Abstract
The skin of patients with an extensive deep burn injury is repaired by a process that leaves a hypertrophic scar without sweat glands and therefore loses the function of perspiration. The aim of this study was to identify whether the key factors related to sweat gland development could directly reprogram fibroblasts into sweat gland-like cells. After introducing the NF-κB and Lef-1 genes into fibroblasts, we found that stably transfected fibroblasts expressed specific markers of sweat glands, including CEA, CK7, CK14 and CK19, both at the protein and mRNA levels. The immunofluorescence staining also showed positive expression of CEA, CK7, CK14 and CK19 in induced fibroblasts, but there were no positive cells in the control groups. The expression of Shh and Cyclin D1, downstream genes of NF-κB and Lef-1, were also significantly increased during regeneration. The induced fibroblasts were implanted into an animal model. Twenty days later, iodine-starch perspiration tests showed that 7 out of the 10 cell-treated paws were positive for perspiration, with a distinctive black point-like area appearing in the center of the paw. Contralateral paws tested negative. Histological examination of skin biopsies from experimental and control paws revealed that sweat glands were fully reconstructed in the test paws, with integral, secretory and ductal portions, but were not present in the control paws. This is the first report of successful reprogramming of fibroblasts into sweat gland-like cells, which will provide a new cell source for sweat gland regeneration in patients with extensive deep burns.
Abbreviations
| CEA | = | carcinoembryonic antigen |
| CK7 | = | cytokeratin 7 |
| CK14 | = | cytokeratin 14 |
| CK19 | = | cytokeratin 19 |
| DMEM | = | Dulbecco's modified Eagle's medium |
| FBS | = | fetal bovine serum |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| Lef-1 | = | lymphoid enhancer-binding factor 1 |
| min | = | minutes |
| NF-κB | = | nuclear factor kappa-light-chain-enhancer of activated B cells |
| Shh | = | sonic hedgehog |
Introduction
The excretory function of skin is devoted to the regulation of body temperature. Under basal conditions, approximately 25% of heat is eliminated by vaporization of sweat excreted from sweat glands.Citation1 Therefore, sweat glands play a significant role in the homeostatic maintenance of body temperature. However, following severe full-thickness burn injury, sweat glands do not regenerate to form their 3D organization because of complete destruction of the duct and secretory cell coiler.Citation2 Although survival rates from even the most severe burns have reached 90% in some countries, most survivors face the problem of the long-term loss of secretory function and endure great pain. Because of the lack of sweat glands in regenerated scar tissue, body temperature regulation is impaired, which greatly affects the patients' quality of life. Therefore, enhancing the regeneration of sweat glands is an important goal in burn treatment.Citation3
Recent rapid advances in regenerative medicine have led us to attempt to reprogram somatic cells into sweat gland cells to regenerate sweat glands.Citation4 Compared to more established methods of cellular derivation, such as embryonic or induced pluripotent stem cell (iPS) directed differentiation, direct reprogramming presents several advantages: lack of tumorigenic risk, fast conversion rate and repair of injured tissues by in vivo reprogramming.Citation5 Compared with stem cells, fibroblasts are the best source for direct reprogramming. Fibroblasts are easily isolated and cultured, they do not instigate a host-versus-graft reaction, and most importantly, they can migrate to the injured sites of skin.Citation6 Thus, fibroblasts might be successfully reprogrammed and induced to acquire a sweat-gland-cell phenotype.
Direct reprogramming of fibroblasts by defined factors has already been reported by many other researchers. In 2010, Vierbuchen successfully reprogrammed fibroblasts into neuronal cells using a cocktail of Ascl1, Brn2 and Myt11.Citation7 In 2012, Song's group reported that mouse cardiac fibroblasts could be reprogrammed into beating cardiac-like myocytes using a defined factor set consisting of GMT and the Hand 2 gene.Citation8 EDA/EDAR/NF-κB and Wnt/β-catenin signaling pathways play an important role in the development of skin appendages (including sweat glands and hair follicles).Citation9 The ectodysplasin (EDA) gene is one of the functional genes that regulate the development of sweat glands. In EDA knockout mice, sweat glands do not regenerate after injury.Citation10,11 NF-κB is a downstream factor of EDA and EDAR signaling and promotes the expression of NF-κB target genes such as Shh, cyclin D1, Dkk4, Fox family genes and keratins.Citation12 These genes are required at different stages in sweat gland development. Lymphoid enhancer-binding factor 1 (Lef-1) is a downstream transcription factor of β-catenin signaling.Citation13 The protein encoded by this gene can bind to a functionally important site in the T-cell receptor-α enhancer, thereby conferring maximal enhancer activity, which may function in hair cell differentiation and follicle morphogenesis.Citation14 In addition, Lef-1 also can enhance activation of the EDA promoter and promote the expression of the EDA gene during ectoderm development.
In this study, we sought to determine whether key developmental sweat gland factors could directly reprogram fibroblasts into sweat gland-like cells in vitro, with the notion that the in vivo environment may ultimately permit further reprogramming. We first constructed a eukaryotic NF-κB and Lef-1 expression vector and transfected it into fibroblasts. Then, we observed the influence of the defined combination of NF-κB and Lef-1 on reprogrammed cells. This study will provide us new insights into the regeneration of sweat gland-like cells by showing a new ideal cell source for cell therapy.
Results
Plasmid construction
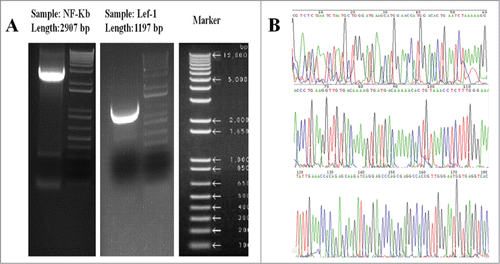
The pcDNA3.1 (+) plasmid was used to construct the transgene vector pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1. The final pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 plasmids were extracted via an alkaline lysis method. The plasmids were identified separately by HindIII/SalI and HindIII/PstI digestion and gel electrophoresis. The products of this digestion are shown in . The enzymes HindIII and SalI specifically recognized and digested the desired sites on pcDNA3.1 (+)-NF-κB. The enzymes HindIII and PstI specifically recognized and digested the desired sites on pcDNA3.1 (+)-Lef-1. One fragment of approximately 3,000 bp was observed after digestion, which is the NF-κB gene. Another fragment of approximately 1,200 bp was observed after digestion for the Lef-1 gene. Meanwhile, the pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 recombinant plasmids were separately isolated from bacteria and analyzed by Invitrogen Biotechnology Co. Ltd (Branch, Shanghai, China). The sequences were identical to the GenBank reference sequences (Serial Number: NM_003998, NM_016269). These results confirmed that the NF-κB and Lef-1 gene fragments were successfully inserted into the pcDNA3.1 (+) plasmid. The recombinant plasmids were then used in the following experiments ().
Figure 1. Plasmid construction. (A) Enzyme digestion analysis of NF-κB and Lef-1 transgene vectors. Markers and (Sample) fragments of pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 plasmid separately digested by HindIII and PstI/SalI. (B) Gene sequencing analysis. This image depicts the gene sequences of the Homo sapiens NF-κB and Lef-1 from GenBank (Serial Number: NM_003998 and NM_016269).
Cell transfection and expression of NF-κB and Lef-1
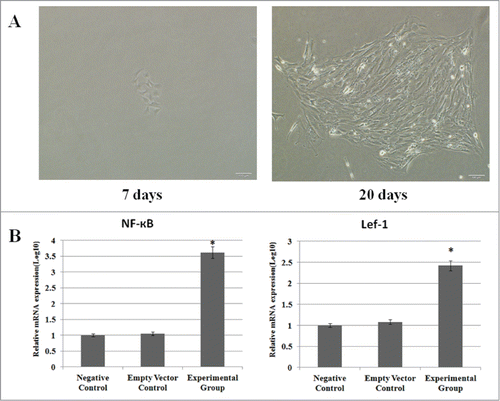
To study the biological functions of NF-κB and Lef-1, the recombinant plasmids were transfected into fibroblasts. Forty-eight hours after transfection, the cells were selected by G418 for 20 days, and G418-resistant clones were propagated and screened for NF-κB and Lef-1 expression. Stably transfected cells were analyzed individually to determine the expression level of NF-κB and Lef-1 using real-time PCR. This analysis confirmed that the fibroblast cells transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 expressed higher levels of NF-κB and Lef-1 at the mRNA levels when compared with the negative control and empty vector control groups ().
Figure 2. Cell transfection and expression of NF-κB and Lef-1. (A) G418-resistant fibroblast cells clone selection. Small colonies were formed in the presence of G418 on the 7th day of selection (left), whereas significantly larger colonies appeared after 20 days of selection (right). (B) The expression of NF-κB and Lef-1 in the three groups. Significant differences in NF-κB and Lef-1 expression between cells transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1, empty vector or negative control are shown (*P < 0.05 negative control vs. the experimental group and empty vector control). All data are normalized to GAPDH and calibrated based on the negative control, whose expression was considered 1 for all genes. The Y-axis is on a logarithmic scale.
Fibroblasts expressed specific markers of sweat glands after reprogramming
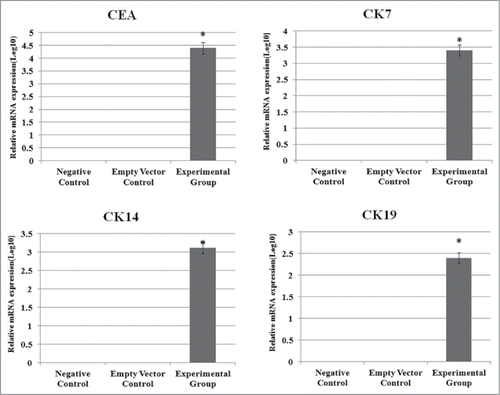
Stably transduced cells were cultured and analyzed individually to determine the expression levels of CEA, CK7, CK14 and CK19 using Western blots and realtime-PCR. Our Western blots and Image J gray scan analysis demonstrated that stably transfected fibroblast cells expressed the specific marker proteins of sweat glands (CEA, CK7, CK14 and CK19) (). The control groups did not express these proteins. Real-time-PCR showed that the stably transfected group had significant expression of sweat gland markers (CEA, CK7, CK14 and CK19), while control cells did not. This analysis confirmed that the Fibroblast cells transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 expressed CEA, CK7, CK14 and CK19 at the mRNA and protein levels ().
Figure 3. The expression of CEA, CK7, CK14 and CK19 proteins is modulated by NF-κB and Lef-1 in fibroblast cells. Equal amounts of cell lysates (containing 50 mg protein) were probed by specific antibodies as described in Materials and Methods Section. β-actin was used as a loading control. The pictures show CEA, CK7, CK14 and CK19 protein. The figure is representative of three separate experiments. The expression profiles of the specific markers and β-actin proteins in the Experimental group (Lane 1), empty vector control group (Lane 2) and negative control group (Lane 3) are shown. A significant difference between transfection with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 and transfection with vector alone or control is shown.
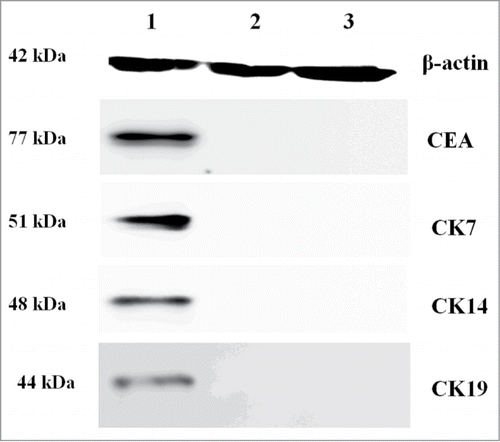
Figure 4. The influence of NF-κB and Lef-1 on CEA, CK7, CK14 and CK19 gene expression. Significant differences in CEA, CK7, CK14 and CK19 expression between cells transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1, empty vector or negative control are shown (*P < 0.05 negative control vs. the experimental group and empty vector control). All data are normalized to GAPDH and calibrated based on the negative control, whose expression was considered 1 for all genes. The Y-axis is on a logarithmic scale.
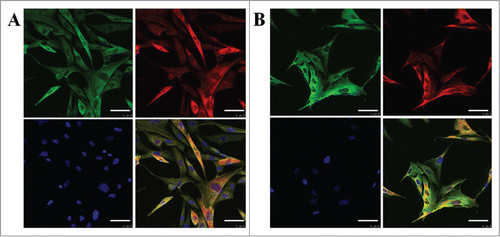
Triple-immunofluorescence staining was conducted to identify CEA, CK7, CK14 and CK19 in fibroblasts to elucidate the direct contribution of the NF-κB and Lef-1 genes to conversion, measured by specific markers of sweat glands. CEA, CK7, CK14 and CK19 staining could be observed in fibroblast cells with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1, compared with the control cells. In fact, CEA, CK7, CK14 and CK19 staining was barely detectable in the control fibroblasts. No significant differences in the DAPI signal were observed between the 3 groups. From this immunofluorescence staining, we conclude that in the stably transfected group, specific markers of sweat glands (CEA, CK7, CK14 and CK19) are more highly expressed than that in the control groups ().
Figure 5. The expression of CK19, CK14, CK7 and CEA in fibroblasts after transfection. Immunofluorescent images of fibroblast cells transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1. Single color and merged images for CEA (green), (red) and nucleus (blue) staining are shown in A. Single color and merged images for CK14 (red), CK19 (green) and nucleus (blue) staining are shown in B. The scale bar represents 50 μm (top row). Representative photomicrographs are shown for three independent experiments. Red and green could not be seen in the control group.
Expression of Shh and Cyclin D1, downstream genes of NF-κB and Lef-1
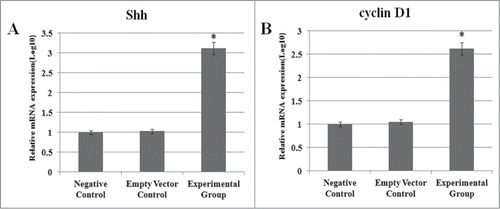
Real-time-PCR showed that the stably transfected group had significantly higher expression of genes downstream of NF-κB and Lef-1 signaling (Shh and cyclin D1) compared to controls ().
Figure 6. Expression of Shh and Cyclin D1. The influence of NF-κB and Lef-1 on downstream gene expression. Significant differences in Shh and cyclin D1 expression between cells transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1, empty vector or negative control are shown (*P < 0.05 negative control vs. the experimental group and empty vector control). All data are normalized to GAPDH and calibrated based on the negative control, whose expression was considered 1 for all genes. The Y-axis is on a logarithmic scale.
Transplanting reprogrammed fibroblasts in animal models
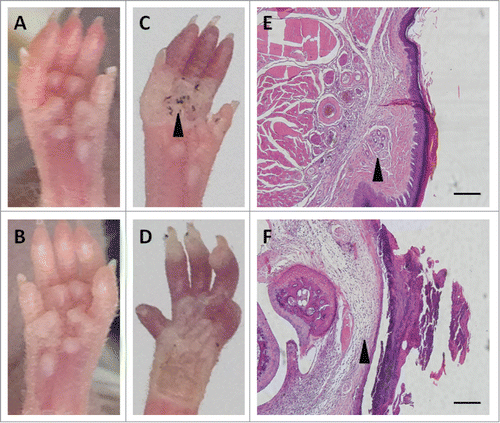
Fibroblasts transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 were implanted in an animal model at a concentration of 106 cells/ml. Twenty days later, an iodine-starch perspiration test was performed on both paws of the mice. Seven out of the 10 cell-treated paws were positive for the perspiration test, with a distinctive blue and black area appearing in the center of the paw. The contralateral paws did not test positive. Histological examination of skin biopsies from experimental and control paws revealed the damaged sweat glands were fully reconstructed in the positive paws, with integral secretory and ductal portions, but they were poorly represented in the control paws. The basal layer connecting the epidermis and sweat gland duct appeared to be intact and continuous in the treated paws, but it was distorted in the control paws. The results demonstrate that transplanted fibroblasts transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 aided in the regeneration of sweat glands ().
Figure 7. Animal model. Contribution of reprogrammed fibroblasts to sweat gland regeneration in nude mice (A, B). Iodine-starch perspiration test was positive in a representative mouse paw (black arrow; C) implanted with reprogrammed fibroblasts but negative in the control scalded paw (D); damaged sweat glands were fully reconstructed with complete secretory and ductal portions in the treated paw (black arrow; E) but poorly reconstructed in the control paw (black arrow; F); Scale bar is 50 μm.
Discussion
We demonstrated that NF-κB and Lef-1 reprogrammed fibroblasts into sweat gland-like cells. We constructed eukaryotic NF-κB and Lef-1 expression vectors and stably transfected them into fibroblasts. After 20 days, some transfected fibroblasts were reprogrammed into sweat gland-like cells expressing CEA, CK7, CK14 and CK19 mRNA and protein. We also found the enhanced expression of Shh and Cyclin D1, downstream genes of NF-κB and Lef-1, which might be involved in the process of reprogramming. Then, we transplanted these sweat gland-like cells into scalded paws of nude mice and found that the paws were positive for a perspiration test and that the damaged sweat glands were fully reconstructed in the positive paws. These findings suggest that fibroblasts reprogrammed with NF-κB and Lef-1 possess the phenotypic and functional characteristics of sweat gland cells.
Cellular differentiation is usually considered an irreversible process during development due to robust lineage commitment. Lineage-specific transcription factors produced during development may strengthen cell type-specific gene expression patterns.Citation15 This view has been reconsidered, as the ability to change the pluripotency of a differentiated cell or to change a cell into an entirely different cell type has been demonstrated. It is possible to induce fully differentiated cells to transform into other cell types by reprogramming.Citation16 Somatic cells can be changed into pluripotent cells by methods such as cell fusion, culture-induced reprogramming and direct reprogramming.Citation17 Specifically, direct reprogramming is a complex process that involves many methods and specific factors.Citation18 Although the application of iPS cell technology has a promising future, obstacles exist. For example, the fidelity and safety of iPS/ESC-derived cells requires assessment before these cells can be used clinically.Citation19,20 Considering the disadvantages and limitations of iPS cell technology, other direct reprogramming methods, which avoid the pluripotent stage, may be preferable.Citation21 Direct reprogramming may replace the iPS cell method. Several studies have revealed that fibroblasts may be a source for cell-based therapy. It is possible to directly reprogram fibroblasts into other cell types using a cocktail of defined factors and microRNAs.Citation22-26 In 2012, Song's group reported that mouse cardiac fibroblasts could be reprogrammed into beating cardiac-like myocytes using a defined factor set consisting of GMT and the Hand 2 gene.Citation8 Torper's study showed that human fibroblasts and astrocytes could be transplanted and converted into neurons when specific genes were activated.Citation27 In addition, Torper also found that mouse astrocytes could be directly reprogrammed into neurons with nuclear expression in vivo.
During sweat gland regeneration, NF-κB and Lef-1 regulate cell proliferation and differentiation through the signaling pathways of EDA and Wnt. NF-κB and Lef-1 play a significant physiological role in the regulation of sweat gland development.Citation28 NF-κB is activated downstream of EDA and EDAR signaling and promotes the expression of genes such as Shh, cyclin D1, Dkk4, Fox family genes and keratins. These genes are required at different stages in sweat gland development. Lymphoid enhancer-binding factor 1 (Lef-1) is a downstream transcription factor of β-catenin signaling. The protein encoded by this gene can bind to a functionally important site in the T-cell receptor-α enhancer, thereby conferring maximal enhancer activity, which may function in hair cell differentiation and follicle morphogenesis.Citation29 In addition, Lef-1 also can enhance the activation of the EDA promoter and promote the expression of the EDA gene during ectoderm development. Activated NF-κB enters the nucleus and promotes the expression of genes such as Shh, cyclin D1, Dkk4, Fox family genes and keratins. These genes are required at different stages in sweat gland development.Citation30,31 The downstream gene Cyclin D1 is a β-catenin target gene whose expression is upregulated during the process of aged epidermal cell dedifferentiation.Citation32 Therefore, we sought to identify whether key developmental sweat gland factors could directly reprogram fibroblasts into sweat gland-like cells in vitro, with the assumption that the in vivo environment may ultimately permit further reprogramming.
In our study of the development of sweat glands and their specific cell markers, it was possible to regain functional sweat glands in healed wounds in survivors of deep burns. The results of the iodine-starch perspiration test clearly demonstrated that the transplanted fibroblasts transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 permitted the regeneration of sweat glands. Although our preliminary result is promising, there are still several questions that need to be addressed. As a normal sweat gland structure was not seen in our biopsy specimens, only a sweat gland-like structure, and we observed the expression of specific cell markers in reprogrammed fibroblasts, it seems logical to suggest that a different eccrine mechanism might exist, which deserves further study.
Experimental results also show that NF-κB and Lef-1 can promote the differentiation of fibroblasts into sweat gland cells, possibly because NF-κB and Lef-1 activate EDA and the Wnt signaling pathway. The interactions between proteins of these 2 pathways may be responsible for the promotion of differentiation. Activated NF-κB and Lef-1 could enter the nucleus and promote the expression of genes such as Shh and cyclin D1, which are required at different stages in sweat gland development. Realtime-PCR showed that the stably transfected fibroblasts significantly upregulated downstream targets of NF-κB and Lef-1 signaling (Shh and cyclin D1) compared with controls. Our preliminary results are consistent with the relevant literature.Citation9,33
However, we did not thoroughly explore many other aspects of the EDA/NF-κh and Wnt/Lef-1 pathways, such as ligand binding sites, receptor activation, desensitization and transportation. We will continue to examine how these proteins affect downstream signaling pathways activated by NF-κF and Lef-1 in our future research. Our work may provide a promising method for sweat gland regeneration and skin tissue engineering.
Materials and Methods
Materials
The following reagents were purchased from Invitrogen Corp. (Invitrogen, Carlsbad, CA): pcDNA3.1 (+), PureLink™ HiPure Plasmid Miniprep Kit, Lipofectamine™ 2000 Transfection Reagent, Opti-MEM® Reduced Serum Medium and TRIzol® Reagent. CEA (ab33562), CK7(ab82253), CK14(ab51054) and CK19(ab53119) antibodies were purchased from Abcam Corp. The Reverse Transcription System was purchased from Promega Corp. (Promega, Madison, WI, USA).
Mice
BALB/c nude mice (No. 01030101) were supplied by the Institute of Experimental Animals, Chinese Academy of Science (CAS). Mice were housed in standard animal cages under controlled temperature and humidity with 12-hour light/dark cycles. Animal handling and experimentation was approved by the Animal Care and Use Review Committee of IOZ, CAS, and the Institute of Biological Products of Beijing.
Cell culture
Fibroblast cells were obtained from ATCC (America Type Culture Collection). Cells were cultured in DMEM (Dulbecco's modified Eagle's medium; Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (GIBCO, Grand Island, NY, USA), 100 U/ml penicillin and 0.1 mg/ml streptomycin. Cells were subcultured every 72 h in a humidified 5% CO2 incubator. Cells from passages 4–6 were used in the experiments.
Vector construction, Cell transfection and G418 selection
We retrieved the NF-κB and Lef-1 gene sequences from GenBank and designed the following primers: NF-κB-F : 5′-AAC AGA GAG GAT TTC GTT TCC G- 3′, NF-κB-R : 5′-TTT GAC CTG AGG GTA AGA CTT CT- 3′, Lef-1-F : 5′-TGC CAA ATA TGA ATA ACG ACC CA- 3′ and Lef-1-R : 5′ -GAG AAA AGT GCT CGT CAC TGT- 3′. For NF-κB, the eukaryotic expression plasmid pcDNA3.1 (+) was digested with the restriction endonucleases HindIII and SalI. For Lef-1, pcDNA3.1(+) was digested with HindIII and PstI. The NF-κB insert and linearized plasmid were joined by T4-DNA ligase to construct the plasmid pcDNA3.1(+)-NF-κB. The Lef-1 insert and linearized plasmid were joined by T4-DNA ligase to construct the plasmid pcDNA3.1(+)-Lef-1. The recombinant plasmids pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 were characterized by PCR, restriction endonuclease digestion and sequencing analysis.
The fibroblast cells were divided into negative control, empty vector control and overexpression groups. Untransfected cells were used as the negative control. Transfection was carried out using Lipofectamine™ 2000 according to the manufacturer's instructions.
To obtain stably transfected cells, at 24 h post-transfection, G418 was added to a final concentration of 0.8 mg/ml, and antibiotic selection was continued for 10 d Cells stably expressing NF-κB and Lef-1 were continuously passaged in DMEM medium supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (10 g/ml) and G418 (0.2 mg/ml). G418-resistant single cell clones were screened and amplified for the following experiments. Control cells were mock-transfected with empty vector and selected in the same manner described above. The expression of CEA, CK7, CK14 and CK19 in fibroblast cells was observed on a confocal laser scanning microscope and measured using Western blots and real-time PCR.
Western blotting
Western blots were performed to detect the expression of CEA, CK7, CK14 and CK19 at the protein level. After transfection and G418 selection, cells were washed 3 times with ice-cold PBS, incubated with lysis buffer (20 mM HEPES, pH 7.5, 200 mM NaCl, 0.2 mM EDTA–Na2, 1% Triton X-100, 0.05% SDS, 0.5 mM DTT, 1 mM Na3VO4, 20 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 10 μg/ml leupeptin) at 4 °C for 30 min and centrifuged at 12000 g for 5 min; the supernatant was stored at −80 °C. The amount of total protein was determined using a bicinchoninic acid (BCA) assay. Equal amounts (20 μg) of total protein from each sample were separated on 12% Bis–Tris gels following the manufacturer's protocol and transferred to PVDF membranes. Membranes were blocked overnight in TBST/5% fat free milk. After incubating with the appropriate antibodies, immunoreactive bands were visualized with enhanced chemiluminescence (ECL) detection reagents on autoradiography films, as described previously.Citation34 The intensity of immunoreactive bands was quantified by image analysis software (ImageJ_1.32J, NIH).
RNA extraction and Realtime-PCR
Cells were washed 2–4 times with PBS prior to RNA isolation. Total RNA was isolated with the Trizol reagent and used for first strand cDNA synthesis with a reverse transcription system. Quantification of gene transcripts was performed in a 7500 Real-Time PCR System using Power SYBR® Green. The primer sequences of the genes, including NF-κB, Lef-1, CEA, CK7, CK14, CK19, Shh and Cyclin D1 are listed in . PCR conditions were one cycle of 94 °C for 2 min followed by 50 cycles of 94 °C for 10 s, a specified annealing temperature for 15 s and 72 °C for 15 s. Amplification was followed by melting curve analysis, which used the following program: one cycle at 65 °C for 1 s, 94 °C for 2 s and 37 °C for 5 s. The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an endogenous reference gene to which the expression of other genes was normalized using the comparative cycle of threshold value.
Table 1. Primers for real time PCR
Immunofluorescence
After transfection and G418 selection, fibroblasts were seeded at 1 × 106 cells/well in 6-well plates (Nunc). The cells were cultured to 70–90% confluence, and then the media in the 6-well plate were removed. The cells were washed 3 times with PBS, fixed with 4% paraformaldehyde for 10 min at 37 °C, and then washed with PBS (pH 7.4) 3 times. Fibroblasts were separately labeled with CEA, CK7, CK14 and CK19 primary antibodies and incubated overnight at 4°C. After several washes with PBS (pH 7.4), cells were incubated with the appropriate Cy3- and FITC-conjugated secondary antibodies for 1 h in the dark at room temperature. To counterstain the nuclei, 4′6-diamidino-2-phenylindole dihydrochloride (DAPI; 0.5 μg/ml in PBS; Molecular Probes) was applied after the secondary antibody. Images were acquired on a fluorescence microscope (Leica) equipped with a charged-coupled-device camera and processed with Axiovision software (Leica MicroImaging).
Transplantation experiments in animals
Fibroblasts transfected with pcDNA3.1(+)-NF-κB and pcDNA3.1(+)-Lef-1 were implanted in an animal model. Full-thickness scald injuries were produced on both paws of the hind legs of 12 athymic BALB/c nude mice. Then, one scalded paw of each mouse received a subcutaneous injection of 1×106 reprogrammed fibroblasts suspended in 150 μl of medium. The contralateral (control) scalded paw received a subcutaneous injection of normal fibroblasts. Twenty days later, an iodine-starch perspiration test was performed on both paws of the mice. Skin biopsies from experimental and control paws were examined by histology.
Statistical analysis
The data were analyzed with SPSS software, Version 12.0 (SPSS Inc., Chicago, IL). All values are expressed as the mean ± standard deviation. The data were analyzed using one-way analysis of variance (ANOVA) and Newman–Keuls–Student's t test. A tied-P value of < 0.05 was considered statistically significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Funding
This study was supported in part by the National Nature Science Foundation of China 81171798, 2014ZD004, 81421064, 81230041), Beijing Municipal Natural Science Foundation (Grant No. 7142124) and the National Basic Science and Development Program (973 Program, 2012CB518105).
References
- Xu Y, Huang S, Ma K, Fu X, Han W, Sheng Z. Promising new potential for mesenchymal stem cells derived from human umbilical cord wharton's jelly: sweat gland cell-like differentiative capacity. J Tissue Eng Regen Med 2012; 6: 645-54; PMID:21916019; http://dx.doi.org/10.1002/term.468
- Li H, Chen L, Zhang M, Tang S, Fu X. Three-dimensional culture and identification of human eccrine sweat glands in matrigel basement membrane matrix. Cell Tissue Res 2013; 354: 897-902; PMID:23996202; http://dx.doi.org/10.1007/s00441-013-1718-3
- Lu CP, Polak L, Rocha AS, Amalia Pasolli H, Chen S, Sharma N. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 2012; 1:136-50; http://dx.doi.org/10.1016/j.cell.2012.04.045
- Plikus MV, Gay DL, Treffeisen E, Wang A, Supapannachart RJ, Cotsarelis G. Epithelial stem cells and implications for wound repair. Cell Dev 2012; 23:946-53
- Salem H, Ismail M, Seify H. Adult Gland Derived Stem Cells (Gdscs); Potentials, Hurdles and Expectations. J Stem Cell Res Ther 2014; 4:2
- Simeonov KP, Uppal H. Direct reprogramming of human fibroblasts to hepatocyte-like cells by synthetic modified mRNAs. PloS One 2014; 9:e100134; PMID:24963715; http://dx.doi.org/10.1371/journal.pone.0100134
- Vierbuchen T, Ostermeier A, Pang ZP, Kokubu Y, Südhof TC, Wernig M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010; 463: 1035-41; PMID:20107439; http://dx.doi.org/10.1038/nature08797
- Song K, Nam YJ, Luo X, Qi X, Tan W, Huang GN, Acharya A, Smith CL, Tallquist MD, Neilson EG et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature 2012; 485: 599-604; PMID:22660318; http://dx.doi.org/10.1038/nature11139
- Schmidt-Ullrich R, Tobin DJ, Lenhard D, Schneider P, Paus R, Scheidereit C. NF-kappaB transmits Eda A1/EdaR signalling to activate Shh and cyclin D1 expression, and controls post-initiation hair placode down growth. Development 2006; 133:1045-57; PMID:16481354; http://dx.doi.org/10.1242/dev.02278
- Zhang YH, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier., Paus R, Piccolo S, Mikkola ML et al. Reciprocal requirements for EDA/EDAR/NF-κB and Wnt/β-catenin signaling pathways in hair follicle induction. Dev Cell 2009; 1: 49-61; http://dx.doi.org/10.1016/j.devcel.2009.05.011
- Klika V, Baker RE, Headon D, Gaffney EA. The Influence of Receptor-Mediated Interactions on Reaction-Diffusion Mechanisms of Cellular Self-organisation. Bull Math Biol 2012;74:935-57; PMID:22072186; http://dx.doi.org/10.1007/s11538-011-9699-4
- Guttridge DC, Albanese C, Reuther JY, Pestell RG, Baldwin AS. NF-κB Controls Cell Growth and Differentiation through Transcriptional Regulation of Cyclin D1. Mol Cell Biol 1999; 19:5785-99; PMID:10409765
- Behrens J. Functional interaction of β-catenin with the transcription factor LEF-1. Nature 1996; 382:638-42; PMID:8757136; http://dx.doi.org/10.1038/382638a0
- Fu Y, Zhu HY, Wu W, Xu JD, Chen TM, Xu B, Qian SX, Li JY, Liu P. Clinical significance of lymphoid enhancer-binding factor 1 expression in acute myeloid leukemia. Leuk Lymphoma 2014; 55:371-7; PMID:23713453; http://dx.doi.org/10.3109/10428194.2013.805759
- Karumbayaram S, Novitch BG, Patterson M, Umbach JA, Richter L, Lindgren A, Conway AE, Clark AT, Goldman SA, Plath K et al. Directed Differentiation of Human- Induced Pluripotent Stem Cells Generates Active Motor Neurons. Stem Cells 2009; 27:806-11; PMID:19350680; http://dx.doi.org/10.1002/stem.31
- Han DW, Tapia N, Hermann A, Hemmer K, Höing S, Araúzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S et al. Direct reprogramming of fibroblasts into neural stem cells by defined factors. Cell Stem Cell 2012; 10: 465-72; PMID:22445517; http://dx.doi.org/10.1016/j.stem.2012.02.021
- Their M, Wörsdörfer P, Lakes YB, Gorris R, Herms S, Opitz T, Seiferling D, Quandel T, Hoffmann P, Nöthen MM, Brüstle O, Edenhofer F. Direct conversion of fibroblasts into stably expandable neural stem cells. Cell Stem Cell 2012; 10: 473-9; PMID:22445518; http://dx.doi.org/10.1016/j.stem.2012.03.003
- Ring KL, Tong LM, Balestra ME, Javier R, Andrews-Zwilling Y, Li G, Walker D, Zhang WR, Kreitzer AC, Huang Y. Direct reprogramming of mouse and human fibroblasts into multipotent neural stem cells with a single factor. Cell Stem Cell 2012; 11:100-9; PMID:22683203; http://dx.doi.org/10.1016/j.stem.2012.05.018
- Smith AW, Hoyne JD, Nguyen PK, McCreedy DA, Aly H, Efimov IR, Rentschler S, Elbert DL. Direct reprogramming of mouse fibroblasts to cardiomyocyte-like cells using Yamanaka factors on engineered poly (ethylene glycol)(PEG) hydrogels. Biomaterials 2013; 34:6559-71; PMID:23773820; http://dx.doi.org/10.1016/j.biomaterials.2013.05.050
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 2005; 19: 489-501; PMID:15713842; http://dx.doi.org/10.1101/gad.1248505
- Li M, Suzuki K, Qu J, Saini P, Dubova I, Yi F, Lee J, Sancho-Martinez I, Liu GH, Izpisua Belmonte JC. Efficient correction of hemoglobinopathy-causing mutations by homologous recombination in integration-free patient iPSCs. Cell Res 2011; 21:1740; PMID:22105484; http://dx.doi.org/10.1038/cr.2011.186
- Feng B, Ng JH, Heng JC, Ng HH. Molecules that promote or enhance reprogramming of somatic cells to induced pluripotent stem cells. Cell Stem Cell 2009; 4: 301-12; PMID:19341620; http://dx.doi.org/10.1016/j.stem.2009.03.005
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell 2006;126: 755-66; PMID:16923394; http://dx.doi.org/10.1016/j.cell.2006.06.052
- Sekiya S, Suzuki A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011; 475: 390-3; PMID:21716291; http://dx.doi.org/10.1038/nature10263
- Nam YJ, Song K, Luo X, Daniel E, Lambeth K, West K, Hill JA, DiMaio JM, Baker LA, Bassel-Duby R et al. Reprogramming of human fibroblasts toward a cardiac fate. Proc Natl Acad Sci 2013; 110:5588-93; http://dx.doi.org/10.1073/pnas.1301019110
- Qian L, Huang Y, Spencer CI, Foley A, Vedantham V, Liu L, Conway SJ, Fu JD, Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature 2012; 485:593-8; PMID:22522929; http://dx.doi.org/10.1038/nature11044
- Torper O, Pfisterer U, Wolf DA, Pereira M, Lau S, Jakobsson J, Björklund A, Grealish S, Parmar M. Generation of induced neurons via direct conversion in vivo. Proc Natl Acad Sci 2013; 110:7038-43; http://dx.doi.org/10.1073/pnas.1303829110
- Sato Y, Rifkin DB. Inhibition of endothelial cell movement by pericytes and smooth muscle cells: activation of a latent transforming growth factor-β 1-like molecule by plasmin during co-culture. J Cell Biol 1989; 109: 309-15; http://dx.doi.org/10.1083/jcb.109.1.309
- Planutiene M, Planutis K, Holcombe RF. Lymphoid enhancer-binding factor 1, a representative of vertebrate-specific Lef1/Tcf1 sub-family, is a Wnt-β-catenin pathway target gene in human endothelial cells which regulates matrix metalloproteinase-2 expression and promotes endothelial cell invasion. Vasc Cell 2011; 3: 28; PMID:22168911; http://dx.doi.org/10.1186/2045-824X-3-28
- Cho SW, Kwak S, Woolley TE, Lee MJ, Kim EJ, Baker RE, Kim HJ, Shin JS, Tickle C, Maini PK et al. Interactions between Shh, Sostdc1 and Wnt signaling and a new feedback loop for spatial patterning of the teeth. Development 2011; 138:1807-16; PMID:21447550; http://dx.doi.org/10.1242/dev.056051
- Zhang RR, Oyajobi BO, Harris SE, Chen D, Tsao C, Deng HW, Zhao M. Wnt/β-catenin signaling activates bone morphogenetic protein 2 expression in osteoblasts. Bone 2013; 52: 145-56; PMID:23032104; http://dx.doi.org/10.1016/j.bone.2012.09.029
- Huelsken J, Vogel R, Erdmann B, Cotsarelis G, Birchmeier W. β-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell 2001; 105:533-45; PMID:11371349; http://dx.doi.org/10.1016/S0092-8674(01)00336-1
- Bera A, Ghosh-Choudhury N, Dey N, Das F, Kasinath BS, Abbouda HE, Choudhurya GG. NF-κB -mediated cyclin d1 expression by microrna-21 influences renal cancer cell proliferation. Cell Signal 2013; 25:2575-86; http://dx.doi.org/10.1016/j.cellsig.2013.08.005
- Lindemann RK, Newbold A, Whitecross KF, Cluse LA, Frew AJ, Ellis L, Williams S, Wiegmans AP, Dear AE, Scott CL. Analysis of the apoptotic and therapeutic activities of histone deacetylase inhibitors by using a mouse model of B cell lymphoma. Proc Natl Acad Sci 2007; 104:8071; PMID:17470784; http://dx.doi.org/10.1073/pnas.0702294104
