ABSTRACT
The relationship between the IL1B-511C>T (rs16944) polymorphism and the risk of developing hematologic malignancies remains controversial. Thus, we performed a meta-analysis to evaluate the association between IL1B-511C>T polymorphism and the risk of developing hematologic malignancies. A comprehensive search was conducted to identify all eligible studies on IL1B-511C>T polymorphism and hematologic malignancies. Twelve case-control studies, with 2,896 cases and 3,716 controls, were selected for the analysis. The overall data failed to indicate a significant association between IL1B-511C>T polymorphism and the risk of hematologic malignancies (OR:1.06, 95% Confidence Interval [CI]: 0.93–1.22). Moreover, non-significant associations were observed in a stratified analysis according to neoplasm type (multiple myeloma, Hodgkin’s lymphoma, and non-Hodgkin’s lymphoma), ethnicity (European and Asian), and Hardy-Weinberg equilibrium. In summary, our results suggest that there is no association between the IL1B-511C>T polymorphism and the risk of hematologic malignancies. As such, further large-scale studies are needed to confirm our findings.
Introduction
Hematologic malignancies are a group of malignant diseases that are derived from myeloid and lymphoid hematopoietic lineagesCitation1 and account for approximately 9% of all cancers. It is also the fourth most frequently diagnosed cancer in both men and women in developed countries.Citation2 According to the World Health Organization (WHO), lymphoid neoplasms are grouped into lymphatic precursor neoplasms; mature B-, T-, and NK-cell neoplasms; and lymphomas. Myeloid neoplasms are subdivided into multiple myeloma (MM) and myelodysplastic syndromes (MDS). Leukemias are classified as either acute or chronic based on the rapidity of proliferation and as myelocytic or lymphocytic based on the cell of origin.Citation3,Citation4 Overall, each leukemia subtypes presents different clinical conditions and each has a specific treatment protocol.
It is well established that chronic inflammation drives tumor progression in multiple types of cancer.Citation5 Increased basal inflammatory status seems to promote mutagenesis through the induction of chronic oxidative stress and subsequent DNA oxidative damage, and elicits epigenetic changes that further promote inflammation.Citation6 In addition, a common feature of many hematologic malignancies is the overproduction of proinflammatory cytokines. Although several cytokines are overexpressed in hematological malignancies, overproduction of tumor necrosis factor-alpha (TNF-α), interleukin 6 (IL-6), and interleukin 1 (IL-1) is most commonly observed in patients, suggesting that these cytokines play a role in the development and/or manifestation of hematologic malignancies.Citation7–9
Evidence supporting a pro-tumorigenic role of IL-1b in all cancer types has been described recently.Citation10 IL-1β plays a pleiotropic role in cancer by modulating gene expression and cytokine production and regulating cellular adhesion and migration, angiogenesis, cancer cell proliferation, and metastasis.Citation10 While acute IL-1β exposure contributes to hematopoietic stem cell (HSC) regeneration after myeloablation and transplantation,Citation11 chronic exposure after infection or injury promotes uncontrolled HSC division and eventual exhaustion of the HSC pool.Citation6,Citation12
IL-1β is a key mediator of carcinogenesis via the promotion of chronic inflammation, and genetic variations with gain function in this gene have been extensively studied in recent years with regards to cancer.Citation13 The Interleukin 1 Beta (IL1B) gene is highly polymorphic, and base transitions between C and T at positions –511 (C>T; dbSNP: rs16944) have been associated with increased IL-1β secretion.Citation14,Citation15 Numerous epidemiological studies have investigated the association between IL1B–511 C>T in many types of cancer;Citation16,Citation17 however, in hematologic malignancies, the results remain unclear and many are inconclusive due to inconsistent findings in individual studies. Therefore, this study performed a meta-analysis to provide accurate data on the association between genetic variation and the risk of hematologic malignancies.
Results
Baseline study characteristics
After careful evaluation of the literature based on the search strategy and eligibility criteria, we identified 11 studies that were included in this meta-analysis. As shown in the flow chart depicting the process of selection of studies (), one study was excluded because it did not describe genotype frequency.Citation18 One study had information on Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL), and for this reason, it was duplicated. Therefore, for statistical analysis, we used 12 studies.
Figure 1. Flowchart for identification, screening and selection of the studies.
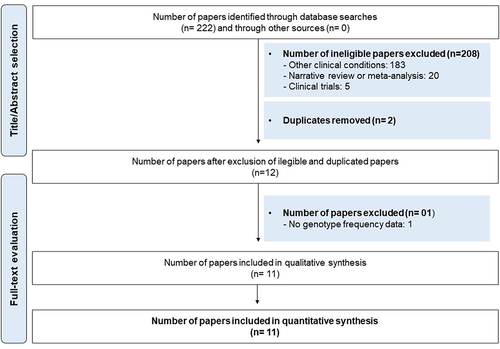
The studies enrolled 2,892 patients diagnosed with a specific hematologic malignancy and compared them with 3,716 cancer-free controls. All studies had a case-control design, were published between 2007 and 2021, and targeted one or more of the following hematologic malignancies: (1) ALL, (1) AML, (1) CLL, (1) CML, (2) HL, (2) NHL, (1) MDS, (2) MM and/or (1) Philadelphia chromosome-negative myeloproliferative neoplasm (MPN).
The detailed characteristics of the data gathered from the 12 case-control studies are summarized in . Eleven studies were in accordance with the Hardy-Weinberg Equilibrium (HWE). However, one was outside the HWE. Six studies involved the Asian population, four involved the European population, one involved the Caucasian population, and one involved the mixed population of Brazil. Most of the studies included adult patients in the case group (91%).
Table 1. Characteristics of studies included in the quantitative synthesis (meta-analysis).
Quality assessment results
The evaluation of the methodological aspects with regard to quality showed that one study had a low score (9 points), nine studies reached a score between 10 and 11 points, and two studies reached a score of 14 points, as shown in .
Meta-analysis of the IL1B–511 C>T polymorphism and its relationship with hematologic malignancies
shows the overall and stratified analyses of this polymorphism according to the type of neoplasm (MM, HL, and NHL), ethnicity (European and Asian), and HWE in allelic and genotypic evaluations. The results of the pooled studies showed that there were no significant associations between the IL1B–511 C>T polymorphism and the risk of hematologic malignancies (OR:1.06; 95% Confidence Interval (CI):0.93–1.22; p = .37) (). Likewise, non-significant associations were found in the stratified analysis by i) type of neoplasm: MM (OR: 1.28, 95% CI: 0.81–2.02, p = .29), HL (OR: 1.02, 95% CI: 0.84–1.24, p = .85) and NHL (OR: 1.00, 95% CI: 0.63–1.61, p = .98), ii) ethnicity: European (OR: 0.86, 95% CI: 0.57–1.30, p = .48) and Asian (OR: 1.09, 95% CI: 0.99–1.20, p = .09) and iii) HWE (OR: 1.05, 95% CI: 0.91–1.20, p = .54). In this study, we used the random-effects statistical model for OR calculation because of the increased heterogeneity value (I2 = 59%, p = .00).
Figure 2. Forest plots for comparison of the (a) mutant allele versus wild-type allele, (b) wild-type allele versus mutant allele, (c) homozygous mutant versus homozygous mutant, and (d) homozygous wild-type versus homozygous mutant in IL1B–511 C>T (rs16944) polymorphism and hematologic malignancies.
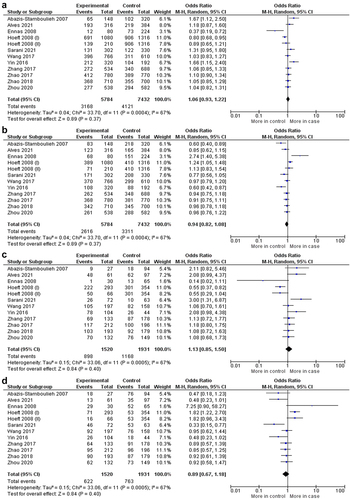
Table 2. Meta-analysis of the association between the IL1B–511 C>T (rs16944) polymorphism and the risk of hematological malignancies (allelic and genotypic comparisons; and stratified analysis).
Publication bias and sensitivity analysis
The results of the evaluation of publication bias showed the absence of apparent asymmetry in the funnel plot graphics for the comparisons of the overall evaluation (Figure S1). These data were supported by the non-significant values from the Begg test and Egger’s linear regression tests, as shown in Table S1. Sensitivity analysis also demonstrated that no single study affected the pooled OR values in the validation of our results.
Discussion
To the best of our knowledge, this is the first meta-analysis to comprehensively evaluate the association between IL1B–511 C>T polymorphism and the risk of hematological malignancies. Single-nucleotide variants (SNVs) is a term that describes a nucleotide variation in the DNA sequence, and is intrinsically associated with drug resistance, disease susceptibility, and ethnic differences.Citation29 Published data regarding the biological functions of IL1B polymorphisms have shown that the IL1B–511 C>T polymorphism strongly influences transcriptional activity only in the context of other IL1B promoter polymorphisms, such as IL-1B −31 C>T.Citation15 Moreover, a C to T single base polymorphism in the promoter of IL1B gene C(−511)→T) has been reported to affect IL-1 and IL-1Ra levels.Citation30
Few studies have demonstrated the role of the IL1B −511 C>T polymorphism in patients with hematologic malignancies. Hence, owing to the inconsistency in results and the limited number of studies available, we performed this meta-analysis. The results of the overall and stratified studies showed that there were no statistically significant associations between IL1B–511 C>T and the risk of hematologic malignancies. In addition, owing to the limited number of studies on mixed and Caucasian ethnicities (n = 1), we could not perform a stratified analysis. Despite this, the use of the suggested guidelines for the evaluation of these studies demonstrated the acceptable quality of studies in this meta-analysis ( and Table S1), which demonstrated the accurate methodological aspects of the studies.
Although our data showed non-significant associations for allelic or genotypic evaluations, in previous reports, the studied alleles and genotypes for the -511 C>T polymorphism in IL1B have been associated with the risk of some hematologic malignancies and clinical implications. The T allele is associated with an increased risk of MM.Citation19 In acute leukemia, the CT and TT genotypes are associated with the risk of pediatric ALL,Citation20 and CT has been associated with a favorable-risk cytogenetic group in AML.Citation24 In chronic leukemia, the T allele has been associated with a lower risk of CLLCitation21 and CT with an early molecular response at 6 months for BCR:ABL in CML.Citation26 Patients with the IL1B–511 C>T polymorphism had a higher score on the International Prognostic Scoring System (IPSS), which might serve as a novel biomarker and potential target for MDS.Citation25 Enhanced IL-1β signaling is a common event in patients with hematological malignancies,Citation13 and knowledge of their genetics and molecular mechanisms will allow us to determine the true potential of IL-1β targeting as a therapy for hematological malignancies and their related complications.
Ethnicity directly influences the incidence of hematologic malignancies in a given population.Citation31 According to previous studies, American descendants are more susceptible to developing Hodgkin and non-Hodgkin lymphoma, multiple myeloma, acute myeloid, and lymphoblastic leukemia,Citation32 while people of South Asian and African descent have the lowest risk.Citation31,Citation33 It is important to note that in our study, Brazilians were exclusively composed of a mixed population, which is characterized by a high degree of admixture from Amerindian, African, and European ancestors.Citation34 In the literature, children of admixture ethnicity had a high risk of developing ALL due to the Amerindian genetic background.Citation35 This corroborates the study by Alves et al. (2021), which described the IL1B–511 C>T polymorphism as a risk factor for ALL in children from the Amazon region,Citation20 which is a region where Amerindian ancestry is predominantly found.Citation36
Although our meta-analysis is the first to approach the association between this polymorphism and hematological malignancies and, as such, brought robust and accurate results with the absence of publication bias, this study has some important limitations that should be cited and discussed: (i) the limited number of studies may explain the non-significant associations observed so far; therefore, a larger sample size is necessary to validate the results, since in a stratified analysis, the number of each subgroup seems to be lower; ii) besides, we were unable to include a global representative population since the frequency of IL1B–511 C>T polymorphism is influenced by population analyzed which in our study was Asian population was predominant, iii) hematologic malignancies are multifactorial diseases caused by the interaction of several factors such as age, sex, infections, exposure to radioactive and/or chemical agents, and ethnicity. Hence, a completed stratified analysis could provide a better understanding of the influence of the ILB −511 C>T polymorphism and its development; however, it was not possible to perform this due to the limited available data in the studies. (iv) More specific analyses might be conducted if individual data were available, such an analysis would have allowed us to adjust for other covariates, such as age, family history, and environmental factors and v) Finally, and our data are accurate and demonstrate non-significant associations. Further studies with larger sample sizes are required to investigate the association between the IL1B–511 C>T polymorphism and hematologic malignancies.
In conclusion, the overall and stratified analyses of this meta-analysis, composed of 12 case-control studies with 2,892 patients and 3,716 cancer-free controls, did not find any evidence of an association between the IL1B–511 C>T polymorphism and the risk of hematologic malignancies.
Materials and methods
Literature search strategy
This meta-analysis followed the Preferred Reporting Items for Systematic Reviews and Meta-Analyses) statement.Citation37 Electronic databases (PubMed, Web of Science, and Google Scholar) were comprehensively searched using the following combined keywords or medical subject headings (MeSH): “polymorphism,” “rs16944,” “inflammasome” and “cancer.” There were no language restrictions in the search strategy, and all studies published before November 30, 2022, were considered. We screened the abstracts of the studies found as well as their references to identify potential additional studies.
Inclusion and exclusion criteria
In this meta-analysis, studies were included according to the following criteria: (1) studies evaluating the association between the IL1B–511 C>T (rs16944) polymorphism and the risk of some hematologic malignancy; (2) case-control studies; and (3) studies with sufficient genotype data in cases and controls to calculate the odds ratio (OR) with 95% confidence intervals (95% CIs). The main reasons for exclusion of studies were as follows: (1) no genotype frequency data and (2) duplication of an earlier publication.
Data extraction
Two researchers (Fabíola Silva Alves Hanna [FSAH] and Felipe Rodolfo Pereira Silva [FRPS]) independently reviewed all the studies and extracted the data using a standardized form. Disagreements were resolved by discussion with the coauthors. The following information was extracted: the first author’s surname, year of publication, disease type, ethnicity, country of origin, study design, age, subject type, number of cases and controls in the sample, and whether the allelic and genotypic frequencies were in Hardy – Weinberg equilibrium (HWE).
Quality score assessment
Two reviewers independently assessed the quality of studies (FSAH and FRPS) according to the scale for quality assessment described by Tian et al. (2016).Citation38 The quality scale was based on the methodological aspects of the included studies, such as the source of cases, source of controls, specimens collected, HWE in controls, and total sample size. These scores were selected on both traditional epidemiological considerations and cancer genetic issues. The scores ranged from 0 to 15, and quality was measured by the variation from low (worst) to high (best) scores (Table S1).
Statistical analysis
The statistical program Review Manager version 5.3 (RevMan, Nordic Cochrane Centre, The Cochrane Collaboration, 2012) was used for the systematic reviews and meta-analyses. Publication bias was evaluated using Comprehensive Meta-analysis statistical software version 3.3.070 (2014).
The presence or absence of true heterogeneity (I2) was calculated using Cochran’s X2 test or chi-squared Q-based statistical test. I2 was also analyzed for heterogeneity by visualizing the funnel plot graph. When the observed value of I2 presented not statistically significant and was defined as mild or moderate (ICitation2 <50%, p > .05), the authors used the fixed-effects model for the pooled odds ratio (OR) calculation. When I2 presented a statistically significant value and was defined as elevated (I2 >50%, p < .05), the random-effects statistical model was used for OR calculations. Statistical significance was set at p < .05. To quantify the exact influence of genetic variation on the risk of disease development, six genetic models were measured using “M” as the mutant allele and “M” allele as the wild-type allele. Therefore, the calculations were composed of allelic comparisons: (I) M versus m, (II) m versus M; genotypic comparisons: (III) MM versus mm, (IV) mm versus MM, and combinations of genotypic variations: (V) MM versus mm + Mm, and (VI) Mm versus MM + mm. In addition, a sensitivity analysis was performed by omitting one study at the time of the pooled OR calculation for the mutant allele in order to detect any type of single interference. A sensitivity analysis was performed by omitting one included study at a time to verify any possible significant changes in the OR value. To assess publication bias, the Begg’s test and Egger’s linear regression test were used to estimate potential publication bias (p < .05). In this meta-analysis, asymmetry of the funnel plot for publication bias was also considered to validate the results of Begg’s test and Egger’s test. All the included studies had dichotomous data expressed as OR with 95% confidence intervals (CI) to verify the possible association between the aforementioned genetic variations and hematologic malignancies.
Author contributions
Study design: FSAH, DSP, FM-G, and AGC. Searched databases and collected full-text papers: FSAH and FRPS. FSAH and FRPS were extracted and analyzed. Statistical analyses: FRPR and ALABL. FSAH and AGC wrote the manuscript All authors have reviewed the final version of the manuscript.
Supplemental Material
Download MS Word (173.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplemental material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/15384047.2024.2382503
Additional information
Funding
References
- Zierhut M, Haen SP, Moehle R, Chan C-C. Hematological Neoplasms. In: Intraocular inflammation. Berlin, Heidelberg: Springer Berlin Heidelberg; 2016. p. 1493–8. doi:10.1007/978-3-540-75387-2_149.
- Smith A, Howell D, Patmore R, Jack A, Roman E. Incidence of haematological malignancy by sub-type: a report from the haematological malignancy research network. Br J Cancer. 2011;105(11):1684–92. doi:10.1038/bjc.2011.450.
- Arber DA, Orazi A, Hasserjian RP, Borowitz MJ, Calvo KR, Kvasnicka HM, Wang SA, Bagg A, Barbui T, Branford S, et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood. 2022;140(11):1200–28. doi:10.1182/blood.2022015850.
- Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, Bejar R, Berti E, Busque L, Chan JKC, et al. The 5th edition of the world health organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36(7):1703–19. doi:10.1038/s41375-022-01613-1.
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74. doi:10.1016/j.cell.2011.02.013.
- Hasselbalch HC. Chronic inflammation as a promotor of mutagenesis in essential thrombocythemia, polycythemia vera and myelofibrosis. A human inflammation model for cancer development? Leuk Res. 2013;37(2):214–20. doi:10.1016/j.leukres.2012.10.020.
- Craver BM, El Alaoui K, Scherber RM, Fleischman AG. The critical role of inflammation in the pathogenesis and progression of myeloid malignancies. Cancers (Basel). 2018;10(4):1–18. doi:10.3390/cancers10040104.
- Myers JS. Proinflammatory cytokines and sickness behavior: implications for depression and cancer-related symptoms. Oncol Nurs Forum. 2008;35(5):802–07. doi:10.1188/08.ONF.802-807.
- Magalhães-Gama F, Kerr MWA, De Araújo ND, Ibiapina HNS, Neves JCF, Hanna FSA, Xabregas LDA, Carvalho MPSS, Alves EB, Tarragô AM, et al. Imbalance of chemokines and cytokines in the bone marrow microenvironment of children with B-Cell acute lymphoblastic leukemia. J Oncol. 2021;2021:1–9. doi:10.1155/2021/5530650.
- Rébé C, Ghiringhelli F. Interleukin-1 β and cancer. Cancers (Basel). 2020;12(7):1791–822. doi:10.3390/cancers12071791.
- Hermouet S, Vilaine M. The JAK2 46/1 haplotype: a marker of inappropriate myelomonocytic response to cytokine stimulation, leading to increased risk of inflammation, myeloid neoplasm, and impaired defense against infection? Haematologica. 2011;96(11):1575–79. doi:10.3324/haematol.2011.055392.
- Pietras EM, Mirantes-Barbeito C, Fong S, Loeffler D, Kovtonyuk LV, Zhang S, Lakshminarasimhan R, Chin CP, Techner J-M, Will B, et al. Chronic interleukin-1 drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol. 2016;18(6):607–18. doi:10.1038/ncb3346.Chronic.
- Arranz L, Arriero M, Del M, Villatoro A. Interleukin-1β as emerging therapeutic target in hematological malignancies and potentially in their complications. Blood Rev. 2017;31(5):306–17. doi:10.1016/j.blre.2017.05.001.
- Hall SK, Perregaux DG, Gabel CA, Woodworth T, Durham LK, Huizinga TWF, Breedveld FC, Seymour AB. Correlation of polymorphic variation in the promoter region of the interleukin-1β gene with secretion of interleukin-1β protein. Arthritis Rheum. 2004;50(6):1976–83. doi:10.1002/art.20310.
- Chen H, Wilkins LM, Aziz N, Cannings C, Wyllie DH, Bingle C, Rogus J, Beck JD, Offenbacher S, Cork MJ, et al. Single nucleotide polymorphisms in the human interleukin-1B gene affect transcription according to haplotype context. Hum Mol Genet. 2006;15(4):519–29. doi:10.1093/hmg/ddi469.
- He B, Zhang Y, Pan Y, Xu Y, Gu L, Chen L, Wang S. Interleukin 1 beta (IL1B) promoter polymorphism and cancer risk: evidence from 47 published studies. Mutagenesis. 2011;26(5):637–42. doi:10.1093/mutage/ger025.
- Xu J, Yin Z, Cao S, Gao W, Liu L, Yin Y, Liu P, Shu Y. Systematic review and meta-analysis on the association between IL-1B polymorphisms and cancer risk. PLOS ONE. 2013;8(5):e63654. doi:10.1371/journal.pone.0063654.
- Zhang C, Han F, Yu J, Hu X, Hua M, Zhong C, Wang R, Zhao X, Shi Y, Ji C, et al. Investigation of NF-κB-94ins/del ATTG and CARD8 (rs2043211) gene polymorphism in acute lymphoblastic leukemia. Front Endocrinol (Lausanne). 2019;10(August):1–9. doi:10.3389/fendo.2019.00501.
- Abazis-Stamboulieh D, Oikonomou P, Papadoulis N, Panayiotidis P, Vrakidou E, Tsezou A. Association of interleukin-1A, interleukin-1B and interleukin-1 receptor antagonist gene polymorphisms with multiple myeloma. Leuk Lymphoma. 2007;48(11):2196–203. doi:10.1080/10428190701615892.
- Alves FS, Xabregas LA, Kerr MWA, Souza GL, Pereira DS, Magalhães-Gama F, Santiago MRR, Garcia NP, Tarragô AM, Ogusku MM, et al. Genetic polymorphisms of inflammasome genes associated with pediatric acute lymphoblastic leukemia and clinical prognosis in the Brazilian Amazon. Sci Rep. 2021;11(1):1–10. doi:10.1038/s41598-021-89310-4.
- Ennas MG, Moore PS, Zucca M, Angelucci E, Cabras MG, Melis M, Gabbas A, Serpe R, Madeddu C, Scarpa A, et al. Interleukin-1B (IL1B) and interleukin-6 (IL6) gene polymorphisms are associated with risk of chronic lymphocytic leukaemia. Hematol Oncol. 2008;26(2):98–103. doi:10.1002/hon.843.
- Hoeft B, Becker N, Deeg E, Beckmann L, Nieters A. Joint effect between regular use of non-steroidal anti-inflammatory drugs, variants in inflammatory genes and risk of lymphoma. Cancer Causes Control. 2008;19(2):163–73. doi:10.1007/s10552-007-9082-9.
- Sarani H, Mollashahi B, Taheri M, Bahari G, Hashemi SM, Hashemi M, Ghavami S. Association between the IL-1A, IL-1B and IL-1R polymorphisms and lymphoma. Nucleosides Nucleotides Nucleic Acids. 2021;40(7):707–19. doi:10.1080/15257770.2021.1929317.
- Wang H, Hua M, Wang S, Yu J, Chen C, Zhao X, Zhang C, Zhong C, Wang R, He N, et al. Genetic polymorphisms of IL-18 rs1946518 and IL-1β rs16944 are associated with prognosis and survival of acute myeloid leukemia. Inflammation Res. 2017;66(3):249–58. doi:10.1007/s00011-016-1012-4.
- Yin C, He N, Li P, Zhang C, Yu J, Hua M, Ji C, Ma D. Polymorphisms of interlukin-1β rs16944 confer susceptibility to myelodysplastic syndromes. Life Sci. 2016;165:109–12. doi:10.1016/j.lfs.2016.09.019.
- Zhang A, Yu J, Yan S, Zhao X, Chen C, Zhou Y, Zhao X, Hua M, Wang R, Zhang C, et al. The genetic polymorphism and expression profiles of NLRP3 inflammasome in patients with chronic myeloid leukemia. Hum Immunol. 2017;79(1):57–62. doi:10.1016/j.humimm.2017.10.013.
- Zhao X, Zhang C, Hua M, Wang R, Zhong C, Yu J, Han F, He N, Zhao Y, Liu G. NLRP3 inflammasome activation plays a carcinogenic role through effector cytokine IL-18 in lymphoma. Oncotarget. 2017;8(65):108571–83. doi:10.18632/oncotarget.21010.
- Zhou Y, Yan S, Liu N, He N, Zhang A, Meng S, Ji C, Ma D, Ye J. Genetic polymorphisms and expression of NLRP3 inflammasome-related genes are associated with Philadelphia chromosome-negative myeloproliferative neoplasms. Hum Immunol. 2020;81(10–11):606–13. doi:10.1016/j.humimm.2020.09.001.
- Somberg JC. Genetic polymorphisms. Am J Ther. 2002;9(4):271. doi:10.1097/00045391-200207000-00001.
- Lai J, Zhou D, Xia S, Shang Y, Zhu J, Pan J, Hua B, Zhu Y, Cui L. Association of interleukin-1 gene cluster polymorphisms with ischemic stroke in a Chinese population. Neurol India. 2006;54(4):366–69. doi:10.4103/0028-3886.28107.
- Shirley MH, Sayeed S, Barnes I, Finlayson A, Ali R. Incidence of haematological malignancies by ethnic group in England, 2001-7. Br J Haematol. 2013;163(4):465–77. doi:10.1111/bjh.12562.
- Kirtane K, Lee SJ. Racial and ethnic disparities in hematologic malignancies. Blood. 2017;130(15):1699–705. doi:10.1182/blood-2017-04-778225.
- Tran HN, Li Y, Udaltsova N, Armstrong MA, Friedman GD, Klatsky AL. Risk of cancer in Asian Americans: a kaiser permanente cohort study. Cancer Causes Control. 2016;27(10):1197–207. doi:10.1007/s10552-016-0798-2.
- Giolo SR, Soler JMP, Greenway SC, Almeida MAA, De Andrade M, Seidman JG, Seidman CE, Krieger JE, Pereira AC. Brazilian urban population genetic structure reveals a high degree of admixture. Eur J Hum Genet. 2012;20(1):111–16. doi:10.1038/ejhg.2011.144.
- Carvalho DC, Wanderley AV, Amador MAT, Fernandes MR, Cavalcante GC, Pantoja KBCC, Mello FAR, de Assumpção PP, Khayat AS, Ribeiro-dos-Santos Â, et al. Amerindian genetic ancestry and INDEL polymorphisms associated with susceptibility of childhood B-cell Leukemia in an admixed population from the Brazilian Amazon. Leukemia Res. 2015;39(11):1239–45. doi:10.1016/j.leukres.2015.08.008.
- Moura RRCA, Balbino VQ, Brandão LAC. Meta-analysis of Brazilian genetic admixture and comparison with other Latin America countries. Am J Hum Biol. 2015;27(5):674–80. doi:10.1002/ajhb.22714.
- Moher D, Liberati A, Tetzlaff J, Altman DG. Reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–69. doi:10.7326/0003-4819-151-4-200908180-00135.
- Tian X, Dai S, Sun J, Jiang S, Jiang Y. Association between TP53 Arg72Pro polymorphism and leukemia risk: a meta-analysis of 14 case-control studies. Sci Rep. 2016;6(October 2015):1–6. doi:10.1038/srep24097.
