ABSTRACT
Lung squamous cell carcinoma (LSCC) is a deadly cancer in the world. Histone demethylase Jmjd2c is a key epigenetic regulator in various tumors, while the molecular mechanism underlying Jmjd2c regulatory in LSCC is still unclear. We used the aldehyde dehydrogenasebright (ALDHbri+) subtype as a research model for cancer stem cells (CSCs) in LSCC and detected the sphere formation ability and the proportion of ALDHbri+ CSCs with Jmjd2c interference and caffeic acid (CA) treatment. Additionally, we carried out bioinformatic analysis on the expression file of Jmjd2c RNAi mice and performed western blotting, qRT-PCR, Co-IP and GST pull-down assays to confirm the bioinformatic findings. Moreover, we generated Jmjd2c-silenced and Jmjd2c-SOX2-silenced ALDHbri+ tumor-bearing BALB/c nude mice to detect the effects on tumor progression. The results showed that Jmjd2c downregulation inhibited the sphere formation and the proportion of ALDHbri+ CSCs. The SOX2 decreased expression significantly in Jmjd2c RNAi mice, and they were positively co-expressed according to the bioinformatic analysis. In addition, SOX2 expression decreased in Jmjd2c shRNA ALDHbri+ CSCs, Jmjd2c and SOX2 proteins interacted with each other. Furthermore, Jmjd2c interference revealed significant blocking effect, and Jmjd2c-SOX2 interference contributed even stronger inhibition on ALDHbri+ tumor progression. The Jmjd2c and SOX2 levels were closely related to the development and prognosis of LSCC patients. This study indicated that Jmjd2c played key roles on maintaining ALDHbri+ CSC activity in LSCC by interacting with transcription factor SOX2. Jmjd2c might be a novel molecule for therapeutic targets and biomarkers in the diagnosis and clinical treatment of lung cancer.
Introduction
Lung cancer is one of the most common malignant tumors in humans, and its morbidity and mortality have been on the rise in recent years.Citation1 LSCC is one of the common subtypes that associates with poor clinical prognosis and lacks available targeted therapy.Citation2,Citation3 It is inevitable that drug resistance developed by LSCC patients to platinum-based chemotherapy leads to tumor recurrence.Citation4 The 5-y survival rate of patients has been hovering at 15%.Citation5 Thus, it is of urgent requirement to explore novel molecules for therapeutic targets and biomarkers in LSCC.
Epigenetic aberrations have been described in the etiology of cancer that chromatin modifiers and remodelers such as DNA methylation, histone modification or microRNA, which disrupt gene expressions, are believed to be strongly related to the development and progression of solid malignancies.Citation6–8 In contrast to genetic changes, epigenetic alterations are pharmaceutically reversible.Citation9,Citation10 Reliable epigenetic markers are targeted for clinical diagnosis and treatment. Jmjd2c, originally cloned from LSCC in 2000, is a member of Jumonji C domains containing proteins (JmjD family), which are a major class of histone lysine demethylases.Citation11–14 Jmjd2c is capable of demethylating tri- or di-methylated histone 3/2 in either K9 or K36 residuals, balancing the methylation state of histones and affecting heterochromatin formation, genomic imprinting and gene expression regulation.Citation15 Jmjd2c was involved in embryonic and stem cell regulation,Citation16 and defects from Jmjd2c function were associated with the genesis and progression of various tumors, such as osteosarcoma, breast cancer, blood neoplasms, prostate carcinomas, and so on.Citation17–20 Furthermore, Jmjd2c was observed overexpressed in lung tumors, and revealed even higher expressions in metastatic lung tissues than that in non-metastatic ones.Citation21 Knockdown of Jmjd2c blocked the migration and invasion of lung cancer cells,Citation21 which indicated that Jmjd2c was a promising anti-lung-cancer target. However, the molecular mechanism underlying Jmjd2c regulation in lung cancers has not yet been clarified.
CSCs are a subpopulation of tumor cells, possessing high self-renewal and differentiation properties, which promote tumor initiation or cause relapses.Citation22,Citation23 CSCs are thought to be the source of all tumor cells in malignant tumors and the reason for the resistance to chemotherapy.Citation24 Thus, reducing CSCs in residual tumors and effectively maintaining chemotherapy sensitivity may become a breakthrough in solving drug resistance, reducing recurrence rate, and improving prognosis for patients.Citation5 Transcription factor sex-determining region Y-box2 (SOX2) is a key regulator in stem cell biology,Citation25 and dysregulation of SOX2 expression induces pluripotent stem cells, drives CSC properties, fuels tumorigenesis, and promotes tumor aggressiveness.Citation26 SOX2 is closely correlated with self-renewal and chemoresistance of CSCs.Citation27
In this study, we used the ALDbri+ subtype as a research model for CSCs in LSCC, and confirmed that Jmjd2c interference suppressed the self-renewal and pluripotent CSC generation. In addition, we elucidated that SOX2 expression was regulated by Jmjd2c, meanwhile they positively co-expressed with each other at protein levels with bioinformatic analysis on the expression profiling of the Jmjd2c RNAi mice, and confirmed these findings in ALDHbri+ CSCs. Moreover, we established Jmjd2c-silenced and Jmjd2c-SOX2-silenced ALDHbri+ tumor-bearing BALB/c nude mice and explained that Jmjd2c silence retarded tumor growth and Jmjd2c-SOX2 silence contributed stronger prevention on tumor progression. Thus, this study demonstrated that Jmjd2c maintained the ALDHbri+ CSC activity with transcription factor SOX2 and acted as a mark in LSCC. Inhibitors targeting Jmjd2c are expected to become anti-tumor drugs for clinical treatment of lung cancer.
Results
Jmjd2c was necessary for the self-renewal and pluripotent ALDHbri+ CSC generation
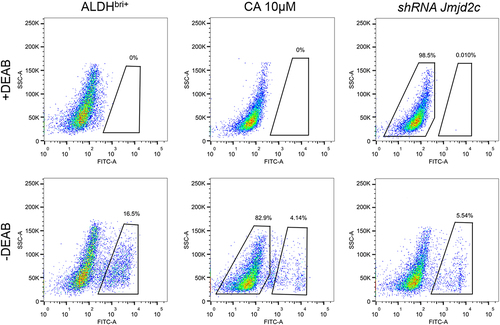
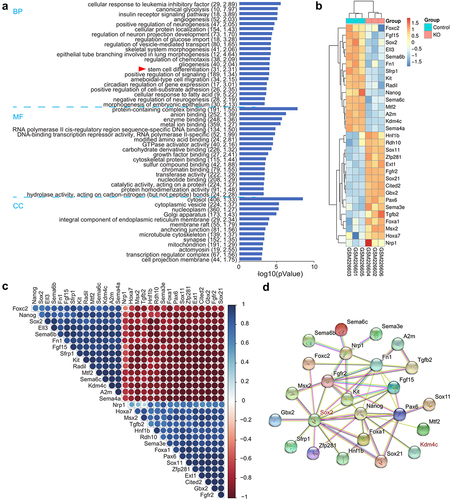
To investigate the function of Jmjd2c on CSC regulation and tumor development, we detected the self-renewal potential of CSCs by the sphere formation assay in Jmjd2c-silenced and CA treated ALDHbri+ CSCs. The results showed that both constructions of Jmjd2c interference significantly reduced the CSC sphere numbers compared to the control group of ALDHbri+ CSCs (**p < 0.01) (), and the following shRNA jmjd2c interference refers to shRNA-2 jmjd2c (Figure S1). In addition, the inhibitor of the Jmjd2c CA treatment significantly reduced the value of spheres/5000 cells, and higher concentration (10 μM, refer to the following CA treatment) revealed stronger inhibition of CSC sphere formation capacity (**p < 0.01) (). Results from aldefluor assay showed that both the Jmjd2c silencing and CA treatment dramatically reduced the ratio of ALDHbri+ CSCs, the values were 5.54% and 4.14%, respectively, compared to the 16.5% in the control (). These results indicated that Jmjd2c was a key regulator of ALDHbri+ CSC activity in lung squamous cell carcinoma.
Figure 1. The clonal sphere-forming ability with Jmjd2c silencing and CA treatment in ALDHbri+ CSCs. (a) The images of the formed spheres. (b) and (c) the quantification of the sphere sizes in a with Jmjd2c silencing (b) and CA treatment (c) in ALDHbri+ CSCs. Data were represented as means±SD, n ≥ 3; **p < 0.01. Bars = 100 μm.
SOX2 was involved in Jmjd2c signaling regulating stem cell differentiation
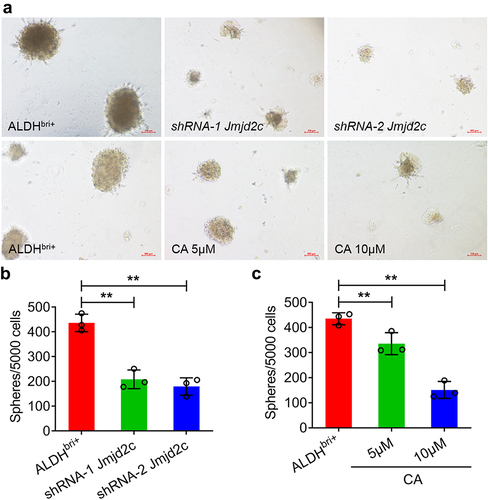
To figure out the molecular mechanism underlying Jmjd2c signaling regulating lung cancer progression, we carried out a bioinformatics analysis on the expression profiling using an array of the Jmjd2c RNAi mice. The volcano plot analysis showed 977 DEGs upregulated and 731 DEGs downregulated, which were depicted in a heatmap (Figure S3). Further GO analysis of the DEGs revealed that 31 genes classified as stem cell differentiation in BP category were enriched significantly (). Among these 31 genes, the heatmap analysis showed 15 genes were downregulated and 16 genes were upregulated, and both SOX2 and Jmjd2c (Kdm4c) were deregulated (). Correlation analysis indicated that SOX2 and Jmjd2c were absolutely positive () and protein–protein interaction network analysis by STRING showed SOX2 and Jmjd2c could interact with each other (). Thus, these results suggested that SOX2 was a crucial regulator involved in Jmjd2c signaling regulating stem cell activity.
Figure 3. The GO and protein-protein association analysis of the Jmjd2c RNAi mice. (a) The GO analysis of BP (biological process), MF (molecular function), and CC (cellular component) for the genes. X-axis: The minus logarithm of the P-value. Y-axis: the GO category. (b) A heatmap showed the 31 DEGs classified to stem cell differentiation in GO category. X axis: sample name; Y axis: gene name. (c) Correlation of the DEGs classified to stem cell differentiation was analyzed. The correlation coefficient ranges from − 1 (red color) to 1 (blue color). The red represented absolute negative correlations, and blue represented absolute positive correlations. (d) The protein–protein interaction network of the DEGs classified to stem cell differentiation was analyzed by STRING.
To further confirm these findings, we performed western blotting and quantitative real-time PCR (qRT-PCR) experiments to detect the SOX2 expressions in Jmjd2c silenced ALDHbri+ CSCs (). The protein expressions of Jmjd2c and SOX2 were both dramatically increased in ALDHbri+ CSCs compared to the CSC controls (**p < 0.01, ***p < 0.001) (). However, they both substantially reduced in Jmjd2c-silenced or CA-treated ALDHbri+ CSCs (**p < 0.01) (). The mRNA expressions from qRT-PCR were consistent with the western blotting results (*p < 0.05, **p < 0.01) (). These results suggested that Jmjd2c regulated the expression of SOX2.
Figure 4. Jmjd2c regulated the expression of SOX2 and they interacted with each other at protein level. (a) The western blotting analysis showed the protein expressions of Jmjd2c and SOX2 with Jmjd2c silencing and CA treatment in ALDHbri+ CSCs. The red triangle indicated the bands of Jmjd2c. GAPDH was applied as internal control. (B) and (C) the quantitative analysis of Jmjd2c (b) and SOX2 (c) protein expressions normalized to GAPDH. (d) and (e) the mRNA expressions of Jmjd2c and SOX2 with Jmjd2c silencing and CA treatment in ALDHbri+ CSCs. (f) and (g) the Co-IP assay (f) and GST-pull down assay (g) showed the interaction of Jmjd2c with SOX2 in ALDHbri+ CSCs. β-tubulin was applied as the internal control. Data were represented as means±SD, n ≥ 3; *p < 0.05, **p < 0.01.
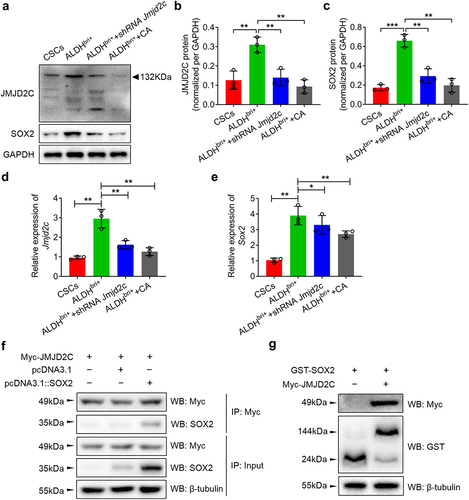
Moreover, the Co-immunoprecipitation (Co-IP) assay indicated that the Myc-Jmjd2c immunoprecipitated the SOX2 proteins (), which was confirmed by the GST pull-down assay that GST-SOX2 fused proteins pulled down the Myc-Jmjd2c proteins (). These results indicated that SOX2 and Jmjd2c interacted with each other at protein levels.
Jmjd2c-SOX2-silenced ALDHbri+ CSCs retarded the tumor progression potently in tumor-bearing BALB/c nude mice
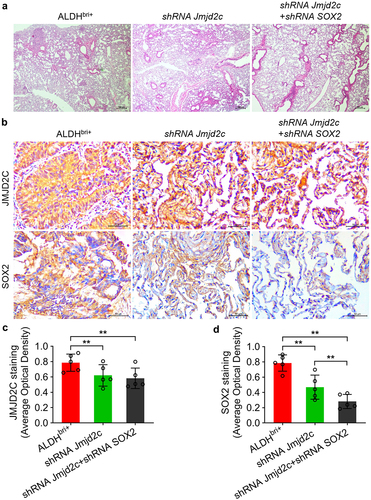
To clarify the biological impact of Jmjd2c-SOX2 signaling on tumor progression in mice, we generated Jmjd2c-silenced and Jmjd2c-SOX2-silenced ALDHbri+ tumor-bearing BALB/c nude mice (, Figure S1 and S2). The observation after 17-d ALDHbri+ CSC injection, the tumor size from Jmjd2c-SOX2-silenced mice was obviously smaller than that of the control group (). The statistical analysis showed that tumor size from Jmjd2c-silenced mice was significantly reduced compared to the ALDHbri+ controls, and the Jmjd2c-SOX2-silenced mice generated even more smaller tumors (**p < 0.01) (). The hematoxylin–eosin (HE) staining of the mouse lung tissues showed that clonogenicity of the Jmjd2c-SOX2-silenced mice was obviously reduced (), which suggested that the micrometastases of the ALDHbri+ CSCs were inhibited by Jmjd2c-SOX2 silencing. In addition, the immunochemistry results showed that the expressions of Jmjd2c and SOX2 decreased in lung tissues of the Jmjd2c-silenced mice (**p < 0.01) (), which was consistent with the western blotting analysis in ALDHbri+ CSCs (). SOX2 expression further declined to even lower levels in Jmjd2c-SOX2-silenced mouse lungs, but not Jmjd2c (**p < 0.01) ().
Figure 5. The tumorigenic capacity of Jmjd2c-silenced and Jmjd2c-SOX2-silenced ALDHbri+ CSCs in BALB/c nude mice. (a) The image of the BALB/c nude mice bearing tumors. (b) The tumors isolated from the groups of a were arranged and displayed. (c) The statistical analysis of the tumor sizes of the three groups within the 17 d. Data were represented as means±SD, n ≥ 3; **p < 0.01.
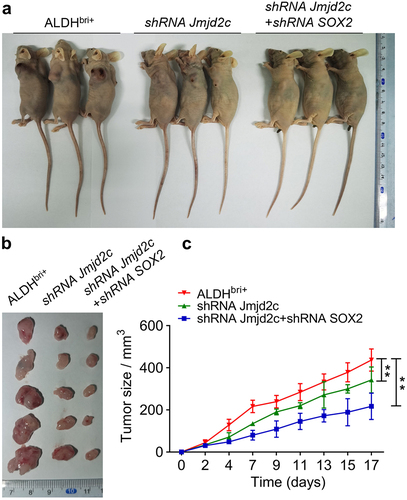
Figure 6. The micrometastases was reduced in mouse lung tissues injected with Jmjd2c-silenced and Jmjd2c-SOX2-silenced ALDHbri+ CSCs. (a) The HE staining results revealed the micrometastases in different groups. (b) The protein expressions of Jmjd2c and SOX2 with Jmjd2c silence or Jmjd2c-SOX2 silence from immunochemistry in mouse lung tissues. (c) and (d) quantification of the results in B. Bars = 100 μm.In A, 50 μm in B. Data were represented as means±SD, n ≥ 3; **p < 0.01.
Jmjd2c and SOX2 expression are closely related to the development and prognosis of LSCC patients
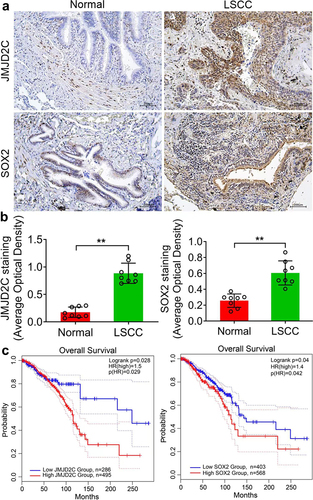
To further determine the role of Jmjd2c and SOX2 in LSCC, we collected patient tissues from the Department of Respiratory and Critical Care Medicine, The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology; 20 samples were from patients were diagnosed with LSCC tissues, and 20 were from paracancerous tissue. Immunohistochemistry showed that in normal and LSCC tissues, the Jmjd2c and SOX2 proteins were mainly located in the cytoplasm (). Furthermore, survival analysis of Jmjd2c and SOX2 was performed using Kaplan Meier-plotter. The results showed that Jmjd2c and SOX2 were negatively associated with the overall survival ().
Figure 7. Jmjd2c and SOX2 expression are closely related to the development and prognosis of LSCC. (a) Immunochemistry results showed the protein expressions of Jmjd2c and SOX2 in normal and LSCC tissues. (b) The quantitative analysis of Jmjd2c and SOX2 expressions in A. (c) Overall survival of Jmjd2c and SOX2 in LSCC based on Kaplan Meier-plotter. The patients were stratified into high-level group and low-level group according to different expression ratio. The data are represented as means ± SD, n ≥ 100,***p < 0.001.
Discussion
Lung cancer is emerging as the leading cause of death and has become a public health issue worldwide.Citation28,Citation29 LSCC is a non-small cell lung cancer (NSCLC), accounting for nearly 40% of lung cancer, but lacks targeted therapy and poor clinical diagnosis.Citation30,Citation31 Jmjd2c was suggested to epigenetically regulate cancer cells and altered Jmjd2c histone methylation regulation led to various tumor initiation and progression.Citation17,Citation31 In this study, Jmjd2c interference or CA treatment significantly inhibited ALDHbri+ CSC sphere formation and reduced the proportion of ALDHbri+ CSCs. These results indicated that Jmjd2c was a key regulator maintaining ALDHbri+ CSC stemness and that the self-renewal ability of ALDHbri+ CSCs was sensitive to the downregulation of Jmjd2c. As CSCs are a population of self-renewal cells with high tumorigenic potency and cancer is viewed as stem cell diseases,Citation32,Citation33 Jmjd2c should be a promising target and inhibition of Jmjd2c might be a choice for treatment of LSCC patients, which was confirmed by the previous uncovers that knockdown of Jmjd2c suppressed the migration and invasion of lung cancer cells.Citation21
Emerging evidence revealed that histone methylation played a crucial role in regulating gene expression and genome stability.Citation34 The JmJC domain was identified as a novel demethylase signature motif, and demethylation conducted by JmjC domain containing protein was conserved from yeast to human.Citation35 In this study, 31 genes classified as stem cell differentiation were enriched in the microarray data of Jmjd2c RNAi mice. Thus, Jmjd2c epigenetically regulated expressions of genes mediating stem cell activity. SOX2 was significantly decreased in Jmjd2c RNAi mice and Jmjd2c silenced ALDHbri+ CSCs. SOX2 was a key player mediating the stemness of various stem cells. It expressed that CSCs and abnormal SOX2 expressions were associated with kinds of cancer types and promoted resistance to chemotherapies.Citation36 Thus, Jmjd2c might maintain the ALDHbri+ CSC stemness through epigenetically regulating SOX2 expressions.
In addition, SOX2 and Jmjd2c were positively co-expressed according to the correlation analysis, and they interacted with each other based on the results from Co-IP assay and GST pull-down assay in ALDHbri+ CSCs. Loose chromatin structures facilitate transcription factors accessing their target DNA sequences, during which epigenetic factors play key roles.Citation37 Jmjc2d was reported to demethylate H3K9me2/3 and H3K36me2/3, two histone modifications involving transcriptional repression.Citation38 Thus, the interaction of Jmjd2c and transcription factor SOX2 might indicate that Jmjd2c relaxed chromatins by balancing the methylation state of histones and facilitated the recruitment of SOX2 to the transcription start site (TSS), affecting a series of downstream gene expressions and maintaining cancer cell stemness in LSCC. In this study, Jmjd2c-SOX2 interfered ALDHbri+ tumor bearing mice revealed weak clonogenicity, inhibiting the micrometastases of the ALDHbri+ CSCs and reduced tumor size. Additionally, Jmjd2c and SOX2 expressions are closely related to the development and prognosis of LSCC according to the results of immunochemistry and overall survival analysis. Therefore, blockade of Jmjd2c-SOX2 signaling might be efficient to control LSCC tumor progression.
In summary, this study suggested that Jmjd2c maintained the ALDHbri+ cancer stemness with transcription factor SOX2 in lung cancers. Inhibitors targeting Jmjd2c is promising to be an anti-tumor drugs for clinical treatment for LSCC patients. Due to the complexity of epigenetic factor regulatory on chromatin structure and gene expression, more investigations are needed in the future.
Materials and methods
Cell culture
The primary ALDHbri+ CSCs of LSCC was obtained as described previously,Citation39 and were inoculated in petri dishes. When the cells proliferated to 80% of the dish, trypsinize them and then cultured in Dulbecco’s modified Eagle medium: nutrient mixture F-12 (DMEM/F12) (Thermo Fisher Scientific, US) with 10% fetal bovine serum (Zhenjiang, China) under 37°C and 5% CO2 conditions. The harvested cells were prepared for the following experiments.
The lentiviral-mediated Jmjd2c-silenced ALDHbri+ CSCs
The PLL3.7-Jmjd2c/SOX2 vectors were transformed to the 293T cells and cultured under 37°C and 5% CO2 conditions for 10 h. Then, add Titer Boost to the mixtures. The harvested viruses were then transformed into the ALDHbri+ CSCs to obtain the Jmjd2c-silenced and Jmjd2c-SOX2-silenced ALDHbri+ CSCs.
Sphere formation assay
Resuspended the ALDHbri+ CSCs, adjusted the cell concentration to 5000 cells/ml, cultured with 24-well plates, and obtained the primary spheres 7 d later. Collected these primary spheres and resuspend the cells in DMEM/F12 and adjusted the cell concentration to 5000 cells/ml again. Cultured these cells in 24-well plates and obtained the secondary spheres another 7 d later. Observed and photographed the spheres under a microscope. Those larger than 40 μm in diameter were counted.
Aldefluor assay
The ALDHbri+ CSCs were prepared same as the above method. The ALDH activity was detected with the ALDEFLOURTM assay kit (NWBiotech, Beijing) following the manufacturer's instruction on a flow cytometer (BD FACSCanto II, BD Biosciences, USA).
Western blotting analysis
The proteins were extracted using RIPA protein extraction reagent (Solarbio, Beijing). Approximately, 30 μg lysis products were subjected to SDS – PAGE, transferred to polyvinylidene fluoride (PVDF) membranes and blocked in 5% milk. The rabbit anti-Jmjd2c and rabbit anti-SOX2 (Abcam, US) were applied in dilution 1:1000. After washed, the membrane was incubated with goat anti-rabbit IgG (Abcam, UK) in dilution 1:5000. The protein bands were visualized with the enhanced chemiluminescence detection method controlled by GAPDH.
qRT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen, US) following the manufacturer’s instructions. The PrimeScript® RT Master Mix Perfect Real Time kit (TAKARA, Japan) and the SYBR Green Master Mix (Applied Biosystems, US) were employed in RNA reverse transcription and qPCR on an instrument (Applied Biosystems 7900HT Real-Time System, US). 2−ΔΔCt method was applied to calculate the relative gene levels normalized to actin.
Co-IP assay
The Jmjd2c and SOX2 proteins were enriched with the Protein A/G Immunoprecipitation Kit (Invitrogen, US). Five milligrams of Myc-Jmjd2c and SOX2 was lysed and used for IP with Myc antibody (Abcam, China). The IP of the interacting protein SOX2 was controlled by Myc-Jmjd2c with empty pcDNA3.1. The input was applied to determine the initial protein levels of Jmjd2c and SOX2 in each group.
GST pull-down assay
Approximately 100 µg of GST-SOX2 fusion protein was incubated with 50 µl of glutathione agarose (Yeasen, China) at 4°C for 1 h. Then, added approximately 100 µg of Myc-Jmjd2c fusion protein to the immobilized GST-SOX2 solutions and incubated at 4°C overnight. Finally, eluted the bound protein with 10 mM glutathione (pH 8.0) and analyzed it by western blotting analysis. Beta-tubulin was used as a loading control.
The construction of subcutaneous tumor-bearing BALB/c nude mice
Eighteen female BALB/c nude mice (16–18 g) were randomly divided into three groups. Mice of one group were injected with 1 × 107 Jmjd2c-silenced ALDHbri+ CSCs each, and another group was injected with 1 × 107 Jmjd2c-SOX2-silenced ALDHbri+ CSCs, controlled by one group injected with equal amount of primary ALDHbri+ CSCs, subcutaneously on the right thigh. Observe the tumor growth and measure tumor volume every 2 d. All the mice were sacrificed with pentobarbital (100 mg/kg) after 17 d and stripped of the subcutaneous tumors in nude mice.
HE staining
The lungs of the tumor-bearing BALB/c nude mice were isolated and fixed with paraformaldehyde (4%), embedded in paraffin, sectioned to 5 μm in thickness and stained with HE solution (Solarbo, Beijing).
Immunochemistry
The LSCC tissues and mouse lung tissues were fixed in 10% formalin and stored in 70% ethanol. Tissues were sectioned to 5 μm in thickness and were deparaffinized and rehydrated in order. Slides were incubated in 3% hydrogen peroxide for 30 min and blocked in serum for 1 h, then incubated with the primary antibodies with 1:1000 in dilution overnight at 4°C. After washed with PBS solutions, the slides were incubated with the secondary antibodies with 1:5000 in dilution for 1 h. The protein expressions were then visualized using 3,3-diaminobenzidine (DAB) chromogen (Sigma-Aldrich, US).
Statistical analysis
GraphPad Prism 5.0 (GraphPad Inc., US) and SPSS 22.0 (IBM Corporation, US) were applied to statistical analysis. Independent Student’s t test or one-way ANOVA were performed to analyze the differences between distinctive groups. All the data were presented as means ± SD, and p < .05 was considered statistically significant.
Authors’ contributions
Conception and design: M.W.; analysis and interpretation of data: Y.H, F.C., L.G. and Y.M.; drafting of the article: M.W. and Y.Z.; revision of the article for important intellectual content and final approval: all authors.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
The authors of this study have read and approved the submitted form of this manuscript and declare that it is not under consideration for publication elsewhere.
Ethics approval and consent to participate
All experimental protocols were approved by the Ethics Committee of the First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology. The study is reported in accordance with ARRIVE guidelines. All methods were performed in accordance with the relevant guidelines and regulations.
Supplemental materials.docx
Download MS Word (742.2 KB)Acknowledgments
This study was funded by the Henan Provincial Program for Medical Science and Technology Research (LHGJ20200569).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2024.2373447
Additional information
Funding
References
- Yuan X, He J, Sun F, Gu J. Effects and interactions of MiR-577 and TSGA10 in regulating esophageal squamous cell carcinoma. Int J Clin Exp Pathol. 2013;6(12):2651–11.
- Li Y, Gu J, Xu F, Zhu Q, Ge D, Lu C. Transcriptomic and functional network features of lung squamous cell carcinoma through integrative analysis of GEO and TCGA data. Sci Rep. 2018;8(1):15834. doi:10.1038/s41598-018-34160-w.
- Chen JW, Dhahbi J. Lung adenocarcinoma and lung squamous cell carcinoma cancer classification, biomarker identification, and gene expression analysis using overlapping feature selection methods. Sci Rep. 2021;11(1):13323. doi:10.1038/s41598-021-92725-8.
- Xue Y, Gao S, Gou J, Yin T, He H, Wang Y, Zhang Y, Tang X, Wu R. Platinum-based chemotherapy in combination with PD-1/PD-L1 inhibitors: preclinical and clinical studies and mechanism of action. Expert Opin Drug Deliv. 2021;18(2):187–203. doi:10.1080/17425247.2021.1825376.
- Croagh D, Thomas RJ, Phillips WA, Kaur P. Esophageal stem cells—a review of their identification and characterization. Stem Cell Rev. 2008;4(4):261–68. doi:10.1007/s12015-008-9031-3.
- Miranda Furtado CL, Dos Santos Luciano MC, Silva Santos RD, Furtado GP, Moraes MO, Pessoa C. Epidrugs: targeting epigenetic marks in cancer treatment. Epigenet. 2019;14(12):1164–76. doi:10.1080/15592294.2019.1640546.
- Zhao L, Duan YT, Lu P, Zhang ZJ, Zheng XK, Wang JL, Feng WS. Epigenetic targets and their inhibitors in cancer therapy. Curr Top Med Chem. 2018;18(28):2395–419. doi:10.2174/1568026619666181224095449.
- Nebbioso A, Tambaro FP, Dell’aversana C, Altucci L, Greally JM. Cancer epigenetics: moving forward. PloS Genet. 2018;14(6):e1007362. doi:10.1371/journal.pgen.1007362.
- Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55.
- Sigalotti L, Fratta E, Coral S, Cortini E, Covre A, Nicolay HJ, Anzalone L, Pezzani L, Di Giacomo AM, Fonsatti E, et al. Epigenetic drugs as pleiotropic agents in cancer treatment: biomolecular aspects and clinical applications. J Cell Physiol. 2007;212(2):330–44. doi:10.1002/jcp.21066.
- Perillo B, Tramontano A, Pezone A, Migliaccio A. LSD1: more than demethylation of histone lysine residues. Experiment Molecul Med. 2020;52(12):1936–47. doi:10.1038/s12276-020-00542-2.
- Yang J, Hu Y, Zhang B, Liang X, Li X. The JMJD family histone demethylases in crosstalk between inflammation and cancer. Front Immunol. 2022;13:881396. doi:10.3389/fimmu.2022.881396.
- Zhang H, Wang J, Huang J, Shi T, Ma X, Luo X, Li X, Li M. Inhibiting Jumoji domain containing protein 3 (JMJD3) prevent neuronal apoptosis from stroke. Exp Neurol. 2018;308:132–42. doi:10.1016/j.expneurol.2018.07.007.
- Yang ZQ, Imoto I, Fukuda Y, Pimkhaokham A, Shimada Y, Imamura M, Sugano S, Nakamura Y, Inazawa J. Identification of a novel gene, GASC1, within an amplicon at 9p23-24 frequently detected in esophageal cancer cell lines. Cancer Res. 2000;60(17):4735–39.
- Hyun K, Jeon J, Park K, Kim J. Writing, erasing and reading histone lysine methylations. Experim Molecul Med. 2017;49(4):e324. doi:10.1038/emm.2017.11.
- Tomaz RA, Harman JL, Karimlou D, Weavers L, Fritsch L, Bou-Kheir T, Bell E, Del Valle Torres I, Niakan KK, Fisher C, et al. Jmjd2c facilitates the assembly of essential enhancer-protein complexes at the onset of embryonic stem cell differentiation. Develop. 2017;144(4):567–79. doi:10.1242/dev.142489.
- Zhang C, Wang Z, Ji Q, Li Q. Histone demethylase JMJD2C: epigenetic regulators in tumors. Oncotarget. 2017;8(53):91723–33. doi:10.18632/oncotarget.19176.
- Li X, Dong S. Histone demethylase JMJD2B and JMJD2C induce fibroblast growth factor 2: mediated tumorigenesis of osteosarcoma. Med Oncol. 2015;32(3):53. doi:10.1007/s12032-015-0503-4.
- An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun D, Chen X, He X. circZMYM2 competed endogenously with miR-335-5p to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem. 2018;51(5):2224–36. doi:10.1159/000495868.
- Hong Q, Yu S, Yang Y, Liu G, Shao Z. A polymorphism in JMJD2C alters the cleavage by caspase-3 and the prognosis of human breast cancer. Oncotarget. 2014;5(13):4779–87. doi:10.18632/oncotarget.2029.
- Li N, Jiang D. Jumonji domain containing 2C promotes cell migration and invasion through modulating CUL4A expression in lung cancer. Biomed Pharmacother. 2017;89:305–15. doi:10.1016/j.biopha.2017.02.014.
- Walcher L, Kistenmacher AK, Suo H, Kitte R, Dluczek S, Strauss A, Blaudszun AR, Yevsa T, Fricke S, Kossatz-Boehlert U. Cancer stem cells—origins and biomarkers: perspectives for targeted personalized therapies. Front Immunol. 2020;11:1280. doi:10.3389/fimmu.2020.01280.
- Kusoglu A, Biray Avci Ç. Cancer stem cells: a brief review of the current status. Gene. 2019;681:80–85. doi:10.1016/j.gene.2018.09.052.
- Dawood S, Austin L, Cristofanilli M. Cancer stem cells: implications for cancer therapy. Oncol (Williston Park). 2014;28(12):1101–7, 1110.
- Porter L, McCaughan F. SOX2 and squamous cancers. Semin Cancer Biol. 2020;67(Pt 1):154–67. doi:10.1016/j.semcancer.2020.05.007.
- Zhu Y, Huang S, Chen S, Chen J, Wang Z, Wang Y, Zheng H. SOX2 promotes chemoresistance, cancer stem cells properties, and epithelial–mesenchymal transition by β-catenin and Beclin1/autophagy signaling in colorectal cancer. Cell Death Dis. 2021;12(5):449. doi:10.1038/s41419-021-03733-5.
- Chen X, Xie R, Gu P, Huang M, Han J, Dong W, Xie W, Wang B, He W, Zhong G, et al. Long noncoding RNA LBCS inhibits self-renewal and chemoresistance of bladder cancer stem cells through epigenetic silencing of SOX2. Clin Cancer Res. 2019;25(4):1389–403. doi:10.1158/1078-0432.CCR-18-1656.
- Wu F, Wang L, Zhou C. Lung cancer in China: current and prospect. Curr Opin Oncol. 2021;33(1):40–46. doi:10.1097/CCO.0000000000000703.
- Hirsch FR, Scagliotti GV, Mulshine JL, Kwon R, Curran WJ Jr., Wu YL, Paz-Ares L. Lung cancer: current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi:10.1016/S0140-6736(16)30958-8.
- Kulasingam V, Diamandis EP. Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol. 2008;5(10):588–99. doi:10.1038/ncponc1187.
- Zhang J, Wang M, Wang J, Wang W. JMJD2C-mediated long non-coding RNA MALAT1/microRNA-503-5p/SEPT2 axis worsens non-small cell lung cancer. Cell Death Dis. 2022;13(1):65. doi:10.1038/s41419-022-04513-5.
- Najafi M, Mortezaee K, Majidpoor J. Cancer stem cell (CSC) resistance drivers. Life Sci. 2019;234:116781. doi:10.1016/j.lfs.2019.116781.
- Peiris-Pages M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Breast Cancer Res. 2016;18(1):55. doi:10.1186/s13058-016-0712-6.
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48(4):491–507. doi:10.1016/j.molcel.2012.11.006.
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439(7078):811–16. doi:10.1038/nature04433.
- Novak D, Huser L, Elton JJ, Umansky V, Altevogt P, Utikal J. SOX2 in development and cancer biology. Semin Cancer Biol. 2020;67(Pt 1):74–82. doi:10.1016/j.semcancer.2019.08.007.
- Funaya S, Aoki F. Regulation of zygotic gene activation by chromatin structure and epigenetic factors. J Reprod Dev. 2017;63(4):359–63. doi:10.1262/jrd.2017-058.
- Lawrence M, Daujat S, Schneider R. Lateral thinking: How histone modifications regulate gene expression. Trends Genet. 2016;32(1):42–56. doi:10.1016/j.tig.2015.10.007.
- Zhuang X, Zhang W, Chen Y, Han X, Li J, Zhang Y, Zhang Y, Zhang S, Liu B. Doxorubicin-enriched, ALDHbr mouse breast cancer stem cells are treatable to oncolytic herpes simplex virus type 1. BMC Cancer. 2012;12(1):549. doi:10.1186/1471-2407-12-549.