In this issue of Cell Cycle, Kozakova et al. report that MAGEA1 stimulates the E3 ubiquitin ligase activity of TRIM31 within a TRIM31-MAGEA1-NSE4 complex.Citation1 The melanoma-associated antigens, MAGEs, represented for long time just a bizarre group of tumor-expressed peptides. Structurally, they share a 170-residue MAGE Homology Domain (MHD) that folds in 2 winged-helix motifs referred to as WH-A and WH-B. The MAGE family counts dozens human genes classified according to their expression patterns into Type I and Type II.Citation2 MAGE II genes are expressed in a wide variety of tissues whereas MAGE I genes are normally restricted to germline and trophoblast cells to become then aberrantly expressed in melanoma, as initially discovered, and in a wide range of other tumor types. It is increasingly clear that type I MAGEs represent not only useful diagnostic-prognostic markers of malignancies but also oncogenes per se.Citation2
The enigmatic function of the MAGE proteins commenced to be unveiled with the relatively recent discovery of their preferred binding partners: RING proteins, the largest class of E3 ubiquitin ligases. Amazingly, the MAGE preference for RING-containing proteins is not exerted through the RING domain but other binding moieties that can differ in the diverse RING E3 partners.Citation3 Among the MAGE RING E3 ligase partners, the TRIpartite Motif (TRIM) proteins are coming into play. TRIM proteins represent a large subfamily of RING E3 ligases that share a common N-terminal module composed of a RING domain, 1 or 2 B-box domain(s), and a Coiled-coil region followed by variable C-terminal domains.Citation4 Up to date, few MAGE-TRIM interactions involving TRIM27, 28, 37, and 69 have been reported, identified either in large interactome screenings or through more targeted approaches.Citation3,5,6
Kozakova et al. provide now evidence that MAGE-TRIM interaction is not a sporadic occurrence and report that TRIM8, TRIM31, and TRIM41 interact with MAGE proteins as well.Citation1 The study reveals specific MAGE-TRIM pairings and confirms that TRIM proteins can bind both MAGE types. When it comes to the implicated domain, contrary to what expected, the 3 TRIM proteins exploit different regions to bind MAGE proteins: TRIM8 and TRIM41 use their C-terminal domain while in the case of TRIM31 the Coiled-coil region is necessary and sufficient for MAGE binding . This finding further endorses the fact that MAGE-RING interactions did not evolve through the recognition of common motifs in the RING partners.
Figure 1. Schematic of TRIM31 within a TRIM-MAGE-NSE4 complex. NSE3-related stimulation of NSE1 E3 ligase activity within the NSE1-NSE3-NSE4 complex is WH-A-binding-dependent; model for MAGEA1-TRIM31-NSE4 enhancement of ubiquitination, mediated by WH-A and Coiled-coil recognition; alternative TRIM-MAGE mode of binding through C-terminus and WH-B. How co-factors (C), substrates (S) and E2 enzymes fit into the model is still unclear.
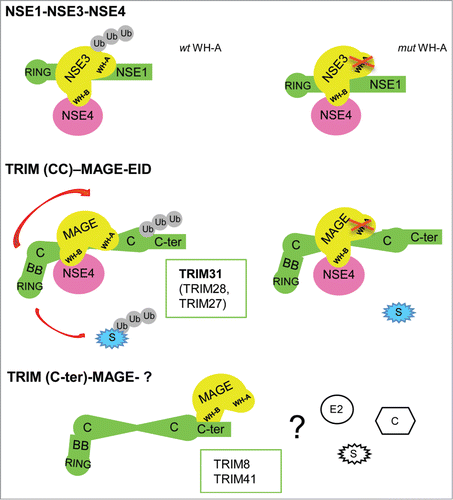
It was reported that MAGEs stimulate the E3 ligase activity of several RING partners among which TRIM28 and TRIM27.Citation3,5,6 Kozakova et al. demonstrate that TRIM31 is mono- and di-ubiquitinated and that co-expression of its MAGE partner, MAGEA1, specifically enhances TRIM31 ligase activity.Citation1 Direct binding is necessary to increase this activity since the MAGEA1 WH-A/L114A, L115A mutant, which interferes with TRIM31 binding, abolishes the ligase activity enhancement (Fig. 1). These results are consistent with those showing that MAGEC2 binds the coiled-coil domain of TRIM28 thus enhancing its E3 ligase activity.Citation3 How the E2 enzymes participate in this framework is still unclear. Conversely, the E3 ubiquitin ligase activity of TRIM8 and TRIM41 is not enhanced by their cognate MAGE. Besides the different TRIM sub-domain involved, TRIM8 and 41 demand MAGE to principally use the WH-B helix to mediate the interaction.Citation1 It is tempting to speculate that the ‘E3 enhancing combination’ necessitates TRIM Coiled-coil and WH-A helix collaboration. It is conceivable that other combinations would lead to traditional E3 ligase (TRIM)-substrate (MAGE) relationships as was proposed for MAGE-D1-Praja interaction (Fig. 1).
To start elucidating MAGEA1-TRIM31 function, the authors demonstrate that TRIM31 not only binds MAGEA1 but it also directly interacts with NSE4a, an EID (E1A-like inhibitor of differentiation) family member. This interaction conceivably occurs in a TRIM31-MAGEA1-NSE4 complex that parallels the NSE1 (RING)-NSE3 (MAGEG1)-NSE4 hetero-trimer, thus implying a TRIM31 role in transcriptional regulation.Citation1 Interestingly, TRIM28 and TRIM27 also interact with EID family proteins, suggesting that TRIM-MAGE-EID complexes evolved from an ancestral NSE1-NSE3-NSE4 trimer through the diversification of RING-containing partners.Citation1,3,7 As both MAGE and TRIM families underwent large expansion in mammals it is attractive to envisage a co-evolution of these genes within the ubiquitination process.
References
- Kozakova L, et al. Cell Cycle 2015; 14(6):920-30; PMID:25590999; http://dx.doi.org//10.1080/15384101.2014.1000112
- Simpson AJG, et al. Nat. Rev. Cancer 2005; 5: 605-625; http://dx.doi.org/10.1038/nrc1669
- Doyle JM, et al. Mol Cell 2010; 39: 963-974; PMID:20864041; http://dx.doi.org/10.1016/j.molcel.2010.08.029
- Reymond A, et al. EMBO J 2001; 20: 2140-2151; PMID:11331580; http://dx.doi.org/10.1093/emboj/20.9.2140
- Yang B, et al. Cancer Res. 2007; 67: 9954-9962; PMID:17942928; http://dx.doi.org/10.1158/0008-5472.CAN-07-1478
- Hao YH, et al. Cell 2013; 152:1051-1064; PMID:23452853; http://dx.doi.org/10.1016/j.cell.2013.01.051
- Hudson JJR, et al. PLoS ONE 2011; 6(2): e17270; PMID:21364888; http://dx.doi.org/10.1371/journal.pone.0017270
