Article title: Hepatocellular alterations and dysregulation of oncogenic pathways in the liver of transgenic mice overexpressing growth hormone
Authors: Johanna G Miquet, Thomas Freund, Carolina S Martinez, Lorena González, María E Díaz, Giannina P Micucci, Elsa Zotta, Ravneet K Boparai, Andrzej Bartke, Daniel Turyn, and Ana I Sotelo
Journal: Cell Cycle
Bibliometrics: Volume 12, Issue 7, Pages 1042–1057
DOI: 10.4161/cc.24026
The appeared incorrectly in print and online. The correct is provided below:
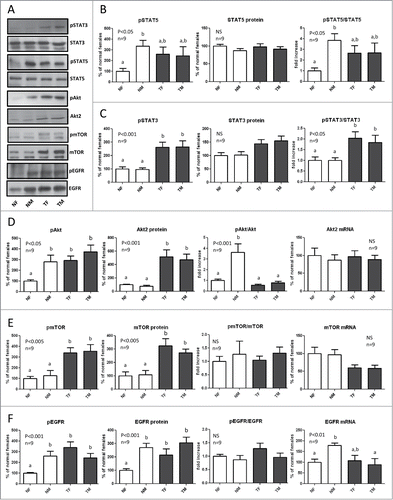
Figure 3. Expression and phosphorylation of STAT5, STAT3, Akt, mTOR and EGFR in the liver of GH-overexpressing transgenic mice and normal controls. Liver extracts from young adult normal female (NF), normal male (NM), GH-transgenic female (TF) and GH-transgenic male (TM) mice were analyzed by immunoblotting to determine the phosphorylation and protein content, or by reverse-transcriptase quantitative PCR (qRT-PCR) to assess mRNA levels. (A) Representative results of immunoblots are shown. (B) sTAT5 phosphorylation at Tyr694/696 (pSTAT5), protein content and phos-phorylation/protein content ratio. STAT5 was also used as a control for equal loading. (C) STAT3 phosphorylation at Tyr705 (pSTAT3), protein content and phosphorylation/protein content ratio. (D) Akt phosphorylation at ser473 (pAkt), Akt2 protein content, phosphorylation/protein content ratio and mRNA levels. (E) mTOR phosphorylation at ser2448 (pmTOR), protein content, phosphorylation/protein content ratio and mRNA levels. (F) EGFR phosphorylation at Tyr845 (pEGFR), protein content, phosphorylation/protein content ratio and mRNA levels. Results are expressed as % of the mean values in normal female mice. Data are the mean ±SEM of 9 sets of different individuals per group (n). Different letters denote significant difference at p < 0.05. NS, not significant.
The authors apologize for any inconvenience caused.
