Abstract
Autophagy-promoting proteins and stimuli are often associated with inhibition of cell proliferation; in this context, we recently described a key role for the pro-autophagic protein AMBRA1. Indeed, AMBRA1, through its direct interaction with the protein phosphatase PP2A, tightly regulates the stability of the oncoprotein and pro-mitotic factor c-Myc. Moreover, the AMBRA1-mediated regulation of c-Myc affects both cell proliferation rate and tumorigenesis. Interestingly, AMBRA1/PP2A activity is under the control of the master regulator of autophagy and cell growth, the protein kinase mTOR. Besides the mechanistic details of this regulation pathway which we dissected previously, any possible interplay(s) between AMBRA1 and its interactor BECLIN 1 was not investigated in this scenario. Here we show that both AMBRA1 and BECLIN 1 affect c-Myc regulation, but through two different pathways. Nevertheless, these two pro-autophagic proteins are, together with PP2A, in the same macromolecular complex, whose functional significance of which will be addressed in future studies.
Introduction
Macroautophagy (hereafter autophagy) is a lysosomal catabolic process, which constitutes a fundamental response against several stresses that the cell has to cope with. Nevertheless, autophagy also participates in the maintenance of cellular homeostasis in physiological conditions.Citation1,2 Because of these multiple autophagy roles, it is not surprising that several cellular pathways impact on autophagy and viceversa.Citation3-5 In this context, an inverse correlation has long been observed between cell proliferation and autophagy.Citation6-8 Along the same lines, it has been shown that the depletion of the pro-autophagic gene AMBRA1,Citation7,9 as well as of the AMBRA1-interactor BECLIN 1,Citation8 is responsible for increased proliferation. AMBRA1 and BECLIN 1 are both components of the PI3KIII complex,Citation7,10,11 which is involved in one of the early steps of autophagy induction, the formation of the autophagy-related vesicle, named autophagosome.Citation2 Besides this common feature, both AMBRA1 and BECLIN 1 participate in autophagy regulation together with other interactors and through several regulatory mechanisms.Citation12-16 Despite their deep characterization in the autophagy pathway, hyperproliferative phenotypes associated to AMBRA1- and BECLIN 1-defective systems were not supported by clear molecular mechanisms until very recently. Indeed, the characterization of AMBRA1 role in cell proliferation has been the focus of one our more recent publications.Citation9 In particular, we found that AMBRA1 interacts with the catalytic subunit of the protein phosphatase PP2A (PP2A-C) through two PP2A binding sites which we first identified and named PXP-motifs.Citation9 Mutation on the PXP-motifs (AMBRA1PXP mutant) disrupts AMBRA1/PP2A-C interaction and results in c-Myc hyperphosphorylation and stabilization; as a consequence, c-Myc driven hyperproliferation and tumorigenesis are observed in AMBRA1-defective systems.Citation9 Interestingly, we also found that the AMBRA1-mediated regulation of c-Myc is under the control of the protein kinase mTOR,Citation9 a master regulator of autophagy and cell growth.Citation17 Nevertheless, autophagy regulation is mainly unaffected by the mutation of AMBRA1 PXP-motifs, this due to this mutant's maintained capability to interact with the main partners identified for AMBRA1 in the autophagy pathway, BECLIN 1 among others.Citation9
However, in our recent work,Citation9 we investigate neither the presence of other AMBRA1 interacting proteins in the AMBRA1/PP2A complex nor the existence of other plausible crosstalk(s) between AMBRA1-related proteins and c-Myc regulation. These two aspects are the focus of this Extra View article.
BECLIN 1 is in a macromolecular complex with AMBRA1 and PP2A-C
As a scaffold protein, AMBRA1 fine-tunes autophagy by bringing together proteins functionally related to the autophagy pathway. Consistently with this evidence, we and other groups identified numerous protein complexes of which AMBRA1 is a component.Citation16 Notably, in our recent publication, we describe a role in the regulation of cell proliferation for a new AMBRA1 complex (AMBRA1/PP2A complex).Citation9 Hitherto, some of the possible interaction(s) between the different AMBRA1 complexes remain unexplored. Indeed, a question remains as whether AMBRA1/PP2A form an independent complex or join a macromolecular complex also related to autophagy.
In order to answer this question, endogenous proteins extracted from HEK293 cells were analyzed by Size-Exclusion Fast-Protein-Liquid-Chromatography (sec-FPLC). The collected protein fractions were then analyzed by Western Blot analysis, using specific antibodies against AMBRA1, PP2A-C and the autophagy-related protein BECLIN 1, a known and well-characterized AMBRA1 interactor.Citation7,10 As shown in , AMBRA1, PP2A-C and BECLIN 1 are co-purified in the same fractions (fractions 23-27), demonstrating the existence of a macromolecular complex comprising all of them, and with a Molecular Weight (MW) of ~700 kDa. The MW of the purified complex suggests that other proteins than AMBRA1, PP2A-C and BECLIN 1 are in the same complex. In addition, as expected, PP2A-C is also purified from fractions free of AMBRA1 (as we previously observedCitation9) and BECLIN 1.
Figure 1. SEC analysis of AMBRA1, BECLIN 1 and PP2A-C complex. (A) Protein extracts from HEK293 cells were separated into different fractions and the presence of AMBRA1, BECLIN 1 and PP2A-C was assessed by Western Blot, using anti-AMBRA1, anti-BECLIN 1 and anti-PP2A-C antibodies. (B) Protein extracts, collected from HEK293 cells upon autophagy induction (EBSS treatment; starvation), were treated as in (A). Of note, variations of AMBRA1 and PP2A-C profiles can be observed upon autophagy induction [(B), fractions 23-27], with respect to the control condition (A). Although the meaning of these observations needs further investigation, upon starvation changes in the composition of the macromolecular complex can be hypothesized, most likely due ambra1, BECLIN 1 and PP2A-C interactors joining or leaving the complex.
![Figure 1. SEC analysis of AMBRA1, BECLIN 1 and PP2A-C complex. (A) Protein extracts from HEK293 cells were separated into different fractions and the presence of AMBRA1, BECLIN 1 and PP2A-C was assessed by Western Blot, using anti-AMBRA1, anti-BECLIN 1 and anti-PP2A-C antibodies. (B) Protein extracts, collected from HEK293 cells upon autophagy induction (EBSS treatment; starvation), were treated as in (A). Of note, variations of AMBRA1 and PP2A-C profiles can be observed upon autophagy induction [(B), fractions 23-27], with respect to the control condition (A). Although the meaning of these observations needs further investigation, upon starvation changes in the composition of the macromolecular complex can be hypothesized, most likely due ambra1, BECLIN 1 and PP2A-C interactors joining or leaving the complex.](/cms/asset/a189ac82-ba99-4d1b-bdb1-aa68a73ec079/kccy_a_1021526_f0001_c.gif)
In order to test whether autophagy induction could affect the interaction between AMBRA1, PP2A-C and BECLIN 1, we performed the same sec-FPLC analysis, using protein extracts from starved cells [cells treated with the starvation medium (EBSS, Earle's Balanced Salt Solution)]. The EBSS is depleted of both amino acids and serum and contains a reduced amount of glucose (1 g/l) in relation to the control medium (4.5 g/l), thus resulting in the activation of several pro-autophagic pathways and in a powerful induction of autophagy. We know that the AMBRA1 interaction with both PP2A-C and BECLIN 1 is not affected by EBSS treatmentCitation7,9; Moreover, it could be hypothesized that AMBRA1/PP2A-C and/or AMBRA1/BECLIN 1 complexes reside in separated pools upon autophagy induction. To verify this hypothesis, fractions collected from starved cells were analyzed by Western Blot analysis. The results, shown in , reveal that AMBRA1, PP2A-C and BECLIN 1 are also in the same multimeric complex upon autophagy induction.
More generally, these results do not clarify whether AMBRA1 is also a scaffold between PP2A-C and BECLIN 1, binding the two proteins at the same time, or whether AMBRA1/PP2A-C and AMBRA1/BECLIN 1 constitute two different sub-complexes joining the same high-molecular-weight multimeric complex. Given that BECLIN 1 and PP2A-C interactions occur on different regions of the AMBRA1 protein,Citation7,9 and that AMBRA1PXP mutant is still able to bind BECLIN 1,Citation9 PP2A-C and BECLIN 1 should not compete for the binding to AMBRA1; for this reason, we cannot rule out the existence of a hetero-trimeric AMBRA1/PP2A-C/BECLIN 1 complex. Nevertheless, we could observe no interaction between BECLIN 1 and PP2A-C by co-immunoprecipitation assay (data not shown), this weakening the hypothesis of a stable AMBRA1/PP2A-C/BECLIN 1 complex.
A recent study shows that a molecular platform, named exocyst, ensures the physical proximity among pro-autophagic complexes, BECLIN 1 among them, making it plausible that AMBRA1 as well is located at the exocyst.Citation18,19 Indeed, we can thus hypothesize that the high-molecular-weight multimeric complex, which we purify in fractions 23-27, is the exocyst itself ().
In any case, an open question remains as to the functional significance of the physical proximity among AMBRA1, PP2A-C and BECLIN 1.
AMBRA1 and BECLIN 1 impact on c-Myc regulation via two different pathways
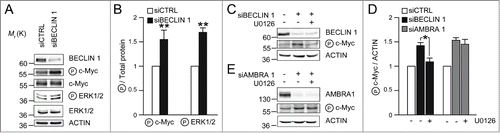
In our previous manuscript, we showed that AMBRA1 participates in c-Myc de-phosphorylation, regardless of BECLIN 1 interaction. Also, we proved AMBRA1PXP mutant fails in regulating c-Myc, even though it is still able to bind BECLIN 1.Citation9 Nevertheless, a link between BECLIN 1 and the EGFR pathway, which indirectly regulates c-Myc phosphorylation, has been previously shown.Citation20 Upon EGFR activation, the c-Myc kinases ERK1/2 are activated, resulting in increased phosphorylation of c-Myc on Ser62 (P c-MycS62).Citation21-23 Upon receptor activation, a sub-complex of PI3KIII, of which BECLIN 1 is a component, regulates EGFR internalization and degradation, by means of the endocytotic pathway.Citation20 Also, a defect in the EGFR down-regulation results in a prolonged propagation of proliferative signals, this a possible cause of neoplastic growth. In their manuscript, Thoresen and co-authors showed that BECLIN 1 participates in EGFR downregulation, and that BECLIN 1 transient knock-down is sufficient to disturb this regulation.Citation20 In this scenario, also c-Myc phosphorylation would be presumably altered, so constituting a possible common target of AMBRA1 and BECLIN 1. In order to verify this hypothesis, we knocked-down BECLIN 1 (siBECLIN 1) in H1975 lung cancer cells which express a constitutive active form of EGFR. As expected, the Western Blot analysis revealed increased P c-MycS62 in siBECLIN 1 cells, as well as an increased activation of the kinases ERK1/2 (), suggesting that BECLIN1 indirectly regulates c-Myc through the EGFR pathway. This evidence is further supported by the data shown in , demonstrating that BECLIN 1-mediated increase of P c-MycS62 is clearly prevented by U0126 treatment (inhibitor of MEK, the ERK1/2 kinase). On the contrary, AMBRA1-mediated regulation of c-Myc is not rescued by ERK1/2 inhibition (), consistent with the involvement of PP2A rather than ERK1/2 in this pathway of regulation.Citation9
Figure 2. BECLIN 1 and AMBRA1 affect c-Myc phosphorylation in ERK1/2 dependent and independent manner, respectively. (A) H1975 lung cancer cells were treated with oligo-interference against BECLIN 1 (siBECLIN 1) or with aspecific oligonucleotides (siCTRL). Protein extracts were analyzed by SDS-PAGE and Western Blot using antibodies against BECLIN 1, P c-MycS62, c-Myc, P ERK1/2, ERK1/2 and ACTIN. (B) Densitometric analysis of the Western Blot shown in (A), performed using the ImageJ software; the average of the values from different experiments related to the control ratio was arbitrarily defined as 1.00. The band density ratio of phosphorylated proteins (P c-MycS62 and P ERK1/2) relative to total proteins (c-Myc and ERK1/2, respectively), are analyzed, with the control ratio arbitrarily defined as 1.00. n = 3 extracts prepared from independent experiments; data are presented as means ± s.d. and significance is *P ≤ 0.05 and **P ≤ 0.005. (C) Cells were treated as in (A) and, where indicated, were incubated with U0126 (5 μM) for 6 hours. (D) Densitometric analysis of the Western Blots shown in (C) and (E) was performed as described in (B). The band density ratio of P c-MycS62 relative to ACTIN is analyzed, with the control ratio arbitrarily defined as 1.00. n = 3 extracts prepared from independent experiments; data are presented as means ± s.d. and significance is *P ≤ 0.05.
Interestingly, Beth Levine's group recently showed that active EGFR phosphorylates BECLIN 1 and inhibits its pro-autophagic activity by preventing its association with VPS34.Citation24 In the light of the data reported by Thoresen and colleagues,Citation20 the effect of EGFR on BECLIN 1 may also represent a negative feedback loop regulating the endocytosis-mediated degradation of EGFR.
Taken overall, these data suggest that the regulation of c-Myc, taking place down-stream of EGFR, is affected by both BECLIN 1/VPS34 and AMBRA1,Citation9,20 through two different pathways.
Concluding remarks and future challenges
The recent literature strongly supports the physiological and pathological relevance of the crosstalks between autophagy and other pathways.Citation1,3 As an example, autophagy-related proteins also affect cell proliferation.Citation7-9 In this context, our results picture the autophagy proteins AMBRA1 and BECLIN 1 participating in a double-lock mechanism of regulation of c-Myc. Although the roles of AMBRA1 and BECLIN 1 in this context could be uncoupled from their roles in autophagy, they could constitute key molecules in the crosstalk between autophagy and cell proliferation regulation. In line with this, we also found that AMBRA1, BECLIN 1 and PP2A are together in a high-molecular-weight multimeric complex. A fascinating possibility is that several signallings are integrated at the same molecular platform, where different sub-complexes could be interconnected and coordinately regulated. Further studies are necessary to address this hypothetical model.
Materials and Methods
Cell culture and reagents
HEK293 cells were cultured as previously described.Citation9 The human lung carcinoma cell line (H1975) was purchased from the American Type Culture Collection (ATCC) and cultured according to the supplier's instructions. For autophagy induction, cells were washed with PBS and cultured for 30 min in Earle's balanced salt solution (EBSS; Sigma-Aldrich). When indicated, cells were incubated in the presence of 5 μM U0126 (Sigma-Aldrich).
Size-Exclusion Fast-Protein-Liquid-Chromatography (sec-FPLC)
Four milligrams of HEK293 cell lysate were injected onto a Superose 6 HR 10/30 Fast Protein Liquid Chromatography (FPLC) gel filtration column (GE Healthcare). Lysate preparation and column equilibration have been previously described.Citation9 Proteins were collected in 500 μl fractions, precipitated with 10% trichloroacetic acid (TCA), and resolved in SDS-PAGE for Western Blot analyses. Gel filtration column was calibrated with the following molecular mass markers: thyroglobulin (669 kDa), apoferritin (443 kDa), β-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), and carbonic anhydrase (29 kDa) (Sigma-Aldrich MW-GF-1000).
RNA interference
Where indicated, cells were transiently transfected with siRNA oligonucleotides using Lipofectamine 2000 (Invitrogen), as indicated by the supplier. RNA interference was performed using the BECLIN 1, AMBRA1 and control siRNA oligonucleotides that were previously described.Citation9,10
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
We wish to thank M. Acuña Villa and L. Fischer for secretarial work, and M. W. Bennett for editorial work. We also thank A. Battistoni for critical reading of the manuscript.
Funding
This work was supported by grants from KBVU (R72-A4408), Lundbeck Foundation (R167-2013-16100), Novo Nordisk Foundation (7559), The Bjarne Saxhof Foundation, AIRC (IG2012), and in part from FISM (2009), the Telethon Foundation (GGP14202), the Italian Ministry of University and Research (FIRB Accordi di Programma 2011) and the Italian Ministry of Health (RF 2009). VC is supported by the Lundbeck Foundation (R165-2013-15982).
References
- Boya P, Reggiori F, Codogno P. Emerging regulation and functions of autophagy. Nat Cell Biol 2013; 15:713–20; PMID:23817233; http://dx.doi.org/10.1038/ncb2788
- Abada A, Elazar Z. Getting ready for building: signaling and autophagosome biogenesis. EMBO Rep 2014; 15:839–52; PMID:25027988; http://dx.doi.org/10.15252/embr.201439076
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell 2010; 40:280–93; PMID:20965422; http://dx.doi.org/10.1016/j.molcel.2010.09.023
- Marino G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Biol 2014; 15:81–94; PMID:24401948; http://dx.doi.org/10.1038/nrm3735
- Cianfanelli V, Cecconi F. Autophagy-dependent NFkappaB regulation. Cell Cycle 2012; 11:436–7; PMID:22262191; http://dx.doi.org/10.4161/cc.11.3.19224
- Neufeld TP. Autophagy and cell growth–the yin and yang of nutrient responses. J Cell Sci 2012; 125:2359–68; PMID:22649254; http://dx.doi.org/10.1242/jcs.103333
- Fimia GM, Stoykova A, Romagnoli A, Giunta L, Di Bartolomeo S, Nardacci R, Corazzari M, Fuoco C, Ucar A, Schwartz P, et al. Ambra1 regulates autophagy and development of the nervous system. Nature 2007; 447:1121–5; PMID:17589504
- Liang XH, Jackson S, Seaman M, Brown K, Kempkes B, Hibshoosh H, Levine B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999; 402:672–6; PMID:10604474; http://dx.doi.org/10.1038/45257
- Cianfanelli V, Fuoco C, Lorente M, Salazar M, Quondamatteo F, Gherardini PF, De Zio D, Nazio F, Antonioli M, D'Orazio M, et al. AMBRA1 links autophagy to cell proliferation and tumorigenesis by promoting c-Myc dephosphorylation and degradation. Nat Cell Biol 2015; 17:20–30; PMID:25438055; http://dx.doi.org/10.1038/ncb3072
- Di Bartolomeo S, Corazzari M, Nazio F, Oliverio S, Lisi G, Antonioli M, Pagliarini V, Matteoni S, Fuoco C, Giunta L, et al. The dynamic interaction of AMBRA1 with the dynein motor complex regulates mammalian autophagy. The J Cell Biol 2010; 191:155–68; PMID:20921139; http://dx.doi.org/10.1083/jcb.201002100
- Funderburk SF, Wang QJ, Yue Z. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond. Trends Cell Biol 2010; 20:355–62; PMID:20356743; http://dx.doi.org/10.1016/j.tcb.2010.03.002
- He C, Levine B. The Beclin 1 interactome. Curr Opin Cell Biol 2010; 22:140–9; PMID:20097051; http://dx.doi.org/10.1016/j.ceb.2010.01.001
- Nazio F, Strappazzon F, Antonioli M, Bielli P, Cianfanelli V, Bordi M, Gretzmeier C, Dengjel J, Piacentini M, Fimia GM, et al. mTOR inhibits autophagy by controlling ULK1 ubiquitylation, self-association and function through AMBRA1 and TRAF6. Nat Cell Biol 2013; 15:406–16; PMID:23524951; http://dx.doi.org/10.1038/ncb2708
- Strappazzon F, Vietri-Rudan M, Campello S, Nazio F, Florenzano F, Fimia GM, Piacentini M, Levine B, Cecconi F. Mitochondrial BCL−2 inhibits AMBRA1-induced autophagy. EMBO J 2011; 30:1195–208; PMID:21358617; http://dx.doi.org/10.1038/emboj.2011.49
- Strappazzon F NF, Corrado M, Cianfanelli V, Romagnoli A, Fimia GM, Campello S, Piacentini M, Campanella M, Cecconi F. AMBRA1 is able to induce mitophagy via LC3 binding and independently of PARKIN and p62. Cell Death Differ 2014; In press; PMID:25215947
- Cianfanelli V, Nazio F, Cecconi F. Connecting autophagy: AMBRA1 and its network of regulation. Mol Cell Oncol 2015; 2(1).
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 2011; 12:21–35; PMID:21157483; http://dx.doi.org/10.1038/nrm3025
- Bodemann BO, Orvedahl A, Cheng T, Ram RR, Ou YH, Formstecher E, Maiti M, Hazelett CC, Wauson EM, Balakireva M, et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 2011; 144:253–67; PMID:21241894; http://dx.doi.org/10.1016/j.cell.2010.12.018
- Farre JC, Subramani S. Rallying the exocyst as an autophagy scaffold. Cell 2011; 144:172–4; PMID:21241888; http://dx.doi.org/10.1016/j.cell.2011.01.005
- Thoresen SB, Pedersen NM, Liestol K, Stenmark H. A phosphatidylinositol 3-kinase class III sub-complex containing VPS15, VPS34, Beclin 1, UVRAG and BIF-1 regulates cytokinesis and degradative endocytic traffic. Exp Cell Res 2010; 316:3368–78; PMID:20643123; http://dx.doi.org/10.1016/j.yexcr.2010.07.008
- Sears R, Nuckolls F, Haura E, Taya Y, Tamai K, Nevins JR. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev 2000; 14:2501–14; PMID:11018017; http://dx.doi.org/10.1101/gad.836800
- Yeh E, Cunningham M, Arnold H, Chasse D, Monteith T, Ivaldi G, Hahn WC, Stukenberg PT, Shenolikar S, Uchida T, et al. A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nat Cell Biol 2004; 6:308–18; PMID:15048125; http://dx.doi.org/10.1038/ncb1110
- Sears RC. The life cycle of C-myc: from synthesis to degradation. Cell Cycle 2004; 3:1133–7; PMID:15467447; http://dx.doi.org/10.4161/cc.3.9.1145
- Wei Y, Zou Z, Becker N, Anderson M, Sumpter R, Xiao G, Kinch L, Koduru P, Christudass CS, Veltri RW, et al. EGFR-mediated Beclin 1 phosphorylation in autophagy suppression, tumor progression, and tumor chemoresistance. Cell 2013; 154:1269–84; PMID:24034250; http://dx.doi.org/10.1016/j.cell.2013.08.015
