ABSTRACT
Prostate cancer has heterogeneous growth patterns, and its prognosis is the poorest when it progresses to a neuroendocrine phenotype. Using bioinformatic analysis, we evaluated RNA expression of neuroendocrine genes in a panel of five different cancer types: prostate adenocarcinoma, breast cancer, kidney chromophobe, kidney renal clear cell carcinoma and kidney renal papillary cell carcinoma. Our results show that specific neuroendocrine genes are significantly dysregulated in these tumors, suggesting that they play an active role in cancer progression. Among others, synaptophysin (SYP), a conventional neuroendocrine marker, is upregulated in prostate adenocarcinoma (PRAD) and breast cancer (BRCA). Our analysis shows that SYP is enriched in small extracellular vesicles (sEVs) derived from plasma of PRAD patients, but it is absent in sEVs derived from plasma of healthy donors. Similarly, classical sEV markers are enriched in sEVs derived from plasma of prostate cancer patients, but weakly detectable in sEVs derived from plasma of healthy donors. Overall, our results pave the way to explore new strategies to diagnose these diseases based on the neuroendocrine gene expression in patient tumors or plasma sEVs.
Introduction
Prostate cancer is the most common cancer among men in the United States, with 299,010 estimated new cases diagnosed and 35,250 estimated deaths attributed to the disease this year.Citation1 Prostate cancer has heterogeneous growth patterns;Citation2 treatment strategies include androgen deprivation therapy.Citation3 If the disease has progressed to androgen independence, it becomes castrate-resistant prostate cancer (CRPC);Citation3 its prognosis is the poorest when it progresses to neuroendocrine prostate cancer (NEPC) phenotype.Citation4
The role of extracellular vesicles (EVs) in cancer has been recently established, as EVs can contribute to cancer progression hallmarks including proliferation, migration, angiogenesis, evasion of cell death, and metastasis.Citation5 EVs are heterogeneous, displaying different biogenesis, and size ranges.Citation6 small EVs (sEVs) may have an endosomal or non-endosomal origin, and a size range of <200 nm.Citation6,Citation7 sEVs are characterized by classical markers such as Syntenin, TSG101, Alix, and the tetraspanins (CD9, CD63, and CD81)6. Syntenin, TSG101, and Alix are part of the ESCRT (endosomal sorting complex required for transport) family of proteins, and play important roles in sEV biogenesis, as these proteins regulate multivesicular body formation and the inward budding of intraluminal vesicles.Citation6 The tetraspanins are also crucially involved in sEV biogenesis and release as they can re-arrange the membrane curvatures and thus promote sEV budding and facilitate release.Citation8,Citation9 sEVs are present in several biological fluids, such as blood (serum or plasma), urine, semen, and milk.Citation10 The content of sEVs can differ between cancer and physiological conditions.Citation11–14
In this study, we performed a bioinformatic analysis of publicly available RNA-seq data of a neuroendocrine gene panel. This panel included twenty-two genes which were chosen based on the work done by Hofsli et al., who analyzed genes differentially expressed in neuroendocrine and non-neuroendocrine cancer cell lines that did not include prostate cancer cells.Citation15 In our study, we used this neuroendocrine gene panel to analyze the differential expression in five different cancer type and normal tissue samples: Prostate adenocarcinoma (PRAD), Breast cancer (BRCA), Kidney Chromophobe (KICH), Kidney renal clear cell (KIRC) carcinoma, and Kidney renal papillary cell (KIRP) carcinoma. BRCA is the most common non-skin cancer in women in the world.Citation16 Renal cell carcinoma (RCC) is a relatively rare and heterogeneous group of tumors with distinct mutations, histologic appearance, and clinical behaviors.Citation17,Citation18 RCC is frequently divided into KIRC, KIRP, and KICH. KIRC is the most common subtype, and it has been correlated with aggressive clinical features.Citation17 KIRP is the second most common RCC subtype, which is also aggressive and has a poor prognosis when metastatic.Citation19 KICH is the third most common RCC subtype and is mainly sporadic and indolent with a favorable clinical outcome.Citation20 The neuroendocrine gene panel includes conventional neuroendocrine protein markers such as synaptophysin (SYP), chromogranin A (CHGA) and enolase 2,Citation15 which are routinely used for the diagnosis of neuroendocrine tumors. SYP is involved in the process of formation, transport, and release of synaptic vesicles and neuronal transmitters.Citation21 SYP is a widely used immunohistochemical marker for neuroendocrine tumor diagnosis.Citation21 CHGA is a neuroendocrine secretory protein involved in sorting and packaging of neurotransmitters into secretory granules.Citation22 CHGA is an important neuroendocrine tumor marker used both in examination of biopsied tumor tissue and as a serum tumor marker. Enolase 2 is an enzyme of the glycolytic pathway and one of the main markers used to diagnose poorly differentiated neuroendocrine tumors.Citation23,Citation24 Additionally, we evaluated other neuroendocrine genes that have also been implicated in the pathogenesis of neuroendocrine cancers, such as MYCN, GDNF Family Receptor Alpha 2 (GFRA2), and the neurofilament protein genes (NEFL, NEFM, NEFH).Citation25–27
In this manuscript, we show that neuroendocrine genes are expressed in patients diagnosed with PRAD and other cancers such as BRCA, KICH, KIRC, and KIRP. We also provide evidence that SYP and sEV classical markers are detected in PRAD patient plasma-derived sEVs but are barely detectable in healthy donor sEVs.
Results
Neuroendocrine genes are differentially expressed across cancer types
It has been shown that PRAD patients have distinct heterogeneous molecular alterations that vary between individuals.Citation2 Therefore, we decided to analyze the expression levels of literature-curated neuroendocrine genes in PRAD patient tissues compared to normal tissues. In order to broaden the scope of the study, we included in the analysis BRCA, KICH, KIRC, and KIRP patient tissues compared to their healthy counterparts. For this analysis, the RNA-seq expression profiling data were downloaded from the Cancer Genome Atlas (TCGA) database. For TCGA RNA-seq analysis, surgical resection biospecimens were collected from patients diagnosed with PRAD, and had not received prior treatment for their disease (chemotherapy, radiotherapy, or hormonal ablation therapy).Citation28 Analysis of TCGA RNA-seq data shows upregulation or downregulation of several genes in cancer versus normal tissues, as shown in , the volcano plots in as well as . These genes appear to encode proteins found in secretory granules, plasma membrane, cytoskeleton, cytosol, and nucleus ().
Figure 1. Differential expression patterns of known neuroendocrine cancer genes across TCGA cancer types. (a) Venn diagram depicting the neuroendocrine genes reported in TCGA (The Cancer Genome Atlas) that are significantly upregulated (FDR: False Discovery Rate <0.05) in one or more indicated cancer types [Prostate adenocarcinoma (PRAD), Breast cancer (BRCA), Kidney Chromophobe (KICH), Kidney renal clear cell (KIRC) carcinoma, and Kidney renal papillary cell (KIRP) carcinoma] compared to normal samples. The table below indicates the genes that are significantly upregulated according to the cancer type. (b) Volcano plots of log2 fold-change (Log FC) in expression of the known neuroendocrine genes (X-axis) with statistical significance (Y-axis) in five cancer types. Significantly downregulated and upregulated genes are represented in blue and red colors, respectively. Grey color represents not significant (NS). The genes with 1.5 fold-change are shown. Y-axis represents mRNA expression (Normalized expression). Significantly (FDR <0.05) differentially expressed genes are shown. P-value < 0.05 are considered significant.
![Figure 1. Differential expression patterns of known neuroendocrine cancer genes across TCGA cancer types. (a) Venn diagram depicting the neuroendocrine genes reported in TCGA (The Cancer Genome Atlas) that are significantly upregulated (FDR: False Discovery Rate <0.05) in one or more indicated cancer types [Prostate adenocarcinoma (PRAD), Breast cancer (BRCA), Kidney Chromophobe (KICH), Kidney renal clear cell (KIRC) carcinoma, and Kidney renal papillary cell (KIRP) carcinoma] compared to normal samples. The table below indicates the genes that are significantly upregulated according to the cancer type. (b) Volcano plots of log2 fold-change (Log FC) in expression of the known neuroendocrine genes (X-axis) with statistical significance (Y-axis) in five cancer types. Significantly downregulated and upregulated genes are represented in blue and red colors, respectively. Grey color represents not significant (NS). The genes with 1.5 fold-change are shown. Y-axis represents mRNA expression (Normalized expression). Significantly (FDR <0.05) differentially expressed genes are shown. P-value < 0.05 are considered significant.](/cms/asset/054df5f7-0213-4244-85a2-d6ab1a317299/kcbt_a_2364433_f0001_oc.jpg)
Figure 2. Differential expression patterns of known downregulated neuroendocrine cancer genes across cancer types reported in TCGA. Venn diagram depicting the neuroendocrine genes that are significantly downregulated (FDR <0.05) in different TCGA cancer types (PRAD, BRCA, KICH, KIRC, and KIRP) compared to normal samples. The table (right panel) indicates the genes that are significantly downregulated according to the cancer type.
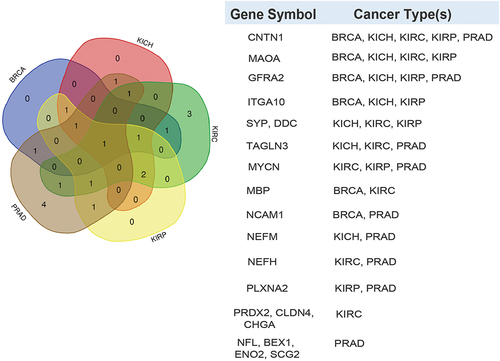
Table 1. Dysregulated proteins encoded by neuroendocrine cancer genes.
Proteins related to synaptic vesicles and secretory granules are differentially expressed across cancer types. For instance, SYP is upregulated in PRAD and BRCA, and downregulated in KICH, KIRC, and KIRP. Secretogranin 2 (SCG2), and Secretogranin 3 (SCG3) are the only genes with statistically significant upregulation in four out of the five cancer types compared to the corresponding normal tissue. SCG2 and SCG3 are upregulated in BRCA, KICH, KIRC, and KIRP, while SCG2 is downregulated in PRAD. CHGA is upregulated in BRCA and downregulated in KIRC. Furthermore, plasma membrane proteins are dysregulated in different cancers. GFRA2, which is significantly upregulated in KIRC, is downregulated in PRAD, BRCA, KICH, and KIRP. Myelin basic protein (MBP) is upregulated in PRAD and KICH while being downregulated in BRCA and KIRC. Additionally, Contactin 1 (CNTN1) is downregulated in all five cancer types. Of note, the cytoskeletal proteins Neurofilament light chain (NEFL), Neurofilament medium chain (NEFM), Neurofilament heavy chain (NEFH), and Transgelin 3 (TAGLN3) are downregulated in PRAD and upregulated in BRCA. In the other cancers, NEFL is upregulated in KIRC and KIRP. NEFM is upregulated in KIRP and downregulated in KICH. In contrast, NEFH is downregulated in KIRC. TAGLN3 is upregulated in KIRP but is downregulated in KIRC and KICH. The cytosolic protein Dopa decarboxylase (DDC) is upregulated in PRAD and BRCA, and downregulated in KICH, KIRC, and KIRP. The transmembrane enzymes Monoamine Oxidase A (MAOA), and Hepsin (HPN) are upregulated in PRAD. In the other cancers, MAOA is downregulated in BRCA, KICH, KIRC, and KIRP, and HPN is upregulated in BRCA. Additionally, the other transmembrane proteins are also distinctly dysregulated in different cancers. Integrin alpha subunit 10 (ITGA10) is downregulated in BRCA, KICH, and KIRP and upregulated in PRAD and KIRC. Plexin A2 (PLXNA2) is downregulated in PRAD and KIRP and upregulated in KIRC. Claudin 4 (CLDN4) is upregulated in BRCA and KIRP and downregulated in KIRC. Neural cell adhesion molecule 1 (NCAM1) is downregulated in PRAD and BRCA and upregulated in KIRC. Finally, the genes encoding for the cytoplasmic/nuclear proteins, Brain expressed X linked 1 (BEX1), MYCN and Enolase 2 (ENO2) are downregulated in PRAD and upregulated in BRCA. In the other cancers, MYCN is downregulated in KIRC and KIRP; ENO2 is upregulated in KIRC, and KIRP. Peroxiredoxin 2 (PRDX2), another protein in this category is upregulated in PRAD, BRCA, and KICH, and is downregulated in KIRC. Collectively, the results show inconsistent dysregulation patterns for most neuroendocrine genes across the cancer types, which is expected due to the high intra-tumor heterogeneity. Citation2,Citation29
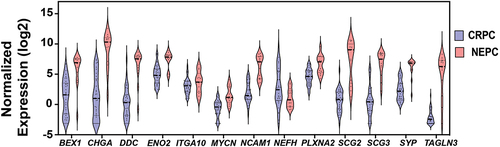
Based on these data, PRAD was selected as an area of specific interest. We then evaluated the levels of these twenty-two genes in CRPC compared to NEPC (). These data show that thirteen genes (SYP, BEX1, CHGA, DDC, ENO2, ITGA10, MYCN, NCAM1, NEFH, PLXNA2, SCG2, SCG3, and TAGLN3) are differentially expressed in NEPC compared to CRPC. Twelve genes are significantly upregulated, and one gene (NEFH) is significantly downregulated in NEPC compared to CRPC. As summarized in , different subgroups of proteins encoded by these genes are significantly dysregulated in PRAD, BRCA, KICH, KIRC or KIRP. Therefore, our data suggest that distinct protein subtypes promote cell differentiation toward a neuroendocrine phenotype in different cancers.
Figure 3. Neuroendocrine gene expression levels in NEPC compared to CRPC. Violin plots represent mRNA expression levels (Y-axis) of neuroendocrine genes (X-axis) that are significantly dysregulated (FDR <0.05) in NEPC compared to CRPC as reported in TCGA. NEPC: neuroendocrine prostate cancer, CRPC: castrate-resistant prostate cancer. P-value < 0.05 are considered significant.
PRAD patient plasma-derived sEVs express higher levels of synaptophysin and sEV classical markers compared to healthy donor sEVs
Among the genes identified, SYP is usually abundant at the protein level in advanced-stage aggressive neuroendocrine in PRAD. Thus, we assessed the levels of SYP in plasma-derived sEVs from PRAD patients and healthy donor plasma sEVs. Furthermore, we also assessed sEV marker levels in plasma sEVs, as it has been shown that sEVs derived from a variety of tumor cells express high levels of sEV classical markers.Citation30,Citation31 Immunoblotting (IB) analysis shows that SYP is absent in all healthy donor plasma sEVs but is expressed in two of the PRAD patient plasma sEVs (). The IB analysis also shows that sEV markers CD63, Syntenin, Alix, and CD9 are enriched in PRAD patient plasma sEVs compared to healthy donor plasma sEVs ().
Figure 4. PRAD patient plasma-derived sEVs are enriched in SYP and sEV classical markers. (a) Immunoblotting (IB) analysis of pooled plasma sEV fractions (Fractions 6–8) from healthy donor (4–13) or PRAD patients (M-Z); 20 μg of protein were loaded per sEV sample. Expression of synaptophysin (SYP), CD63, Syntenin, Alix, and CD9 was analyzed. (b) Nanoparticle tracking analysis (NTA) of size and concentration of pooled sEVs (Fractions 6–8) from healthy donor plasma (7 + 8 + 9) or PRAD patient plasma (R+S+T).
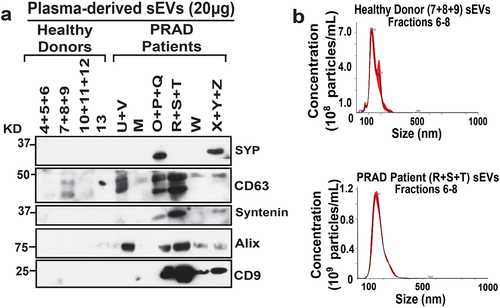
Nanoparticle tracking analysis (NTA) shows that the average size of healthy donor and PRAD patient-derived plasma sEVs is ~ 150-160 nm, falling within the expected size range of sEVs ().
We analyzed the plasma-derived sEVs by IB, and our results show that SYP and sEV markers are enriched in patient plasma sEVs. Then, we reviewed groups, which are a five‐tier grade system based on Gleason score grouping and different histology definitions,Citation32 the pathological state at diagnosis, and prostate-specific antigen (PSA) levels, and found no correlation with these parameters (). Patients diagnosed with T2N0M0 (tumor confined within prostate, no regional lymph node metastasis and no distant metastasis)Citation33 express different levels of CD63, Syntenin, Alix, and CD9 compared to patients diagnosed with more advanced PRAD, T3BN0M0 (tumor with extracapsular extension-seminal vesicle invasion, with no regional nodal or metastatic spread).Citation33 Overall, these data show that SYP and classical sEV marker expression is variable but, in most cases, enriched in PRAD patient plasma sEVs as compared to healthy donor sEVs.
Table 2. Synaptophysin and sEV marker expression in PRAD patient derived plasma sEVs.
Discussion
In this study, we show expression of neuroendocrine genes in PRAD as well as BRCA, KIRC, KICH, and KIRP. Our results pave the way for exploring new perspectives on the functions of neuroendocrine genes in these cancers. Among other genes, we show that SYP mRNA levels are increased in PRAD and SYP protein levels are enriched in PRAD patient plasma-derived sEVs, although absent in healthy donor sEVs.
We analyzed RNA-seq data from PRAD, BRCA, KICH, KIRC, and KIRP patient tissues compared to normal tissues to establish a consistent pattern of up- or downregulation of the twenty-two neuroendocrine genes to shed light on the heterogeneity of these genes’ dysregulation patterns across the cancer types. SYP, a well-established marker for neuroendocrine cancers,Citation4,Citation34,Citation35 is upregulated in PRAD and BRCA, but downregulated in KIRC, KICH and KIRP. Studies have shown that the expression of SYP in PRAD patient tissues is rare since neuroendocrine markers are abundant only when the disease advances to a highly aggressive neuroendocrine stage after androgen deprivation therapy.Citation4,Citation35,Citation36 However, our bioinformatic analysis indicates that SYP may support the pro-tumorigenic progression of non-neuroendocrine cancers, such as PRAD. Aligning with these results, although some sEVs did not express SYP, it appears that sEVs derived from one PRAD patient with a less aggressive pathological state (T2N0M0), grade group (2) or Gleason score (3 + 4) do not differ in SYP expression compared to plasma sEVs derived from one PRAD patient with more aggressive pathological state (T3BN0M0), grade group (4–5) or Gleason score (8–9). Our data suggest that SYP is highly enriched in sEVs derived from PRAD cells and that these sEVs circulate in the bloodstream. As a result, SYP expression in sEVs derived from PRAD patient plasma sEVs could be instrumental as a prognostic tool for the early detection of PRAD. There are no studies about SYP expression in cancer patient derived EVs. Few studies address the presence of SYP in EVs, but not in the context of cancer.Citation37,Citation38 A study investigating the N-glycoproteome of urine-derived EVs (isolated via differential ultracentrifugation) showed that SYP is expressed and N-glycosylated in urine-derived EVs.Citation37 SYP has also been shown to be decreased in EVs (isolated by polymer precipitation, ExoQuick) derived from Alzheimer’s disease or frontotemporal dementia compared to healthy controls.Citation38 Additional studies screening for SYP in patient plasma-derived sEVs must be investigated in the future.
Furthermore, the discovery that twelve genes have significantly increased expression in NEPC versus CRPC strengthens the relationship of these genes to neuroendocrine transformation and pathogenesis. Overall, our bioinformatic analysis results show that the dysregulation of the neuroendocrine genes in cancer tissues could play a key role in treatment resistance and tumor progression in select cancer types.
Our data show that the classical sEV markers Alix, Syntenin as well as the tetraspanins CD9 and CD63 are enriched in sEVs derived from PRAD patient plasma compared to sEVs derived from healthy donor plasma. This result aligns with our previous studies showing that PRAD patient derived sEVs are enriched in CD9 compared to plasma sEVs from healthy donors.Citation31 Furthermore, a study has demonstrated that plasma-derived EVs from pancreatic or lung cancer patients are enriched in classical sEV markers such as CD9, HSPA8, Alix and HSP90AB1 compared to the healthy control counterparts.Citation30 Another publication revealed that Alix is enriched in plasma-derived EVs from pancreatic cancer, thus showing its potential to be a biomarker to distinguish early from advanced stage pancreatic cancer.Citation39 Similarly, Alix is able to discriminate between plasma-derived EVs from melanoma patients compared to healthy donor plasma EVs.Citation40 Moreover, another group has shown that Syntenin is important for increased secretion of sEVs derived from human lung cancer cells, which, in turn, positively regulates endothelial cell migration and tube formation.Citation41 Additionally, CD63 has been shown to have increased expression in PRAD patient tissue (grade group 3–5) compared to patients with benign hyperplasia.Citation42 Since Alix and Syntenin are part of the molecular machinery responsible for sEV biogenesis,Citation6 and the tetraspanins can modulate membrane curvature to enable sEV release,Citation8 we can speculate that PRAD patient plasma-derived sEVs are released from prostate cancer cells at a higher degree compared to healthy donor cells and that EV subpopulations may arise as PRAD progresses. These findings synergize with a recently published paper by the Beltran group,Citation43 which showed that DNA methylation on specific genomic regions can distinguish NEPC patients from CRPC patients. Thus, they developed a neuroendocrine detection and monitoring (NEMO) assay, in which they used a plasma cell-free DNA-targeted methylation panel that allowed them to stratify and differentiate CRPC from NEPC patients. As a result, it may be possible that cell-free DNA in plasma originates from plasma-derived sEVs, and thus, both represent valuable tools for noninvasive liquid biopsies. Along the same lines, a study showed that BRCA patient plasma-derived EVs or cell-free DNA are both informative biomarker sources in BRCA, specifically for HER2 positive patients.Citation44 Moreover, the data demonstrated that combining EV and cell-free DNA assessments increased detection performance.Citation44 Therefore, we can speculate that aligning findings from plasma-derived EV content and cell-free DNA from PRAD and NEPC patients may increase the performance of liquid biopsies.
Our results also show that sEVs derived from PRAD patients with a less aggressive form of PRAD (grade group 2) do not significantly differ in sEV marker expression compared to sEVs derived from more advanced (grade group 3–5) PRAD patients; however, the presence of sEV markers vary among them. Therefore, we speculate that sEV marker expression starts in less aggressive forms of PRAD and increases as PRAD progresses. Overall, we provide novel evidence in which a neuroendocrine gene, such as SYP or patient plasma sEV content, can be used as markers for diagnostic purposes in PRAD. Future studies based on proteomic analysis of PRAD and NEPC patient tumors and sEVs will provide a comprehensive understanding of the role of PRAD derived sEVs in promoting cancer progression.
Materials and methods
Evaluation of differentially expressed neuroendocrine genes across cancer types
The expression levels of literature-curated neuroendocrine cancer marker genes were evaluated in Prostate adenocarcinoma (PRAD), Breast cancer (BRCA), Kidney chromophobe (KICH), Kidney renal clear cell (KIRC) carcinoma, and Kidney renal papillary cell (KIRP) carcinoma patient samples compared to the normal samples. For this analysis, the RNA-seq expression profiling data were downloaded from the Cancer Genome Atlas (TCGA) database using the R/Bioconductor package TCGAbiolinks.Citation45 The read counts were imported to the R package edgeRCitation46 to normalize the data and determine differentially expressed genes. Neuroendocrine genes were identified with significant differential expression in: PRAD (n = 502) compared to normal (n = 52) tissue samples, BRCA (n = 1118) compared to normal (n = 113) tissue samples, KICH (n = 66) compared to normal (n = 25) tissue samples, KIRC (n = 541) compared to normal (n = 72) tissue samples, KIRP (n = 290) compared to normal (n = 32) tissue samples. The obtained differential expression P-values were further adjusted by the Benjamini-Hochberg (BH) method.Citation47 Neuroendocrine genes with an adjusted p value < 0. 05 were considered significantly differentially expressed in the respective cancer type compared to its normal samples. Neuroendocrine genes were identified with significant differential expression in neuroendocrine prostate cancer (NEPC) (n = 13) compared to PRAD (n = 34) samples using the above edgeR analysis pipeline. For this analysis, the RNA-seq expression profiling data were obtained directly from the author upon request.Citation48
Human samples
Blood samples for the isolation of plasma-derived sEVs were obtained from diagnosed PRAD patients (n = 13). Biological samples were obtained through a biorepository under an honest broker policy upon patient consent prior to any treatment. Specimens were de-identified in accordance with guidelines established by the Institutional Review Board (IRB), protocol 20D.826. Data collected included pathological stage, Gleason score, grade group, and metastasis at the time of radical prostatectomy; prostate-specific antigen (PSA) was recorded pre-prostatectomy.
Blood samples for the isolation of plasma-derived sEVs were obtained from healthy donors (n = 10). Written consent was documented in accordance with the IRB approved protocol 19D.011.
Plasma isolation
Human blood was withdrawn via venipuncture. Coagulation was prevented by treatment with Acid Citrate Dextrose (ACD) [trisodium citrate (22.0 g/L), citric acid (8.0 g/L) and dextrose (24.5 g/L)]. Anticoagulated blood was centrifuged at 100 x g for 20 minutes at room temperature with acceleration 1 and brake 0 to separate plasma.
sEV isolation
EV isolation by differential ultracentrifugation of healthy donor and diagnosed PRAD patient plasma was performed as previously described.Citation31,Citation49,Citation50
sEV isolation from the pellet obtained after ultracentrifugation was further processed by iodixanol density gradient (IDG) ultracentrifugation as previously described.Citation49–51 All the data presented in this study were generated using IDG sEVs.
NTA
NTA was performed as previously described.Citation50,Citation51 Briefly, pooled plasma sEVs (fractions 6–8) were diluted in PBS and analyzed using NanoSight NS300. A total of three videos (30 seconds each) were captured. Data were collected at room temperature (25◦C). Data analysis was performed using NTA software version 3.1.54.
IB analysis
IB analysis was performed as previously described.Citation50–52 For IB, sEVs and cells were lysed using RIPA buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% SDS, 1% NP-40, and 1% sodium deoxycholate) and protease inhibitors were added. The following primary rabbit Ab was used: Syntenin (Abcam, ab133267). The following primary mouse Abs were used: SYP (Invitrogen MA-1-213), Alix (BIO-RAD, MCA2493), CD63 (Abcam, ab193349), or CD9 (Santa Cruz, sc13118).
The secondary HRP-linked Abs were: anti-rabbit IgG (Cell Signaling, 7074S) or anti-mouse IgG (Cell Signaling, 7076S). For protein visualization, WesternBrightTM ECL HRP (horseradish peroxidase) substrate kits (Advansta) were used.
Author contributions
L.R.L., R.M. and W.K.K. supervised the study and contributed to manuscript writing. N.M.N. performed the biochemical and NTA analysis, prepared figures and contributed to manuscript writing. A.K. performed searches related to the bioinformatic analysis and contributed to manuscript writing. N.M.N. and A.T. analyzed biological samples of healthy volunteers, generated, and characterized PRAD patient plasma sEVs. W.K.K. supervised the team which provided PRAD patient plasma. R.M. and O.A. performed the bioinformatic analysis. C.E.V., M.W.P., A.D.A., and R.K. provided technical support. A.H. contributed to with PRAD patient stratification according to the pathology reports. D.C.H., J.Y., and J.Z. reviewed the results of the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgments
The authors would like to thank S. Yadav and U. Naik for their support and S. Cremin, S. Herder, and S. Hinojosa for providing discarded, de-identified prostate cancer patients’ plasma (Thomas Jefferson Biobank repository). We thank Md. Niamat Hossain and Camila Bach, for their help in reviewing the manuscript. We also thank Lauren Langbein for her help with editing the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used for the bioinformatics analysis are publicly available. The additional data that support the findings of this study are available from the corresponding authors L.R.L., R.M. or W.K.K. upon reasonable request.
Additional information
Funding
References
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics, 2024. CA Cancer J Clin. 2024;74(1):12–9. doi:10.3322/caac.21820.
- Haffner MC, Zwart W, Roudier MP, True LD, Nelson WG, Epstein JI, De Marzo AM, Nelson PS, Yegnasubramanian S. Genomic and phenotypic heterogeneity in prostate cancer. Nat Rev Urol. 2021;18(2):79–92. doi:10.1038/s41585-020-00400-w.
- Beer TM, Armstrong AJ, Rathkopf DE, Loriot Y, Sternberg CN, Higano CS, Iversen P, Bhattacharya S, Carles J, Chowdhury S, et al. Enzalutamide in metastatic prostate cancer before chemotherapy. N Engl J Med. 2014;371(5):424–33. doi:10.1056/NEJMoa1405095.
- Sainio M, Visakorpi T, Tolonen T, Ilvesaro J, Bova GS. Expression of neuroendocrine differentiation markers in lethal metastatic castration-resistant prostate cancer. Pathol Res Pract. 2018;214(6):848–56. doi:10.1016/j.prp.2018.04.015.
- Moller A, Lobb RJ. The evolving translational potential of small extracellular vesicles in cancer. Nat Rev Cancer. 2020;20(12):697–709. doi:10.1038/s41568-020-00299-w.
- Mathieu M, Martin-Jaular L, Lavieu G, Thery C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. 2019;21(1):9–17. doi:10.1038/s41556-018-0250-9.
- Welsh JA, Goberdhan DCI, O’Driscoll L, Buzas EI, Blenkiron C, Bussolati B, Cai H, Di Vizio D, Driedonks TAP, Erdbrügger U, et al. Minimal information for studies of extracellular vesicles (MISEV2023): from basic to advanced approaches. J Extracell Vesicles. 2024;13(2):e12404. doi:10.1002/jev2.12404.
- Jankovicova J, Secova P, Michalkova K, Antalikova JT. Tetraspanins, more than markers of extracellular vesicles in reproduction. Int J Mol Sci. 2020;21(20):7568. doi:10.3390/ijms21207568.
- Santos MF, Rappa G, Fontana S, Karbanová J, Aalam F, Tai D, Li Z, Pucci M, Alessandro R, Morimoto C. et al. Anti-human CD9 fab fragment antibody blocks the extracellular vesicle-mediated increase in Malignancy of colon cancer cells. Cells. 2022;11(16):2474. doi:10.3390/cells11162474.
- Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478). doi:10.1126/science.aau6977.
- DeRita RM, Zerlanko B, Singh A, Lu H, Iozzo RV, Benovic JL, Languino LR. c-Src, insulin-like growth factor I receptor, G-Protein-coupled receptor kinases and focal adhesion kinase are enriched into prostate cancer cell exosomes. J Cell Biochem. 2017;118(1):66–73. doi:10.1002/jcb.25611.
- Krishn SR, Salem I, Quaglia F, Naranjo NM, Agarwal E, Liu Q, Sarker S, Kopenhaver J, McCue PA, Weinreb PH. et al. The αvβ6 integrin in cancer cell-derived small extracellular vesicles enhances angiogenesis. J Extracell Vesicles. 2020;9(1):1763594. doi:10.1080/20013078.2020.1763594.
- Osti D, Del Bene M, Rappa G, Santos M, Matafora V, Richichi C, Faletti S, Beznoussenko GV, Mironov A, Bachi A, et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin Cancer Res. 2019;25(1):266–76. doi:10.1158/1078-0432.CCR-18-1941.
- Block T, Zezulinski D, Kaplan DE, Lu J, Zanine S, Zhan T, Doria C, Sayeed A. Circulating messenger RNA variants as a potential biomarker for surveillance of hepatocellular carcinoma. Front Oncol. 2022;12:963641. doi:10.3389/fonc.2022.963641.
- Hofsli E, Wheeler TE, Langaas M, Laegreid A, Thommesen L. Identification of novel neuroendocrine-specific tumour genes. Br J Cancer. 2008;99(8):1330–39. doi:10.1038/sj.bjc.6604565.
- Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321(3):288–300. doi:10.1001/jama.2018.19323.
- Hsieh JJ, Purdue MP, Signoretti S, Swanton C, Albiges L, Schmidinger M, Heng DY, Larkin J, Ficarra V. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3(1):17009. doi:10.1038/nrdp.2017.9.
- Haake SM, Rathmell WK. Renal cancer subtypes: should we be lumping or splitting for therapeutic decision making? Cancer. 2017;123(2):200–09. doi:10.1002/cncr.30314.
- Mendhiratta N, Muraki P, Sisk AE Jr., Shuch B. Papillary renal cell carcinoma: review. Urol Oncol. 2021;39(6):327–37. doi:10.1016/j.urolonc.2021.04.013.
- Alaghehbandan R, Przybycin CG, Verkarre V, Mehra R. Chromophobe renal cell carcinoma: novel molecular insights and clinicopathologic updates. Asian J Urol. 2022;9(1):1–11. doi:10.1016/j.ajur.2021.11.010.
- Stridsberg M. The use of chromogranin, synaptophysin and islet amyloid polypeptide as markers for neuroendocrine tumours. Ups J Med Sci. 1995;100(3):169–99. doi:10.3109/03009739509178905.
- Louthan O. Chromogranin a in physiology and oncology. Folia Biol (Praha). 2011;57(5):173–81.
- Soh MA, Garrett SH, Somji S, Dunlevy JR, Zhou XD, Sens MA, Bathula CS, Allen C, Sens DA. Arsenic, cadmium and neuron specific enolase (ENO2, γ-enolase) expression in breast cancer. Cancer Cell Int. 2011;11(1):41. doi:10.1186/1475-2867-11-41.
- Korse CM, Taal BG, Vincent A, van Velthuysen MLF, Baas P, Buning-Kager JCGM, Linders TC, Bonfrer JMG. Choice of tumour markers in patients with neuroendocrine tumours is dependent on the histological grade. A marker study of chromogranin A, neuron specific enolase, progastrin-releasing peptide and cytokeratin fragments. Eur J Cancer. 2012;48(5):662–71. doi:10.1016/j.ejca.2011.08.012.
- Astarloa R, Sanchez-Franco F, Cacicedo L, Garcia-Villanueva M. Differential expression of neurofilament triplet proteins in carcinoid tumours: an immunohistochemical study. Br J Cancer. 1991;63(5):715–18. doi:10.1038/bjc.1991.161.
- Lee JK, Phillips JW, Smith BA, Park JW, Stoyanova T, McCaffrey EF, Baertsch R, Sokolov A, Meyerowitz JG, Mathis C, et al. N-Myc drives neuroendocrine prostate cancer initiated from human prostate epithelial cells. Cancer Cell. 2016;29(4):536–47. doi:10.1016/j.ccell.2016.03.001.
- Li Z, Xie J, Fei Y, Gao P, Xie Q, Gao W, Xu Z. GDNF family receptor alpha 2 promotes neuroblastoma cell proliferation by interacting with PTEN. Biochem Biophys Res Commun. 2019;510(3):339–44. doi:10.1016/j.bbrc.2018.12.169.
- Cancer Genome Atlas Research N, Ahn J, Akbani R, Ally A, Amin S, Andry C, Annala M, Aprikian A, Armenia J, Arora A, et al. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–25. doi:10.1016/j.cell.2015.10.025.
- Bowes AL, Tarabichi M, Pillay N, Van Loo P. Leveraging single-cell sequencing to unravel intratumour heterogeneity and tumour evolution in human cancers. J Pathol. 2022;257(4):466–78. doi:10.1002/path.5914.
- Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, et al. Extracellular Vesicle and particle biomarkers define multiple human cancers. Cell. 2020;182(4):1044–61.e18. doi:10.1016/j.cell.2020.07.009.
- Krishn SR, Singh A, Bowler N, Duffy AN, Friedman A, Fedele C, Kurtoglu S, Tripathi SK, Wang K, Hawkins A, et al. Prostate cancer sheds the αvβ3 integrin in vivo through exosomes. Matrix Biol. 2019;77:41–57. doi:10.1016/j.matbio.2018.08.004.
- Matoso A, Epstein JI. Grading of prostate cancer: past, present, and future. Curr Urol Rep. 2016;17(3):25. doi:10.1007/s11934-016-0576-4.
- Cheng L, Montironi R, Bostwick DG, Lopez-Beltran A, Berney DM. Staging of prostate cancer. Histopathology. 2012;60(1):87–117. doi:10.1111/j.1365-2559.2011.04025.x.
- Bery F, Cancel M, Chantôme A, Guibon R, Bruyère F, Rozet F, Mahéo K, Fromont G. The calcium-sensing receptor is a marker and potential driver of neuroendocrine differentiation in prostate cancer. Cancers (Basel). 2020;12(4):860. doi:10.3390/cancers12040860.
- Yamada Y, Beltran H. Clinical and biological features of neuroendocrine prostate cancer. Curr Oncol Rep. 2021;23(2):15. doi:10.1007/s11912-020-01003-9.
- Bhinder B, Ferguson A, Sigouros M, Uppal M, Elsaeed AG, Bareja R, Alnajar H, Eng KW, Conteduca V, Sboner A, et al. Immunogenomic landscape of neuroendocrine prostate cancer. Clin Cancer Res. 2023;29(15):2933–43. doi:10.1158/1078-0432.CCR-22-3743.
- Saraswat M, Joenväära S, Musante L, Peltoniemi H, Holthofer H, Renkonen R. N-linked (N-) glycoproteomics of urinary exosomes [Corrected]. Mol Cell Proteomics: mCP. 2015;14(2):263–76. doi:10.1074/mcp.M114.040345.
- Goetzl EJ, Kapogiannis D, Schwartz JB, Lobach IV, Goetzl L, Abner EL, Jicha GA, Karydas AM, Boxer A, Miller BL, et al. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer’s disease. FASEB J. 2016;30(12):4141–48. doi:10.1096/fj.201600816R.
- Yang J, Zhang Y, Gao X, Yuan Y, Zhao J, Zhou S, Wang H, Wang L, Xu G, Li X, et al. Plasma-derived exosomal ALIX as a novel biomarker for diagnosis and classification of pancreatic cancer. Front Oncol. 2021;11:628346. doi:10.3389/fonc.2021.628346.
- Pietrowska M, Zebrowska A, Gawin M, Marczak L, Sharma P, Mondal S, Mika J, Polańska J, Ferrone S, Kirkwood JM, et al. Proteomic profile of melanoma cell-derived small extracellular vesicles in patients’ plasma: a potential correlate of melanoma progression. J Extracell Vesicles. 2021;10(4):e12063. doi:10.1002/jev2.12063.
- Kim O, Hwangbo C, Tran PT, Lee JH. Syntenin-1-mediated small extracellular vesicles promotes cell growth, migration, and angiogenesis by increasing onco-miRnas secretion in lung cancer cells. Cell Death Dis. 2022;13(2):122. doi:10.1038/s41419-022-04594-2.
- Folkmanis K, Junk E, Merdane E, Folkmane I, Folkmanis V, Ivanovs I, Eglitis J, Jakubovskis M, Laabs S, Isajevs S, et al. Clinicopathological significance of exosomal proteins CD9 and CD63 and DNA mismatch repair proteins in prostate adenocarcinoma and benign hyperplasia. Diagnostics (Basel). 2022;12(2):287. doi:10.3390/diagnostics12020287.
- Franceschini GM, Quaini O, Mizuno K, Orlando F, Ciani Y, Ku S-Y, Sigouros M, Rothmann E, Alonso A, Benelli M, et al. Noninvasive detection of neuroendocrine prostate cancer through targeted cell-free DNA methylation. Cancer Discov. 2024;14:424–45. doi:10.1158/2159-8290.CD-23-0754.
- Mugoni V, Ciani Y, Quaini O, Tomasini S, Notarangelo M, Vannuccini F, Marinelli A, Leonardi E, Pontalti S, Martinelli A. et al. Integrating extracellular vesicle and circulating cell-free DNA analysis using a single plasma aliquot improves the detection of HER2 positivity in breast cancer patients. J Extracell Biol. 2023;2(9):e108. doi:10.1002/jex2.108.
- Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, Sabedot TS, Malta TM, Pagnotta SM, Castiglioni I, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res. 2016;44(8):e71. doi:10.1093/nar/gkv1507.
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. doi:10.1093/bioinformatics/btp616.
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57(1):289–300. doi:10.1111/j.2517-6161.1995.tb02031.x.
- Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BVSK, Varambally S, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22(3):298–305. doi:10.1038/nm.4045.
- Salem I, Naranjo NM, Singh A, DeRita R, Krishn SR, Sirman LS, Quaglia F, Duffy A, Bowler N, Sayeed A, et al. Methods for extracellular vesicle isolation from cancer cells. Cancer Drug Resist. 2020;3:1–14. doi:10.20517/cdr.2019.118.
- Testa A, Quaglia F, Naranjo NM, Verrillo CE, Shields CD, Lin S, Pickles MW, Hamza DF, Von Schalscha T, Cheresh DA, et al. Targeting the αVβ3/NgR2 pathway in neuroendocrine prostate cancer. Matrix Biol. 2023;124:49–62. doi:10.1016/j.matbio.2023.11.003.
- Naranjo NM, Salem I, Harris MA, Languino LR. IFIT3 (Interferon Induced Protein with Tetratricopeptide Repeats 3) modulates STAT1 expression in small extracellular vesicles. Biochem J. 2021;478(21):3905–21. doi:10.1042/BCJ20210580.
- Quaglia F, Krishn SR, Sossey-Alaoui K, Rana PS, Pluskota E, Park PH, Shields CD, Lin S, McCue P, Kossenkov AV, et al. The NOGO receptor NgR2, a novel αVβ3 integrin effector, induces neuroendocrine differentiation in prostate cancer. Sci Rep. 2022;12(1):18879. doi:10.1038/s41598-022-21711-5.
