ABSTRACT
Despite advances in targeted therapies, primary and acquired resistance make the treatment of colorectal cancer (CRC) a pressing issue to be resolved. According to reports, the development of CRC is linked to miRNA dysregulation. Multiple studies have demonstrated that miR-135b-5p has an aberrant expression level between CRC tissues and adjacent tissues. However, it is unclear whether there is a correlation between miR-135b-5p and cetuximab (CTx) resistance in CRC. Use the GEO database to measure miR-135b-5p expression in CRC. Additionally, RT-qPCR was applied to ascertain the production level of miR-135b-5p in three human CRC cells and NCM460 cells. The capacity of cells to migrate and invade was examined utilizing the wound-healing and transwell assays, while the CCK-8 assay served for evaluating cell viability, as well as colony formation assays for proliferation. The expected target protein of miR-135b-5p in CRC cell cetuximab resistance has been investigated using western blot. Suppression of miR-135b-5p could increase the CTx sensitivity of CTx-resistant CRC cells, as manifested by the attenuation of proliferation, migration, and invasion ability. Mechanistic studies revealed miR-135b-5p regulates the epithelial-to-mesenchymal transition (EMT) process and Wnt/β-catenin signaling pathway through downgulating FOXN3. In short, knockdowning miR-135b-5p could increase FOXN3 expression in CRC cells, promote the EMT process, and simultaneously activate the Wnt/β-catenin signaling pathway to elevate CTx resistance in CRC cells.
Introduction
Malignant tumors and serious infectious diseases will be the leading causes of death, the development of medical technology has decreased the mortality rate brought on by human diseases. Colorectal cancer, the second most lethal and third most common disease in the world, which kills 0.9 million people each year, and damages human health.Citation1 The incidence of CRC is increasing in many nations as a result of changes in people’s dietary practices and lifestyles.Citation2 Despite targeted therapy and immunotherapy have considerably boosted patient survival rates, a significant concern still exists due to medication resistance, which causes patients to become unresponsive.Citation3
MicroRNAs (miRNAs) are a class of short, endogenous, and conserved non-coding RNAs,Citation4 whose synthesis process involves several steps: first, pre-miRNA is produced by the gene encoding the miRNA,Citation5 then Drosha enzyme induces the cleave of pre-miRNA to obtain pre-miRNA with a hairpin structure under the action of polymerase II. The pre-miRNA is cut off by RIIIDa and RIIIDb enzymes with 3’− and 5’ strands, respectively, obtaining two mature miRNAs. Other proteins will combine with mature miRNAs to form miRNA-induced silencing complexes (MiRISC), which cause degradation or translational inhibition of target mRNAs within the cell by combining to the 3’-UTR of mRNAs, thereby mediating multiple abnormal proteins expression in cells.Citation6
Nearly 60% of human protein-coding gene expression is regulated by miRNAs, therefore miRNAs were also regarded as of great importance in the development of tumors and multiple biological processes, such as cell division, death, inflammation, and proliferation.Citation7 For instance, miR-135b-5p is aberrantly expression in renal cell carcinomaCitation8 and pancreatic cancer.Citation9 It is understood the mechanism of miR-135b-5p in CRC, so the new study shows that target miR-135b-5p are significant in CRC. MiR-135b-5p, as a prognostic miRNA, is significantly associated with triple-negative breast cancer (TNBC), which provides a new therapeutic target for the treatment and prognosis of TNBC.Citation10 MiR-135b-5p contributes to the malignant potential of ovarian cancer cells by targeting DUSP5.Citation11
Accumulating evidence highlights that miR-135b-5p is also closely related to the development of drug resistance in tumors. Astragaloside IV (AS-IV) has certain effects on miR-135b-5p during liver cancer development. Cell surface PD-L1 levels decrease and PD-L1-associated immune suppression is alleviated by (AS-IV) via the miR-135b-5p/CNDP1 pathway.Citation12 CTx, an FDA-approved first-line therapy for KRAS wild-type metastatic CRC that targets the EGFR, offers hope for patients with advanced metastatic CRC.Citation13 However, several clinical studies have demonstrated that patients, both with KRAS wild-type patients and those with KRAS mutations,Citation14 have resistance to CTx,Citation15 significantly limits the therapeutic impact of cetuximab. How miR-135b-5p regulates CTx resistance in CRC remains unstudied. As a result, it is essential to explore the causes of CTx resistance in CRC patients.
According to the GEO database, we chose miR-135b-5p as our research target since it is significantly expressed in CRC tissues. In contrast to CTx-sensitive cells and healthy intestinal epithelial cells, we discovered that miR-135b-5p expression is significantly higher in SW480, and HT29 cells that are resistant to CTx. We aimed to investigate the aberrant expression of miR-135b-5p in the development of CRC and resistance to CTx, the underlying molecular mechanisms, and establish a theoretical framework for anticipating and overcoming CTx resistance in CRC.
Materials and methods
Cell Culture
CTx-resistant cell lines SW480, HT29, and a normal intestinal epithelial cell NCM460 were bought from the Cell Bank of the Chinese Academy of Science (Shanghai, China). CTx-sensitive cell lines Caco2 were kindly gifted by M.D Huihui Zheng (Southern Medical University, Guangzhou, China). Caco2 cells are cultured in MEM, while SW480 and NCM460 in high-glucose DMEM, HT29 in McCoy’s 5A medium. In addition, all base medium adds 10% FBS and 1% penicillin/streptomycin antibiotics. Cells were then placed in an incubator with a setup procedure of 5% CO2 at 37°C.
Cell Transfection
RiboBio (Guangzhou, China) synthesized miR-135b-5p mimics, inhibitors, mimics NC, and inhibitors NC. PcDNA3.1 vector for FOXN3 overexpression and related control plasmids were synthesized by Keygen (Nanjing, China). SW480 and HT29 cells were transfected with Lipofectamine 3000 and Opti-MEM following the manufacturer’s instructions.
RT-qPCR
The Total RNA Isolation Kit (Vazyme, Nanjing, China) was applied to isolate total RNA from cells. Subsequently, 1 μg total RNA was reverse transcribed to cDNA by a miRNA 1st Strand cDNA Synthesis Kit and HiScript III All-in-one RT SuperMix Perfect for qPCR (both from Vazyme, Nanjing, China), respectively. Reverse transcription products were performed to detect gene expression on an ABI7500 instrument using the miRNA Universal SYBR® qPCR Master Mix and the Taq Pro Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China). The target gene expression was processed and analyzed via the 2−ΔΔCt algorithm and U6 or GAPDH employed as endogenous controls. All primer sequences are provided in .
Table 1. Primers sequence of relative genes.
Cell Counting Kit-8 assay
After being digested, a density of 5 ~ 8 × 103 of transfected HT29 and SW480 cells were laid on 96-well plates, respectively. Add CCK-8 reagent (Beyotime, Shanghai, China) following the appropriate procedure, and it was incubated at 37°C. At 450 nm, examine the inhibition rate of CTx on cells under a Microplate Reader (Thermo Fisher, CA, USA) after 1 h.
Colony formation assay
After transfection, cells were stimulated with 20 μg/ml CTx 24 h, then incubated under 5% CO2 at 37°C for further 2 weeks. Change the medium as necessary midway. Four percent paraformaldehyde is used to fix cells for 20 min and crystal violet for 15 min. Wash carefully and dry naturally at room temperature. Count and record the colonies.
Wound-healing assay
When CRC cells had grown to 90% confluence in 6-well plates, a 200 μL pipette tip was used to scrape cells perpendicular to the plate. Subsequently, wash the cells twice with PBS and then add a medium containing CTx and 2% FBS. At the same location, pictures were taken at 0 and 48 h after scratching.
Transwell assay
Using serum-free medium to suspend HT29 and SW480 cells, respectively, 1 × 105 cells/200 μl were added to the upper compartment of a Transwell chamber (Labselect, Beijing, China), and 600 μl of medium containing 10% serum was added to the lower compartment. The chamber was taken out and cleaned with PBS after being incubated for 48 h. Fix the stained cells with 4% paraformaldehyde and crystal violet, respectively. Then the cells in the upper chamber were then scraped. Images of the invaded cells in the lower compartment were captured with a Nikon microscope.
Western Blotting
CRC cells were cleaned with pre-cooled PBS and lysed using RIPA buffer containing phenylmethylsulfonyl fluoride. Proteins are separate proteins quantified by a BCA assay kit (Beyotime, Shanghai, China). Transfer the purified protein to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). Use 5% nonfat milk powder for blocking for two hours at 25°C and incubate with primary antibodies at 4°C all night. Then clean by TBST and keep incubating with secondary antibodies at room temperature for 1 h. The PVDF membranes were then infiltrated with enhanced chemiluminescence (ECL) (Proteinbio, Nanjing, China) to measure the level of protein expression on the FCM chemiluminescence. The primary antibodies used in this experiment included anti-E-cadherin (Proteintech, #20874–1-AP, 1:5000), anti-Vimentin (Proteintech, #10366–1-AP, 1:5000), anti-C-myc (Proteintech, #10828–1-AP, 1:5000), anti-beta catenin (Proteintech, #51067–2-AP, 1:5000), anti-Cyclin D1 (Proteintech, #26939–1-AP, 1:5000), anti-FOXN3 (Abcam, #ab129453, 1:1000).
Statistical analysis
Results were provided as median values ± SEM from at least three times. The Student’s t-test was used to analyze the data with the assistance of GraphPad Prism 9.0 software, with a p-value of 0.05 was considered significant.
Results
miR-135b-5p is high expression level in CRC.
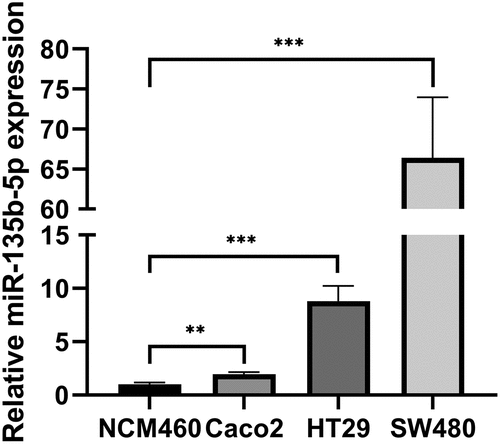
According to studies, we found that in a number of malignancies, miR-135b-5p expression exhibits dysregulation. The GEO databases (GSE72281, GSE156732, and GSE156719) were applied to reveal that miR-135b-5p is higher expression level in CRC tissues compared to adjacent tissues (). According to previous studies, we have known that there were CRC cell lines resisting cetuximab.Citation16 There were different levels of miR-135b-5p expression level from Caco2, HT29, SW480, and NCM460. RT-qPCR assay suggested that miR-135b-5p may be relevant to CTx resistance to CRC cells since it was dramatically elevated in CRC cells and expressed more highly in CTx-resistant cells than in CTx-sensitive cells (). Therefore, we chose the CTx-resistant cell lines HT29 and SW480 cells for additional research to explore further the relationship between miR-135b-5p and CTx resistance.
miR-135b-5p promotes CTx resistance in CRC cells in vitro.
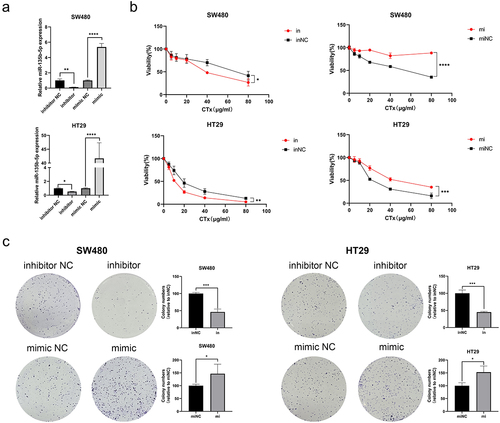
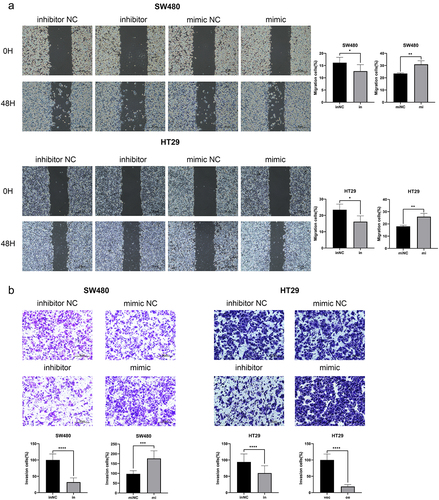
After upregulating and knockdowning miR-135b-5p expression, respectively, in SW480 and HT29, check transfection effectiveness using RT-qPCR (). To figure out how miR-135b-5p worked in CRC, the CCK-8 experiment was used to assess the proliferation activity of SW480 and HT29 cells following CTx therapy. The cell viability curve plotting showed that overexpression of miR-135b-5p significantly promoted the viability of SW480 and HT29 cells after cultured with CTx, while miR-135b-5p inhibition suppressed cell viability (). This suggests that upregulating miR-135b-5p can increase resistance to CTx in CRC, and inhibition of miR-135b-5p can reverse this effect. Colony formation assay supported the outcome of the CCK-8 assay suggested that knockdowning miR-135b-5p considerably reduced SW480 and HT29 cell colony formation capacity, while miR-135b-5p upregulation greatly increased (). Wound healing assay implied that the miR-135b-5p upregulated CRC cells exhibited better migration activity, but the migration activity in miR-135b-5p knockdown cells was weaker (). In addition, miR-135b-5p was shown to be able to enhance the capacity of CRC cells for invasion in transwell assay (). We can draw a conclusion from the above results that upregulation of miR-135b-5p can considerably support CRC cell resistance to CTx, as evidenced by the improvement of typical tumor cell behaviors such as cell proliferation and invasion when cultured with CTx.
MiR-135b-5p expression was negatively correlated with FOXN3.
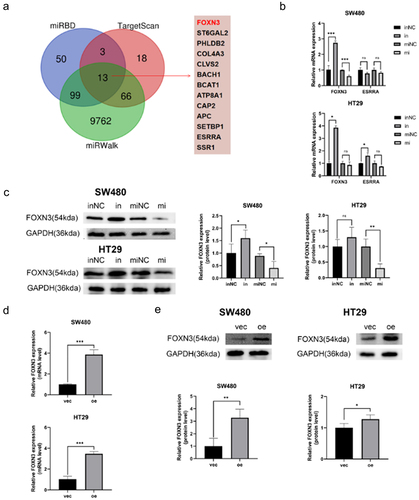
In order to clarify the mechanism by which miR-135b-5p regulates CTx-resistance, three databases, including TargetScan, miRDB, and miRWalk, were utilized to predict the potential target genes of miR-135b-5p. Thirteen promising mRNA were chosen (). Previous researches have demonstrated that APC, FOXN3, and ESRRA expression were downregulated in tumors among these 13 putative targets and the others were upregulated. We selected FOXN3 and ESRRA for additional verification because the inactivating mutation of the APC gene has now been identified as a biomarker for CRC.Citation17 After transfecting SW480 and HT29 cells with miR-135b-5p inhibitors and mimic, use RT-qPCR to evaluate FOXN3 and ESRRA expression levels, respectively. The results showed that FOXN3 and ESRRA expression levels were negatively correlated with miR-135b-5p. To some extent, the correlation of FOXN3 is more significant (). Further, FOXN3 protein expression was measured by western blot assay when SW480 and HT29 cells were knockdown or upregulated for miR-135b-5p expression, our results were consistent with RT-qPCR (). The findings demonstrated that miR-135b-5p overexpression dramatically lowered FOXN3 expression in SW480 and HT29 cells, while miR-135b-5p inhibition considerably boosted it. It can be seen from the result that miR-135b-5p regulates FOXN3 in CRC.
Figure 4. miR-135b-5p suppressed FOXN3 expression. (A) The possible target mRNAs were predicted by online database. B Detect the relative mRNA expression in SW480 and HT29 cells after upregulating or knockdowning miR-135b-5p by RT-qPCR. (C) Western blot analysis of FOXN3 protein expression in SW480 and HT29 cells after upregulating or knocking down miR-135b-5p. (D, E) Evaluate the FOXN3 expression levels by RT-qPCR and western blotting after transfecting with the FOXN3 plasmid and pcDNA3.1(+) vector, respectively.
FOXN3 sensitizes CRC cells to CTx.
We created FOXN3 overexpression cell lines, and RT-qPCR and western blot were assessed transfection effectiveness (). How FOXN3 reacts to CTx resistance in CRC was later demonstrated by a series of in vitro experiments. The CCK-8 experiment demonstrated that the overexpression of FOXN3 greatly increased the sensitivity of CTx in SW480 and HT29 cells (). Upregulation of FOXN3 can inhibit the resistance of HT29 and SW480 cells to CTx with weakened cell proliferation (), migration (), invasion ability (). Therefore, we concluded that suppressing the expression of FOXN3 might increase CTx resistance in CRC cells.
Figure 5. FOXN3 sensitizes CRC cells to CTx. (A) A proliferation assay was used to monitor the growth of SW480 and HT29 cells after transfecting with the FOXN3 plasmid and pcDNA3.1(+) vector when treated with CTx. (B) Experiments with colony formation demonstrated that FOXN3 enhances the effect of CTx and inhibits the colony formation ability. (C, D) Wound-healing assay and transwell assay were carried out to determine the impact of FOXN3 on SW480 and HT29 cells migratory and invasion ability under the action of CTx.
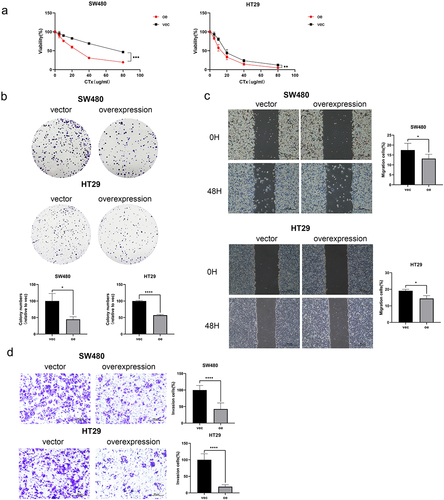
miR-135b-5p/FOXN3 activates the EMT process and Wnt/β-catenin signaling pathway.
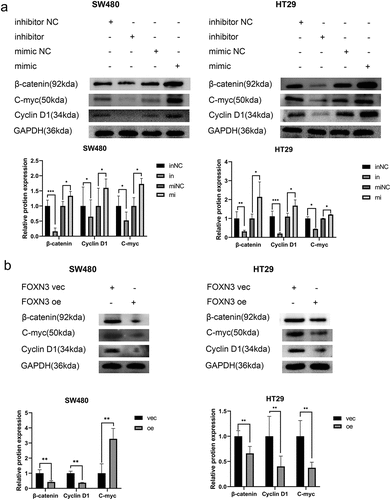
The aforementioned experiments suggested that miR-135b-5p/FOXN3 could regulate CRC CTx resistance in CRC cells. EMT process is typically relevant to tumor growth and invasion. As expected, in the background of miR-135b-5p overexpression, Vimentin was upregulated but E-cadherin downregulated significantly. On the contrary, Vimentin was downregulated, but E-cadherin was upregulated when suppressing miR-135b-5 in SW480 and HT29 cells (). And more notably, FOXN3 regulating relationship is opposite to miR-135b-5p, while E-cadherin was upregulated in FOXN3 overexpression group in comparison with the respective NC significantly (). There are other tumor progression-related pathways involved in tumor progression. For example, in diffuse large B-cell lymphoma and thyroid cancer, respectively, Zhao CCCitation18 and Zhao CCitation19 discovered that miR-135b-5p and FOXN3 are both implicated in tumor development through the Wnt/β-catenin signaling pathway. Does activating the Wnt/β-Catenin signaling pathway increase CTx resistance in CRC cells? The result turned out to be correct. Compared to the respective negative control, β-catenin, C-myc, and cyclin D1 were all significantly upregulated when miR-135b-5p was overexpressed (). On the contrary, downregulation occurs when FOXN3 is upregulated, leading to the downregulation of β-catenin, C-myc, and cyclin D1 (). In short, miR-135b-5p/FOXN3 may facilitate the EMT process and activate the Wnt/β-catenin signaling pathway to contribute to tumorigenesis.
Figure 6. miR-135b-5p/FOXN3 activates the EMT process. (A) E-cadherin, Vimentin expression were detected by western blotting. (B) E-cadherin and Vimentin protein expression in SW480 and HT29 cells after transfection with FOXN3 plasmid or pcDNA3.1(+) vector by western blot analysis.
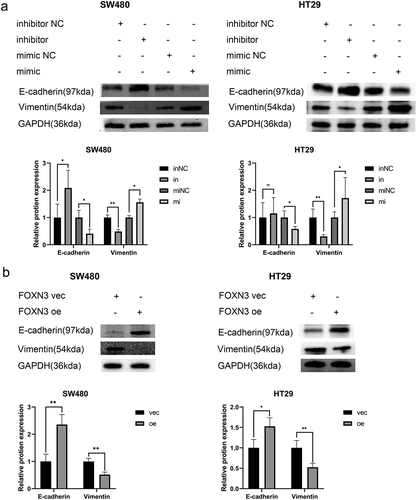
Figure 7. miR-135b-5p/FOXN3 activates the Wnt/β-catenin signaling pathway. (A) β-catenin, c-myc, and cyclin D1 protein expression determined by Western blotting. (B) Western blot analysis was used to assess β-catenin, c-myc, and cyclin D1 protein expression in CRC cells transfected with the FOXN3 plasmid or pcDNA3.1(+) vector.
Discussion
As the third most prevalent malignant tumor in the world, colorectal cancer has had a significant negative impact on human life and health.Citation20 Despite the ongoing development of new anti-cancer medications and the public’s focus on early tumor diagnosis, the incidence and prevalence of CRC continue to rise in many countries. Clinical studies have demonstrated that CTx, which was approved as a first-line treatment for metastatic CRC, can significantly improve the prognosis of patients.Citation21–24 However, the presence of drug resistance is also a significant issue and challenge in clinical applications. LncRNA, circRNA, miRNA, and other non-coding RNAs have steadily come into people’s field of view with the advancement of sequencing technology. Numerous miRNAs have been discovered to regulate tumor development after more than 20 years of research,Citation25 opening up new possibilities for clinical diagnosis and innovative treatment development. Many miRNA-targeted medications are currently undergoing clinical trials,Citation26 demonstrating that the study of miRNA can offer a new theoretical foundation for pharmaceuticals. Numerous studies have demonstrated that miRNAs perform a cancer-suppressing or cancer-promoting role and are different expression levels between various tumor tissues and adjacent tissues significantly.Citation27
With further study of tumor mechanism, members of the miR-135b family have been relative to the progression of numerous tumors, including lung malignancy,Citation28 gastric carcinoma,Citation29 breast cancer,Citation30 melanoma,Citation31 and colorectal cancers.Citation32 There is study has noted that miR-135b is upregulated in triple-negative breast cancer tissues and cells. Notably, in particular, it is markedly relevant to lymph node metastasis and TNM staging of patientsCitation33; If the APC gene is deleted in CRC cells, it will lead to miR-135b overexpression, which then targets a variety of tumor suppressor genes, further inducing inactivation mutations of APC genes.Citation34 According to additional research, upregulating miR-135b-5p causes the tumor to be resistant to oxaliplatin treatment.Citation32
MiRNA functions by targeting the 3’ UTR region of mRNA within the cell, suppressing post-transcriptional translation of mRNA to regulate gene expression and thus participate in life activities in the body. FOXN3 (also known as CHES1), a member of the FOX protein family, is a transcriptional inhibitor that inhibits many DNA damage to activate the yeast checkpoint mutations.Citation35 FOXN3 exists in multiple human organs widely and regulates many important physiological and pathological processes in the body. Database analysis indicated that FOXN3 was significantly downregulated in 15 malignant cancers, including lung cancer, breast cancer, glioma cancer, melanoma cancer, etc.Citation19,Citation36–38 Through a series of in vitro functional studies, we discovered that FOXN3 was related to the resistance of SW480 and HT29 cells to CTx in our study. Following FOXN3 overexpression, CRC cell activity significantly decreased with increasing administration concentration of cetuximab, and higher FOXN3 expression inhibited CRC cell migration and invasion in response to drug stimulation. According to the aforementioned findings, there is a strong correlation between FOXN3 expression and the resistance of CRC cells to CTx resistance.
We examined E-cadherin and vimentin expression as a result of the EMT process largely responsible for the migration and invasion of tumors. MiR-135b-5p overexpression made Vimentin upregulated but E-cadherin downregulated significantly. On the contrary, Vimentin was downregulated, and E-cadherin was upregulated when suppressing miR-135b-5 in SW480 and HT29 cells. Interestingly, FOXN3 expression is opposite to miR-135b-5p. MiR-135b-5p promote SW480 and HT29 cells to CTx resistance by suppressing the expression of FOXN3. As a result of the aforementioned findings, miR-135b-5p may be able to control expression of FOXN3, and promote the EMT process in CRC cells, which contributes to increased CTx resistance in CRC.
There are several key tumor-related signaling pathways that are related to tumor progression: MAPK signaling pathway,Citation39 Wnt/β-catenin signaling pathway,Citation40 PI3K/AKT signaling pathway,Citation41 NF-κB signaling pathway,Citation42 etc. The growth process of tumors depends on multiple signaling pathways, where miRNA and mRNA also play crucial roles through molecular signaling pathways in tumor cells. These pathways are crucial for maintaining tissue homeostasis, and an imbalance in the signaling pathways results from the expression of abnormal signaling molecules. This can lead to the formation of tumor cells, as well as the growth and metastasis of tumors. In diffuse large B-cell lymphoma, lnc SMAD5-AS1 upregulate APCs expression and inhibits cancer cell proliferation as an endogenous competitor of miR-135b-5p, in addition, it inhibits the cancer cell proliferation via Wnt/β-catenin signaling pathway,Citation18 FOXN3 can mediate tumor cell proliferation and invasion through the Wnt/β-catenin signaling pathway in melanoma,Citation43 thyroid cancer,Citation19 and gallbladder cancer.Citation44 In our study, we measured the expression of related proteins of the Wnt/β-catenin signaling, like β-catenin, Cyclin D1 and C-myc. Moreover, the expression of β-catenin, Cyclin D1 and C-myc was regulated by miR-135b-5p. All findings imply that miR-135b-5p is able to be a promising treatment target to improve the cure rate of CRC.
In conclusion, the research revealed that miR-135b-5p acts as a promising diagnostic and treatment target for CTx resistance in CRC. miR-135b-5p expression was negatively correlated with FOXN3 in CRC cells. Additionally, miR-135b-5p promotes the EMT process in the disease, and activates the Wnt/β-catenin signaling pathway to facilitate cetuximab resistance.
Author contributions
Study design: CZ and CP; data collection: CP and YN; data analysis: CP, XL, and YY; cellular experiments: CP, YN, and LF; manuscript review and editing: CP, CZ, XL, and YY, figure and manuscript proofing: CP and XL and YY; funding acquisition: CZ. All the authors read, reviewed, and approved the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The datasets used or analyzed in this study are available from the corresponding authors upon reasonable request.
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin, 2021, 71(3): 209–11.10.3322/caac.21660.
- Brenner H, Kloor M, Pox CP. Colorectal cancer. The Lancet, 2014, 383(9927): 1490–502. 10.1016/S0140-6736(13)61649-9.
- Ciardiello F, Ciardiello D, Martini G, Napolitano S, Tabernero J, Cervantes A. Clinical management of metastatic colorectal cancer in the era of precision medicine. CA Cancer J Clin, 2022, 72(4): 372–401.10.3322/caac.21728.
- Chen L, Heikkinen L, Wang C, Yang Y, Sun H, Wong G. Trends in the development of miRNA bioinformatics tools. Brief Bioinform, 2019, 20(5): 1836–52.10.1093/bib/bby054.
- Achkar NP, Cambiagno DA. Manavella PA. miRNA Biogenesis: a Dynamic Pathway. Trends Plant Sci, 2016, 21(12): 1034–44.10.1016/j.tplants.2016.09.003.
- Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature Structural & Molecular Biology, 2012, 19(6): 586–93.10.1038/nsmb.2296.
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRnas: biology, functions, therapeutics, and analysis methods. J Cell Physiol, 2019, 234(5): 5451–65. 10.1002/jcp.27486.
- Zhang Y, Zhu YY, Chen Y, Zhang L, Wang R, Ding X, Zhang H, Zhang CY, Zhang C, Gu WJ, et al. Urinary-derived extracellular vesicle microRNAs as non-invasive diagnostic biomarkers for early-stage renal cell carcinoma. Clin Chim Acta, 2024, 552: 117672. 10.1016/j.cca.2023.117672.
- Liu D, Jin Y, Wu J, Zhu H, Ye D. MiR-135b-5p is an oncogene in pancreatic cancer to regulate GPRC5A expression by targeting transcription factor KLF4. Cell Death Discov, 2022, 8(1): 23.
- Wang Y, Wang J, Jiang J, Zhang W, Sun L, Ge Q, Li C, Li X, Li X, Shi S. Identification of cuproptosis-related miRNAs in triple-negative breast cancer and analysis of the miRNA-mRNA regulatory network. Heliyon, 2024, 10(7): e28242. 10.1016/j.heliyon.2024.e28242.
- Yu S, Yu M, Chen J, Tang H, Gong W, Tan H. Circ_0000471 suppresses the progression of ovarian cancer through mediating mir-135b-5p/dusp5 axis. Am J Reprod Immunol, 2023, 89(4): e13651.10.1111/aji.13651.
- Ma Y, Li Y, Wu T, Li Y, Wang Q. Astragaloside IV Attenuates programmed death-ligand 1-mediated immunosuppression during liver cancer development via the miR-135b-5p/CNDP1 axis. Cancers (Basel), 2023, 15(20). 5048 10.3390/cancers15205048.
- Fornasier G, Francescon S, Baldo P. An update of efficacy and safety of cetuximab in metastatic colorectal cancer: a narrative review. Adv Ther, 2018, 35(10): 1497–509.10.1007/s12325-018-0791-0.
- Cremolini C, Rossini D, Dell’aquila E, Lonardi S, Conca E, Del Re M, Busico A, Pietrantonio F, Danesi R. Aprile G, et al., Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol, 2019, 5(3): 343–50. 10.1001/jamaoncol.2018.5080.
- Santini D, Vincenzi B, Addeo R, Garufi C, Masi G, Scartozzi M, Mancuso A, Frezza AM, Venditti O, Imperatori M, et al., Cetuximab rechallenge in metastatic colorectal cancer patients: how to come away from acquired resistance? Ann Oncol, 2012, 23(9): 2313–18. 10.1093/annonc/mdr623.
- Lu Y, Zhao X, Liu Q, Li C, Graves-Deal R, Cao Z, Singh B, Franklin JL, Wang J, Hu H, et al., lncRNA MIR100HG-derived miR-100 and miR-125b mediate cetuximab resistance via Wnt/β-catenin signaling. Nat Med, 2017, 23(11): 1331–41. 10.1038/nm.4424.
- Zhang L, Shay JW. Multiple roles of APC and its therapeutic implications in colorectal cancer. J Natl Cancer Inst, 2017, 109(8).10.1093/jnci/djw332.
- Zhao CC, Jiao Y, Zhang YY, Ning J, Zhang YR, Xu J, Wei W, Kang-Sheng G. Lnc SMAD5-AS1 as ceRNA inhibit proliferation of diffuse large B cell lymphoma via Wnt/β-catenin pathway by sponging miR-135b-5p to elevate expression of APC. Cell Death Disease, 2019, 10(4): 252.10.1038/s41419-019-1479-3.
- Zhao C, Mo L, Li C, Han S, Zhao W, Liu L. FOXN3 suppresses the growth and invasion of papillary thyroid cancer through the inactivation of Wnt/β-catenin pathway. Mol Cell Endocrinol, 2020, 515: 110925.10.1016/j.mce.2020.110925.
- Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl Oncol, 2021, 14(10): 101174.10.1016/j.tranon.2021.101174.
- Martinelli E, Martini G, Famiglietti V, Troiani T, Napolitano S, Pietrantonio F, Ciardiello D, Terminiello M, Borrelli C. Vitiello PP, et al., Cetuximab rechallenge plus avelumab in pretreated patients with ras wild-type metastatic colorectal cancer: the phase 2 single-arm clinical CAVE Trial. JAMA Oncol, 2021, 7(10): 1529–35. 10.1001/jamaoncol.2021.2915.
- Tabernero J, Grothey A, Van Cutsem E, Yaeger R, Wasan H, Yoshino T, Desai J, Ciardiello F, Loupakis F, Hong YS, et al., Encorafenib plus cetuximab as a new standard of care for previously treated braf v600e–mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON study. J Clin Oncol, 2021, 39(4): 273–84. 10.1200/JCO.20.02088.
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al., Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med, 2004, 351(4): 337–45. 10.1056/NEJMoa033025.
- Jonker DJ, O’callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, Berry SR, Krahn M, Price T, Simes RJ, et al., Cetuximab for the treatment of colorectal cancer. N Engl J Med, 2007, 357(20): 2040–48. 10.1056/NEJMoa071834.
- Huang X, Zhu X, Yu Y, Zhu W, Jin L, Zhang X, Li S, Zou P, Xie C, Cui R. Dissecting miRNA signature in colorectal cancer progression and metastasis. Cancer Lett, 2021, 501: 66–82.10.1016/j.canlet.2020.12.025.
- Diener C, Keller A, Meese E. Emerging concepts of miRNA therapeutics: from cells to clinic. Trends Genet, 2022, 38(6): 613–26.10.1016/j.tig.2022.02.006.
- Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, Song SJ. Regulatory mechanism of MicroRNA expression in cancer. Int J Mol Sci, 2020, 21(5).10.3390/ijms21051723.
- Zhao J, Wang X, Mi Z, Jiang X, Sun L, Zheng B, Wang J, Meng M, Zhang L. Wang Z, et al., STAT3/miR-135b/NF-κB axis confers aggressiveness and unfavorable prognosis in non-small-cell lung cancer. Cell Death Dis, 2021, 12(5): 493. 10.1038/s41419-021-03773-x.
- Li J, Sun L, Chen Y, Zhu J, Shen J, Wang J, Gu Y, Zhang G, Wang M. Shi T, et al., Gastric cancer-derived exosomal miR-135b-5p impairs the function of Vγ9Vδ2 T cells by targeting specificity protein 1. Cancer Immunol Immunother, 2022, 71(2): 311–25. 10.1007/s00262-021-02991-8.
- Li Z, Qin Y, Chen P, Luo Q, Shi H, Jiang X. miR‑135b‑5p enhances the sensitivity of HER‑2 positive breast cancer to trastuzumab via binding to cyclin D2. Int J Mol Med, 2020, 46(4): 1514–24.10.3892/ijmm.2020.4681.
- Zhang XH, Xin ZM., Zhang, Q., Chen, X. MiR-135b-5p inhibits the progression of malignant melanoma cells by targeting RBX1. Eur Rev Med Pharmacol Sci, 2420, 20(1): 1309–15. 10.1186/s12935-020-01386-6.
- Wang H, Wang X, Zhang H, Deng T, Liu R, Liu Y, Li H, Bai M, Ning T. Wang J, et al. The HSF1/miR-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the MUL1/ULK1 pathway in colorectal cancer. Oncogene, 2021, 40(28): 4695–708. 10.1038/s41388-021-01898-z.
- Lv ZD, Xin HN, Yang ZC, Wang WJ, Dong JJ, Jin LY, Li FN. miR-135b promotes proliferation and metastasis by targeting APC in triple-negative breast cancer. J Cell Physiol, 2019, 234(7): 10819–26.10.1002/jcp.27906.
- Xiang S, Fang J, Wang S, Deng B, Zhu L. MicroRNA‑135b regulates the stability of PTEN and promotes glycolysis by targeting USP13 in human colorectal cancers. Oncol Rep, 2015, 33(3): 1342–48. 10.3892/or.2014.3694.
- Kong X, Zhai J, Yan C, Song Y, Wang J, Bai X, Brown JaL, Fang Y. Recent advances in understanding FOXN3 in breast cancer, and other malignancies. Front Oncol, 2019, 9: 234.10.3389/fonc.2019.00234.
- Li W, Zhang Z, Liu X, Cheng X, Zhang Y, Han X, Zhang Y, Liu S, Yang J, Xu B, et al., The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J Clin Invest, 2017, 127(9): 3421–40. 10.1172/JCI94233.
- Sun J, Li H, Huo Q, Cui M, Ge C, Zhao F, Tian H, Chen T, Yao M, Li J. The transcription factor FOXN3 inhibits cell proliferation by downregulating E2F5 expression in hepatocellular carcinoma cells. Oncotarget, 2016, 7(28): 43534–45.10.18632/oncotarget.9780.
- Yu W, Diao Y, Zhang Y, Shi Y, Lv X, Zhang C, Zhang K, Yao W, Huang D, Zhang J. Bioinformatic analysis of FOXN3 expression and prognostic value in pancreatic cancer. Front Oncol query, 2022, 12: 1008100. 10.3389/fonc.2022.1008100.
- Lee S, Rauch J, Kolch W. Targeting MAPK signaling in cancer: mechanisms of drug resistance and sensitivity. Int J Mol Sci, 2020, 21(3). 1102 10.3390/ijms21031102.
- Zhang Y, Wang X. Targeting the Wnt/β-catenin signaling pathway in cancer. J Hematol Oncol, 2020, 13(1): 165.10.1186/s13045-020-00990-3.
- Dong S, Liang S, Cheng Z, Zhang X, Luo L, Li L, Zhang W, Li S, Xu Q. Zhong M, et al., ROS/PI3K/Akt and Wnt/β-catenin signalings activate HIF-1α-induced metabolic reprogramming to impart 5-fluorouracil resistance in colorectal cancer. J Exp Clin Cancer Res, 2022, 41(1): 15. 10.1186/s13046-021-02229-6.
- Patel M, Horgan PG, Mcmillan DC, Edwards J. NF-κB pathways in the development and progression of colorectal cancer. Transl Res, 2018, 197: 43–56.10.1016/j.trsl.2018.02.002.
- Sun M, Ma X, Tu C, Wang X, Qu J, Wang S, Xiao S. MicroRNA-378 regulates epithelial-mesenchymal transition and metastasis of melanoma by inhibiting FOXN3 expression through the Wnt/β-catenin pathway. Cell Biol Int, 2019, 43(10): 1113–24. 10.1002/cbin.11027.
- Zhang J, Hu Z, Wen C, Liao Q, He B, Peng J, Tang X, Chen Z, Xie Y. MicroRNA-182 promotes epithelial-mesenchymal transition by targeting FOXN3 in gallbladder cancer. Oncol Lett, 2021, 21(3): 200.10.3892/ol.2021.12461.