO/G1 cells in MCF-7 and MDA-MB-231 cells. (d-e) Flow cytometry was used to detect apoptosis of BC cells; miR-1296-5 (+) promoted cell apoptosis. Overexpressing TRIM14 [TRIM14 (+)] reversed them. Data were presented as the mean ± SD, n = 3. ▲p < .05, ▲▲p < .01 vs. the Control group; ★★p < .01 vs. the miR-1296-5p (+) group.](/cms/asset/1c6e51da-9511-4b50-ae6e-057c5f5640ab/kcbt_a_2360768_f0006_oc.jpg)
2.5. TRIM14 overexpression promotes the cell viability and migration in BC cells with circ_RPPH1 silencing
To clarify that TRIM14 acts as a molecule of circ_RPPH1-regulated the malignant phenotype of BC cells, TRIM14 gene and proteins were increased in BC cells with circ_RPPH1 (-) silencing (Supplementary figure S3a). Cell counting kit-8 (CCK-8) and cell wound healing assay were used. TRIM14 overexpression was found to promote the cell viability and migration rats in BC cells with circ_RPPH1 silencing (Supplementary figure S3b,c).
2.6. Circ_RPPH1 promotes tumor progression of BC in mice
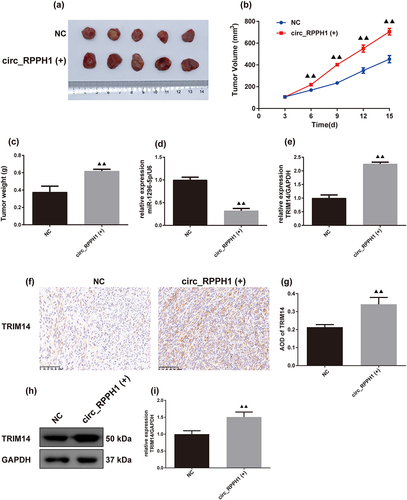
In , the circ_RPPH1 (+) promoted the size and weight of BC in mice. The RT-qPCR results in revealed that circ_RPPH1 (+) decreased the mRNA expression of miR-1296-5p and increased TRIM14 expression. The results in the IHC and Western blot () showed TRIM14 expression in BC tumor and found that circ_RPPH1 (+) increased TRIM14 expression levels.
Figure 7. Circ_RPPH1 overexpression promoted BC progression in mice. (a-c) the BC tumor size and weight in mice (n = 5). The tumor volume = length × width2/2. (d, e) the levels of miR-1296-5p and TRIM14 mRNA were detected by real-time quantitative PCR (n = 3). (f, g) Immunohistochemistry used to observe TRIM14 expression in BC tumor (n = 3, scale bar = 100 μm). (h, i) Western blot was used to detect TRIM14 expression levels in BC tumor. (h) Representative protein bands of TRIM14; (i) the semi-quantitative statistical analysis results of Western blot (n = 3). Data were presented as the mean ± SD. ▲▲p < .01 vs. the Control group. circ_RPPH1(+) group indicated circ_RPPH1 overexpression group.
3. Discussion
At present, circ_RPPH1 plays an important role in BC pathogenesisCitation16,Citation22 and is helpful for BC diagnosis and treatment. CircRNAs up-regulated the expression of TRIM14 via targeting miR-1296-5p to promote BC growth.Citation21 By examining the circ_RPPH1 level in 80 BC tissues of patients, we validated that the circ_RPPH1 level was highly expressed in BC tissues. It was expected to become a potential biomarker and target of BC. Our study confirmed the high expression of circ_RPPH1 in BC, and circ_RPPH1 promoted TRIM14 by inhibiting miR-1296-5p to facilitate BC progression.
BC is a significant threat to women’s lives and health. Although most women have heard of BC and about half are aware of the risk factors for BC, not all women are knowledgeable about BC warning signs or symptoms.Citation23 In this study, clinical BC samples from female patients of different ages showed that circ_RPPH1 expression distribution was different in the TNM staging. The in vitro study verified the high expression of circ_RPPH1 in BC cells. Circ_RPPH1 promoted the tumor growth of BC in vivo. This indicated that circ_RPPH1 was important in BC progression and may serve as a biomarker for disease diagnosis of BC.
Research has shown that circRNAs function through competing endogenous RNA (ceRNA) in BC cell mobility.Citation24 Circ_RPPH1 was conducted as ceRNA to reduce the inhibitory roles of miR-146b-3p on E2F2 to promote BC.Citation16 Circ_RPPH1 knockdown restrained glioma growth through binding to miR-627-5p/miR-663a.Citation25 Moreover, circ_RPPH1 accelerated the aggravation of BC by miR-542-3p/ARHGAP1 axis.Citation22 In this study, circ_RPPH1 affected miR-1296-5p through the ceRNA to regulate TRIM14 and the BC cell mobility had a significant promotion. Therole of miR-1296-5p in BC is still ambiguous, miR-1296-5p was reported to inhibit the gastric cancer via regulating EGFR and CDK6.Citation26 Moreover, miR-1296-5p also inhibited the growth of human osteosarcoma by regulating notch receptor 2.Citation27 TRIM14 is highly expressed in tumors, including gastric cancer,Citation28 tongue cancer,Citation29 gliomaCitation30 and BC,Citation31 etc. Our study confirmed that TRIM14 was a target for miR-1296-5p. We prove that miR-1296-5p plays an antineoplastic role in BC and circ_RPPH1/miR-1296-5p knockdown promotes tumor progression in BC by activating TRIM14, suggesting that the promoting effect in BC of circ_RPPH1 was involved in regulating the miR-1296-5p/TRIM14 axis.
However, our study has some limitations, including the combination mode of circ_RPPH1 and miR-1296-5p. Additionally, if the effects of silencing miR-296-5p and overexpressing TRIM14 in BC cells can be detected separately, the evidence chain of this study will be more complete. In the future, we will probe the effects and mechanisms of circ_RPPH1 in BC in more detail. Moreover, the further study of the circRNA-miRNA-mRNA axis in BC will help the establishment of BC regulatory network and strengthen the mechanism study on anti-BC.
4. Conclusion
In conclusion, circ_RPPH1 promoted the proliferation, invasion, and migration of BC by inhibiting miR-1296-5p and thereby up-regulating TRIM14. Animal experiments confirmed that the oncogenic role of circ_RPPH1 overexpression in BC progression. Circ_RPPH1 could be used as a promising biomarker and therapeutic target for study and treatment of BC.
5. Material and methods
5.1. Clinical samples and analysis
UALCAN was used to analyze circ_RPPH1 expression in primary tumor of BRCA compared to the normal tissues (https://ualcan.path.uab.edu/cgi-bin/TCGAExResultNew2.pl?genenam=RPPH1&ctype=BRCA). BC (n = 15) and para-carcinoma (n = 65) tissues were collected from 80 patients recruited from Ningbo No.2 Hospital. The pa-carcinoma tissues are the adjacent normal tissues, and its isolation method refers to previous research.Citation32 Patients range in age from 28 to 84. Informed consent was obtained from all patients. Ethics approval was obtained from Ningbo No.2 Hospital.
Clinical information was recorded by physicians using a standardized form, including the relation between circ_RPPH1 expression and clinical indications: age, tumor size, pausimenia or not, lymph node metastasis or not, TNM staging, estrogen receptor (ER), progesterone receptor (PR) as well as human epidermal growth factor receptor-2 (HER2).
5.2. Cell culture
MCF-7 cells and MDA-MB-231 cells are both human BC cells. MCF-7 cell lines (iCell-h129) and MDA-MB-231 cell lines (iCell-h133) were obtained from Shanghai iCell Bioscience Inc (China). MCF-7 cells were cultured in a DMEM medium containing 10% fetal bovine serum (FBS), and 1% penicillin-streptomycin solution and incubated in a cell incubator with a condition of 5% CO2, 37°C. In addition, MDA-MB-231 cells were also cultured in the same medium but maintained air culturing without CO2 to avoid cytotoxicity.
5.3. Lentivirus, small interfering RNA, and plasmid transfection and cell grouping
Vector lentivirus and circ_RPPH1 lentivirus were purchased from GeneChem (Shanghai, China). Briefly, the two BC cells (2 × 105 cells/well) were inoculated into 6-well plates and cultured until they reached 70–80% confluence. A complete medium containing virion and viral infection booster fluid was then used to infect cells for 24 h, followed by puromycin (ST551, Beyotime, China) selected for one week. Circ_RPPH1(GOSE0351480), circ_RPPH1 small interfering RNA (siRNA) (GIDE0364124), miR-1296-5p siRNA (GMDE0364125), control siRNA (P22091400), plasmids encoding miR-1296-5p (GMUE0364119) or TRIM14 cDNA (GOSE0364118) were purchased from GeneChem, Shanghai, China and were used to transfect cells for 6 h using a lipofectamine 3000 kit (L3000–008, Invitrogen, USA) according to the manufacturer’s instructions. After transfection, fresh medium was replaced and cultured for 24 h.
Overexpression and knockdown of genes were indicated as (+) and (-), respectively. First, MCF-7 and MDA-MB-231 cells were divided into three groups: control, NC and circ_RPPH1 (+) to evaluate the impact of circ_RPPH1 overexpression on BC malignant phenotype. Additionally, four groups were created to analyze the inhibitory effects of circ_RPPH1 knockdown on BC via miR-1296-5p: control, NC, circ_RPPH1 silencing [circ_RPPH1 (-)], and circ_RPPH1 (-) + miR-1296-5p silencing [miR-1296-5p (-)]. Moreover, to evaluate if TRIM14 activation antagonizes the effects of miR-1296-5p overexpression on BC, these cells were divided into Control, NC, miR-1296-5p overexpression [miR-1296-5p (+)], and miR-1296-5p (+) + TRIM14 overexpression [TRIM14 (+)] groups. Additionally, circ_RPPH1 (-) and circ_RPPH1 (-) + TRIM14 (+) groups were used to clarify that TRIM14 is a downstream molecule of circ_RPPH1 regulating cell viability and migration of BC cells.
5.4. Detection of luciferase activity
The 2 × 104 cells were seeded in 96-well plates for a dual-luciferase reporter gene assay. Luciferase was linked to circ_RPPH1 wild-type (WT) and circ_RPPH1 Mut or TRIM14 WT and TRIM14 Mut reporter vector or miR-1296-5p mimics or miR-NC, then transfected into MDA-MB-231 cells. After transfection, a dual-luciferase reporter gene assay system was applied. 20 µL collected cell lysate of groups was added in a black label plate. A firefly luciferase reaction fluid was added and mixed, followed by detecting the activity of firefly luciferase within 30 min at 550 nm. A Renilla luciferase reaction fluid was then added in each well and mixed, followed by detecting the activity of Renilla luciferase within 30 min at 480 nm.
5.5. Detection of the gene expression of circ_RPPH1, miR-1296-5p, and TRIM14
The gene expression of circ_RPPH1, miR-1296-5p, and TRIM14 in BC human tissues, cells and mice tissues was examined using RT-qPCR. After a pre-processing of the samples, Buffer RLT, RNase-free ddH2O and proteinase K solution were added to extract total RNA. The retro transcriptional response system was prepared for reverse transcriptional reaction. Then, a response system was prepared for Real-time fluorescence quantitative PCR reaction and carried out on the Roche LightCycler 96 RT-qPCR instrument (LightCycler® 96, Roche, Switzerland). GAPDH was the internal reference. The relative levels of target genes were measured by the 2−ΔΔCt. The primer sequences were shown in .
Table 2. The primer sequences.
5.6. Detection of cell proliferation
The cell proliferation of BC cells was detected using CCK-8 and a cell cloning formation assay. For the CCK-8 assay, two cells were inoculated into 96-well plates of 2.5 × 103 cells/well and cultured in the cell incubator (37 °C) with their respective culture conditions for 24 h. Then, the lentiviral infection or plasmid transfection was performed for 6 h. The medium was changed and cultured for 24 h again. 10 μL CCK-8 solution (C0039, Beyotime, China) was added and co-cultured for 2 h. The OD value was captured using a microplate reader (CMaxPlus, MD, USA) at 450 nm. For the cell cloning formation assay, the transfected cells were inoculated into 24-well plates of 2.5 × 105 cells/well. A DMEM medium containing 30% FBS was replaced the original medium. Every three days, the medium was changed. The clone size was observed under an optical microscope (AE2000, Motic, China). After two weeks, the cells were photographed. 4% paraformaldehyde was added to fix cells at 4°C for 1 h and followed by washing with PBS. Then, crystal violet dye was applied in wells to stain cells. Washed with PBS, the colony images were captured using a camera (850D, Canon Ltd., Japan). Finally, the colony number was counted microscopically.
5.7. Detection of cell migration
The cell migration capacity of BC cells was detected using a cell wound healing assay. The transfected cells were inoculated into 6-well plates of 5 × 105 cells/well and cultured in their respective culture conditions. When the cells were covered with the plates of 80%, a straight line was drawn evenly perpendicular to the bottom of the plate hole using a pipette tip. A serum-free medium was added for 24 h culture. The scratch was photographed using the optical microscope. Image J (Image-J, NIH, Germany) was used to analyze the cell migration rate.
5.8. Detection of cell invasion
The cell invasion of BC cells was detected using Transwell assay. The cells were resuspended to 2 × 104 cells/mL. Cell suspension containing 4 × 103 cells was added into the upper chamber of the Transwell chamber, and medium containing 10% FBS was added into the lower chamber, followed by culturing for 24 h in the cell incubator (37 °C). 4% paraformaldehyde was used to fix cells in lower chamber for 0.5 h. The crystal violet dye was used to stain cells. Finally, the cell images were photographed using the camera and the number of invaded cells was counted.
5.9. Detection of cell cycle
The FCM was applied to detect the cell cycle of BC cells. The infected or transfected cells were inoculated into 6-well plates of 5 × 105 cells/well. After 24 h culture, the cells were then collected and resuspended. DNA Staining solution (containing propidium iodide (PI) and RNase A 550,825, BD Pharmingen, USA) and permeabilization solution were added and co-incubated at 25°C for 30 min away from light. The cells were collected, and the cell cycle was detected using a flow cytometer (C6, BD, USA).
5.10. Detection of cell apoptosis
The cell apoptosis of BC cells was detected using FCM. After culture, the cell density was 1 × 106 cells/mL, and the binding buffer was added and mixed. Annexin V-FITC and PI were added for 15 min incubation away from light. Finally, the binding buffer was added again, and the flow cytometry was used for the detection of cell apoptosis rate within 60 min.
5.11. Subcutaneous tumorigenic model of BC cells in nude mice
Female BALB/c nude mice (8 weeks old, 18 g weight) were purchased from Beijing Vital River Laboratory Animal Technology Co., Ltd (China) of animal license permit No.: SCXK (Jing) 2016–0006. The nude mice were housed in the animal room with the SPF barrier system of Zhejiang Eyoung Pharmaceutical Research and Development Co., Ltd (China). The SPF animal room was under a 12-h dark/light cycle, room temperature of 23 ± 2°C and humidity of 60 ± 5%. All animal experiments were performed under the Experimental Animal Ethics Committee of Zhejiang Eyoung Pharmaceutical Research and Development Co., Ltd of License No. SYXK (Zhe) 2021–0033.
The nude mice were divided into two groups: NC and circ_RPPH1 overexpression [circ_RPPH1(+)]. MDA-MB-231 cells with circ_RPPH1 lentivirus and control lentivirus transfection were resuspended in normal saline. MDA-MB-231 cells (3 × 106 cells) were inoculated in the left axillary of each mouse. The tumor size was determined every three days. After 15 d of subcutaneous tumor injection, the mice were euthanized with CO2, and the tumor tissues of mice were separated and weighed. The separated tumors were photographed.
5.12. Detection of the expression of TRIM14 in BC cells or tumor tissues in mice
Immunohistochemistry (IHC) and Western blot were performed to detect the level of TRIM14 in cells or tumor tissues. For the IHC assay of TRIM14, the paraffin-embed blocks of separated tumor tissues were cut into 5-μm sections. The sections were baked, dewaxed, and hydrated. Following by blocking with 3% H2O2 solution and repairing with boiling antigen repairing solution (1 mmol Tris-EDTA, pH = 9.0), 5% BSA was incubated with the sections for 20 min at 25°C. The primary antibody of TRIM14 (1:100, DF12490, Affinity, USA) diluted in antibody diluent was then added and co-incubated for 12 h at 4°C. The next day, the second antibody (1:5000, ab97080, Abcam, USA) diluted in antibody diluent was added and co-incubated for 0.5 h at 37°C. Following staining with DBA solution and hematoxylin dye, the sections were dehydrated and transparentized. Finally, neutral balsam was used to seal the sections and the IHC images were photographed using a microscope.
For the Western blot assay, the total protein in cells or tissues was extracted using Lysis buffer. A BCA kit was then used to measure the protein concentration and the protein concentration of each group was unified. 5 × loading buffer was added and boiled to degenerate protein. 10% SDS-PAGE was prepared, and the protein samples were added into the sample holes respectively and electrophoresed using an electrophoresis apparatus (EPS300, Tianneng, China). When the protein entered the best separation part, transferred it to the PVDF membrane electrophoretically. The membrane was washed with TBST for 3 times and blocked with 5% skimmed milk powder. The membrane was then washed with TBST again and immediately transferred to primary antibodies of TRIM14 (1:1000), GAPDH (1:10000, AF7021, Affinity, USA) diluted in antibody diluent and co-incubated for 12 h at 4°C. The membrane was then washed and immediately co-incubated with the second antibody diluent (1:6000, #7074, Cell Signaling Technology, USA) for 90 min at 25°C. The membrane was washed and transferred into a chemiluminescence apparatus (610020-9Q, Shanghai Qinxiang Scientific Instrument Co., LTD, China) and ECL reagent was dropped on it. The protein band images were captured using chemi capture Software and analyzed using Image-J.
5.13. Statistical analysis
All the experiments above were repeated at least three times. SPSS 20.0 (SPSS, Chicago, USA) was used for data analysis. One-way analysis of variance (ANOVA) was used for quantitative data analysis, followed by Tukey’s posttest with normal distribution data. When the distribution was normal, but the variance was not uniform, Dunnett’s T3 test was used. All data were expressed as mean ± standard deviation (SD). p < .05 or p < .01 was defined as a significant difference between groups.
Authors’ contributions
Conception and design of the research: Jing Jiang and Xujun Li
Acquisition of data: Qidong Ge and Long Sun
Analysis and interpretation of data: Jing Jiang and Shenghong Shi
Statistical analysis: Wei Zhang and Chao Li
Drafting the manuscript: Jing Jiang
Revision of the manuscript for important intellectual content: Jing Jiang and Xujun Li
Approval of final version of manuscript to be published: All authors
Agreement to be accountable for all aspects of the work ensuring that questions related to the accuracy or integrity of the work are appropriately investigated and resolved: Jing Jiang, Shenghong Shi, Wei Zhang, Chao Li, Long Sun, Qidong Ge and Xujun Li.
Ethics
Ethics approval was obtained from Ningbo No.2 Hospital in any experiment with human. All animal experiments were performed under the Experimental Animal Ethics Committee of Zhejiang Eyoung Pharmaceutical Research and Development Co., Ltd of License No. SYXK (Zhe) 2021–0033.
supplementary files.docx
Download MS Word (1.8 MB)Acknowledgments
The authors greatly appreciate the efforts of all the authors.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data presented in this study are available upon reasonable request from the corresponding author.
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2024.2360768
Additional information
Funding
References
- Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, Safari E, Farahmand L. Breast cancer: Biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84:106535. doi:10.1016/j.intimp.2020.106535.
- Wilkinson L, Gathani T. Understanding breast cancer as a global health concern. Br J Radiol. 2022;95(1130):20211033. doi:10.1259/bjr.20211033.
- Hamdan D, Nguyen TT, Leboeuf C, Meles S, Janin A, Bousquet G. Genomics applied to the treatment of breast cancer. Oncotarget. 2019;10(46):4786–14. doi:10.18632/oncotarget.27102.
- Nelson AJO, America GCON. Controversies regarding mammography, breast self-examination, and clinical breast examination. Obstet Gynecol Clin North Am. 2013;40(3):413–427. doi:10.1016/j.ogc.2013.05.001.
- Kashyap D, Pal D, Sharma R, Garg VK, Goel N, Koundal D, Zaguia A, Koundal S, Belay A. Global increase in breast cancer incidence: Risk factors and preventive measures. Biomed Res Int. 2022;2022:1–16. doi:10.1155/2022/9605439.
- Fiorica J. Breast cancer screening, mammography, and other modalities. Clin Obstet Gynecol. 2016;59(4):688–709. doi: 10.1097/GRF.0000000000000246.
- Cserni G, Chmielik E, Cserni B, Tot T. The new TNM-based staging of breast cancer. Virchows Arch. 2018;472(5):697–703. doi:10.1007/s00428-018-2301-9.
- Fisusi F, Akala E. 2019. Drug combinations in breast cancer therapy. Pharm Nanotechnol. 7(1):3–23. doi: 10.2174/2211738507666190122111224.
- Haussmann J, Corradini S, Nestle-Kraemling C, Bölke E, Njanang FJD, Tamaskovics B, Orth K, Ruckhaeberle E, Fehm T, Mohrmann S, et al. 2020. Recent advances in radiotherapy of breast cancer. Radiat Oncol. 15(1):71. doi: 10.1186/s13014-020-01501-x.
- Yin L, Duan J-J, Bian X-W, Yu S-C. 2020. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 22(1):61. doi: 10.1186/s13058-020-01296-5.
- Huang Y, Zheng W, Ji C, Wang X, Yu Y, Deng X, Zhou X, Fang L. Circular RNA circRPPH1 promotes breast cancer progression via circRPPH1-miR-512-5p-STAT1 axis. Cell Death Discov. 2021;7(1):376. doi:10.1038/s41420-021-00771-y.
- Zhao C, Li L, Li Z, Xu J, Yang Q, Shi P, Zhang K, Jiang R. A novel circular RNA hsa_circRPPH1_015 exerts an oncogenic role in breast cancer by impairing miRNA-326-mediated ELK1 inhibition. Front Oncol. 2020;10:906. doi:10.3389/fonc.2020.00906.
- Li J, Li Y, Cheng H. Circ-RPPH1 knockdown retards breast cancer progression via miR-328-3p-mediated suppression of HMGA2. Clin Breast Cancer. 2022;22(3):e286–e295. doi:10.1016/j.clbc.2021.08.009.
- Xie J, Ye F, Deng X, Tang Y, Liang J-Y, Huang X, Sun Y, Tang H, Lei J, Zheng S, et al. Circular RNA: A promising new star of vaccine. J Transl Int Med. 2023;11(4):372–381. doi:10.2478/jtim-2023-0122.
- Yang L, Liu Z, Ma J, Wang H, Gao D, Zhang C, Ma Q. CircRPPH1 serves as a sponge for miR-296-5p to enhance progression of breast cancer by regulating FOXP4 expression. Am J Transl Res. 2021;13(7):7556–7573.
- Feng H, Sun S-Z, Cheng F, Zhang N-Q. Mediation of circ_RPPH1 on miR-146b-3p/E2F2 pathway to hinder the growth and metastasis of breast carcinoma cells. Aging (Albany NY). 2021;13(16):20552–20568. doi:10.18632/aging.203439.
- Yuan Y, Anbalagan D, Lee LH, Samy RP, Shanmugam MK, Kumar AP, Sethi G, Lobie PE, Lim LHK. ANXA1 inhibits miRNA-196a in a negative feedback loop through NF-kB and c-Myc to reduce breast cancer proliferation. Oncotarget. 2016;7(19):27007–27020. doi:10.18632/oncotarget.8875.
- Kristensen L, Jakobsen T, Hager H, Kjems J. The emerging roles of circRnas in cancer and oncology. Nat Rev Clin Oncol. 2022;19(3):188–206. doi:10.1038/s41571-021-00585-y.
- Wang P, Qu H, Wang L, Hu Z. Silencing of circUSPL1 represses breast cancer progression by targeting miR-1296-5p/MTA1 axis. Thorac Cancer. 2023;14(22):2198–2209. doi:10.1111/1759-7714.15007.
- Chen G, He M, Yin Y, Yan T, Cheng W, Huang Z, Zhang L, Zhang H, Liu P, Zhu W, et al. miR-1296-5p decreases ERBB2 expression to inhibit the cell proliferation in ERBB2-positive breast cancer. Cancer Cell Int. 2017;17(1):95. doi:10.1186/s12935-017-0466-y.
- Xie F, Xiong Y, Yan J, Wang L, Yan W. Circular RNA circ_0048764 promotes the development of breast cancer by regulating microRNA-1296-5p/tripartite motif containing 14 axis. Bioengineered. 2022;13(2):1963–1974. doi:10.1080/21655979.2021.1995990.
- Qi L, Sun B, Yang B, Lu S. CircRNA RPPH1 facilitates the aggravation of breast cancer development by regulating miR-542-3p/ARHGAP1 pathway. Cancer Biother Radiopharm. 2022;37(8):708–719. doi:10.1089/cbr.2020.4381.
- Rahman S, Al–Marzouki A, Otim M, Khalil Khayat NEH, Yousuf R, Rahman P. Awareness about breast cancer and breast self-examination among female students at the University of Sharjah: A cross-sectional study. Asian Pac J Cancer Prev. 2019;20(6):1901–1908. doi:10.31557/APJCP.2019.20.6.1901.
- Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–358. doi:10.1016/j.cell.2011.07.014.
- Chen W, Yu X, Wang N, Jing J, Li R, Lian M. Circ_RPPH1 regulates glioma cell malignancy by binding to miR-627-5p/miR-663a to induce SDC1 expression. Metab Brain Dis. 2022;37(4):1231–1245. doi:10.1007/s11011-022-00965-y.
- Jia Y, Zhao L-M, Bai H-Y, Zhang C, Dai S-L, Lv H-L, Shan B-E. The tumor-suppressive function of miR-1296-5p by targeting EGFR and CDK6 in gastric cancer. Biosci Rep. 2019;39(1):BSR20181556. doi:10.1042/BSR20181556.
- Wang L, Hu K, Chao Y, Wang X. MicroRNA-1296-5p suppresses the proliferation, migration, and invasion of human osteosarcoma cells by targeting NOTCH2. J Cell Biochem. 2020;121(2):2038–2046. doi:10.1002/jcb.29438.
- Xiao F, Ouyang B, Zou J, Yang Y, Yi L, Yan H. Trim14 promotes autophagy and chemotherapy resistance of gastric cancer cells by regulating AMPK/mTOR pathway. Drug Dev Res. 2020;81(5):544–550. doi:10.1002/ddr.21650.
- Su X, Wang J, Chen W, Li Z, Fu X, Yang A. Overexpression of TRIM14 promotes tongue squamous cell carcinoma aggressiveness by activating the NF-κB signaling pathway. Oncotarget. 2016;7(9):9939–9950. doi:10.18632/oncotarget.6941.
- Tan Z, Song L, Wu W, Zhou Y, Zhu J, Wu G, Cao L, Song J, Li J, Zhang W. TRIM14 promotes chemoresistance in gliomas by activating Wnt/β-catenin signaling via stabilizing Dvl2. Oncogene. 2018;37(40):5403–5415. doi:10.1038/s41388-018-0344-7.
- Hu G, Pen W, Wang M. TRIM14 promotes breast cancer cell proliferation by inhibiting apoptosis. Oncol Res. 2019;27(4):439–447. doi:10.3727/096504018X15214994641786.
- Yao X, Tu Y, Xu Y, Guo Y, Yao F, Zhang X. Endoplasmic reticulum stress-induced exosomal miR-27a-3p promotes immune escape in breast cancer via regulating PD-L1 expression in macrophages. J Cell Mol Med. 2020;24(17):9560–9573. doi:10.1111/jcmm.15367.
