Abstract
Meiosis is a specialized two-step cell division responsible for genome haploidization and the generation of genetic diversity during gametogenesis. An integral and distinctive feature of the meiotic program is the evolutionarily conserved initiation of homologous recombination (HR) by the developmentally programmed induction of DNA double-strand breaks (DSBs). The inherently dangerous but essential act of DSB formation is subject to multiple forms of stringent and self-corrective regulation that collectively ensure fruitful and appropriate levels of genetic exchange without risk to cellular survival. Within this article we focus upon an emerging element of this control—spatial regulation—detailing recent advances made in understanding how DSBs are evenly distributed across the genome, and present a unified view of the underlying patterning mechanisms employed.
Introduction
A unique feature of the meiotic cell cycle is the generation of programmed DNA double-strand breaks (DSBs) during early prophase I by the evolutionarily conserved topoisomerase-like enzyme, Spo11.Citation1 DSBs initiate the process of homologous recombination (HR)—a fundamental DNA repair process integral to the exchange of genetic information and the segregation of homologous chromosomes during the first meiotic nuclear division.Citation2 Remarkably, the distribution of meiotic DSBs across the genome is not random and is, instead, subject to control at multiple levels. At fine scale, DSBs are concentrated within discrete, scattered and non-randomly distributed regions of permissiveness described as DSB hotspots—of which there are over ∼3600 within the S. cerevisiae genome and ∼10,000-40,000 within mammals.Citation3–7 In recent years, an ever-growing collection of factors have been shown to influence the designation of a hotspot through various means—including recruitment of Spo11 and the promotion of cleavage susceptibility (Section on Hotspot designation).Citation8 Beyond this, multiple layers of active regulation appear to exist that not only quantitatively constrain the number of DSBs forming per cell (~150-200 in S. cerevisiae; for a review see Ref. 2 and Ref. 9), but also ensure that those DSBs that do occur are distributed more evenly across all chromatids—a process referred to here as spatial regulation (Sections on Reactive regulation and Proactive regulation).Citation9 These mechanisms do not operate in isolation but rather coalesce into a multifaceted system, progressively layering over one another to guide the DSB distribution both proactively and reactively in DSB-independent and -dependent manners respectively ().
Figure 1. Hierarchical DSB patterning. Regulation of DSB position during prophase I is achieved by means of a hierarchical collection of processes operating via three major nodes: hotspot designation, proactive regulation, and reactive regulation. Rather than acting in isolation, these processes interconnect — sculpting the final DSB distribution with a high degree of complexity (see text for further details). Further to those spatial mechanisms outlined in this review, meiotic recombination is additionally subject to temporal regulation (reviewed in Ref. 2 and Ref. 9), ensuring, for example, that DSB formation occurs post-replication and that the process is ultimately shutdown by homologous chromosome synapsis. Differences in the usage and timing of replication origin activity, and in the relative efficiency of homolog engagement, may therefore permit such processes to contribute, in a generalized manner, to spatial regulation.
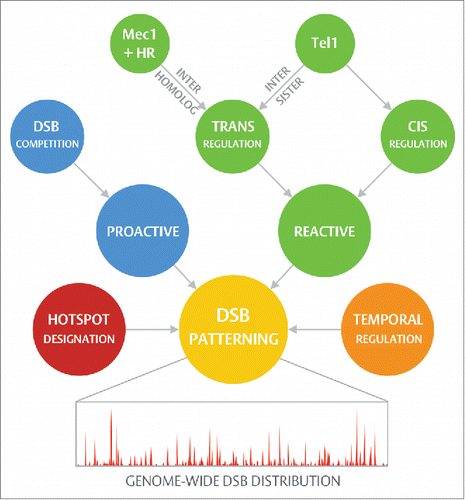
Understanding how cells utilize this hierarchy of processes to spatially guide DSB formation is of critical importance: not only can this “DSB patterning” system potentially protect the genomic integrity of the germline by suppressing aberrant or excessive DSB formation, but it also constructs a foundation—the genome-wide DSB distribution—upon which all downstream processes build, thereby influencing not just the identity of recombinant chromosomes arising from a given individual, but also the rates and distribution of genetic change arising long-term within a population. Within this article we seek to place recent work revealing a role for Tel1ATM in the spatial regulation of DSB formation into this wider context, and construct a generalized framework for how and why this emerging “patterning” system shapes the meiotic recombination landscape throughout prophase I.
Hotspot designation
Spo11 itself possesses only moderate ability to discriminate between DNA sequences,Citation4,10,11 and yet, preferential formation of DSBs within discrete windows of opportunityCitation4 (DSB hotspots) distributed non-randomly across the genome, while not universally conserved, is a distinctive feature of meiosis in many organisms.Citation2,8 The historical analysis of recombination and advent of high-resolution mapping technology has revealed a wealth of information in answer to this apparent contradiction.Citation12 Remarkably, a molecular system to explicitly govern and direct hotspot designation does not appear to be essential, but rather meiotic recombination is able to “piggyback” upon factors embedded within the organizational code of chromosomes—whose primary functions are to orchestrate unrelated cellular processes including gene regulation, transcription and DNA replication.Citation4,8 Within many species, no single factor is key and instead it is the co-occurrence of certain “gatekeeper” factors in a specific fashion that seemingly unlocks the potential for a region to accommodate DSB formation. The dependency of DSB formation upon factors not specifically designed to guide recombination may also go some way toward explaining a certain peculiarity of meiosis: while a handful of common principles exist, no universal, cross-species mechanism underpins hotspot designation and distinctions in strategy are observed between species as well as across evolutionary classes (for an extensive review see Ref. 8) ( – Left).Citation1,8,13
Figure 2. Meiotic hotspot designation. Left - Gatekeeper factors - predictors of recombination. Hotspot designation differs significantly between species. While lower eukaryotes (S. pombe and S. cerevisiae) rely upon a set of passive, low-impact factors, higher eukaryotes (H. sapiens and M. musculus) utilize the multi-functional histone-trimethyltransferase, PRDM9, to guide recombination through the binding of PRDM9 consensus sequences [see text for further details]. Outside of these well-characterized systems, several further organisms display a number of unique properties. Within the canine lineage (C. familiaris), PRDM9 is unexpectedly non-functional—having inactivated between ∼7-9Mya—with GC-richness instead serving as a robust predictor of Spo11-activity.Citation77–79 In contrast to the majority of model organisms, insects (D. melanogaster) and worms (C. elegans) appear devoid of traditional hotspots—consistent with the co-localization of short, repeating sequences with sites of recombination.Citation80–82 A role for non-PRDM9 sequence motifs within recombination, however, does not preclude the existence of hotspots, as noted within A. thaliana and S. pombe.Citation7,83-86 Right - Layers of hotspot designation within S. cerevisiae. Canonical hotspot designation, as seen within S. cerevisiae, requires the co-occurrence of several factors in a specific fashion in order to unlock the potential for a region to initiate recombination [see text for further details].
![Figure 2. Meiotic hotspot designation. Left - Gatekeeper factors - predictors of recombination. Hotspot designation differs significantly between species. While lower eukaryotes (S. pombe and S. cerevisiae) rely upon a set of passive, low-impact factors, higher eukaryotes (H. sapiens and M. musculus) utilize the multi-functional histone-trimethyltransferase, PRDM9, to guide recombination through the binding of PRDM9 consensus sequences [see text for further details]. Outside of these well-characterized systems, several further organisms display a number of unique properties. Within the canine lineage (C. familiaris), PRDM9 is unexpectedly non-functional—having inactivated between ∼7-9Mya—with GC-richness instead serving as a robust predictor of Spo11-activity.Citation77–79 In contrast to the majority of model organisms, insects (D. melanogaster) and worms (C. elegans) appear devoid of traditional hotspots—consistent with the co-localization of short, repeating sequences with sites of recombination.Citation80–82 A role for non-PRDM9 sequence motifs within recombination, however, does not preclude the existence of hotspots, as noted within A. thaliana and S. pombe.Citation7,83-86 Right - Layers of hotspot designation within S. cerevisiae. Canonical hotspot designation, as seen within S. cerevisiae, requires the co-occurrence of several factors in a specific fashion in order to unlock the potential for a region to initiate recombination [see text for further details].](/cms/asset/4c507b3f-2f6c-4fd6-9eb5-2ec3378b9c81/kccy_a_1093709_f0002_c.gif)
Many of the concepts surrounding hotspot designation (outlined above) are particularly well illustrated within the yeast, S. cerevisiae, which relies upon a hierarchical collection of low-impact factors ( – Right).Citation1,4,8 Of particular prominence is the striking correlation of yeast hotspots with regions of nucleosomal depletion (NDRs), a genomic feature primarily associated with promoter regions.Citation4,14,15 However, a significant proportion of detectable NDRs are not associated with robust DSB activity, revealing an insufficiency of chromatin accessibility as an isolated gatekeeper.Citation4 Furthermore, incomplete correlations are observed between Spo11 binding sites and subsequent DSB positions within S. cerevisiae (50-55% overlap),Citation16 Spo11-fusion constructs are incapable of inducing DSB-formation at all targeted lociCitation17,18 and the localization of Spo11 to meiotic chromosomes appears to be a distinct process from that of Spo11 activation,Citation11 collectively suggesting that gatekeeper factors not only facilitate simplistic substrate-enzyme interaction but also create an environment favorable for catalysis.
The influence of gatekeeper factors may also extend beyond that of local effects. At low resolution, S. cerevisiae hotspots themselves cluster, organizing each chromatid into periodic trough and peak sub-domains of recombination potentialCitation4,19,20 (see – Right Top)—an observation that may reflect a non-uniformity in gene density and the impact gene organization seemingly exerts over both hotspot position and chromatin structure (see below). An influence over the latter could be of particular importance: meiotic chromosomes display a unique and functional architecture; self-organizing into linear arrays of protruding chromatin loops, each basally attached to a proteinaceous axis via AT-rich association sites (see - Right).Citation21–23 Within this structural arrangement, hotspots predominantly reside within loop regions while, rather counterintuitively, the machinery essential for the regulation and enzymatic induction of DSBs is bound to the axis.Citation21,24-26 To explain this discrepancy, the tethered-loop axis model proposes that Spp1, a PHD finger domain protein that interacts with both H3K4me3—enriched at S. cerevisiae hotspots—and axial factors, bridges the two entities together and effectively “tethers” the loop to the axis for DSB formation.Citation27–30 The observation that axis proteins are enriched at the 3′ end of S. cerevisiae genes, while strong hotspots preferentially populate transcriptionally divergent intergenic regions at the 5′ end of genes, suggests that the anti-correlation between axis site and hotspot is, in part, driven by the underlying organization of genes and the associated distribution of markers.Citation4,31,32 In addition, components of the axis may serve as active repressors of, or steric occluders to, DSB formation. Indeed, induction of DSBs proximal to Rec8 binding sites—an axial protein thought to demarcate loop boundariesCitation33,34—is notably inefficient,Citation35 and removal of Rec8 profoundly alters both Spo11 binding patterns and DSB distribution.Citation16,21,31,36 In this manner, the placement of genes may not only constitute a gross organizer of meiotic hotspot position, but also a regulator of hotspot usage.
In striking contrast to S. cerevisiae, hotspot designation within mammals (H. sapiens and M. musculus) relies heavily upon a single protein: the rapidly evolving histone trimethyltransferase and C2H2 zinc finger domain factor, PRDM9.Citation5,8,37–39 PRDM9 has emerged as a “swiss army knife” of mammalian hotspot designation, and may be more appropriately thought of as a gatekeeper-organizer. PRDM9 directs hotspot designation by depositing H3K4me3 markersCitation6,40,41 and potentially recruiting Spo11 machinery,Citation8 both of which promote the required co-occurrence of factors around a consensus DNA sequence specified by the PRDM9 zinc finger motif. The identities of these PRDM9 consensus sequences are predominantly dictated by the allelic variant of its repetitive zinc finger array, of which ∼30 have been identified within H. sapiens,Citation42 allowing differing allelic combinations to produce unique DSB distributions.Citation5,38,40,43,44 Interestingly, analysis of hotspot locations within M. musculus PRDM9−/− mutants uncovered a reversion toward S. cerevisiae-like mechanics, with events instead concentrating within H3K4me3-laden promoter regions.Citation43 Such an observation reveals the yeast system to be an ancestral means of determining recombination position (overwritten by the development of PRDM9) as well as a passive system that has persisted despite the evolution of an alternative, dominant method.
Despite the ability of hotspot designation to guide meiotic recombination toward certain sites, the process forms only one part of the patterning system under consideration (see ). Within any given meiotic cell DSBs form in only a subset of hotspots, and as inferred by post-meiotic analysis of recombination, wild-type DSB distributions within individual S. cerevisiae cells do not follow models describing their random, independent placementCitation45 (M. Crawford, T. J. Cooper, and M. J. Neale unpub. obs.). Thus, while it is clear that a great many factors collectively influence the spatial patterning of DSBs as assayed within a population of cells, further layers of regulation exist to control DSB formation on a per-cell basis. Any such additional regulation could conceivably function in one of two distinct ways: (i) reactively — directly activated by or in response to DSBs forming or (ii) proactively — activated independently of DSB formation. A potentially distinguishing feature of reactive regulation over that of proactive is an inability to grossly impact upon population average data due to the low frequencies at which even the strongest hotspots are cleaved (∼10-15%). Interestingly, recent work suggests both forms of regulation function in parallel during meiosis, and within the following sections we aim to use these concepts to explore the constituents and evolution of these extra regulatory layers.
Reactive regulation
Cis and trans interference
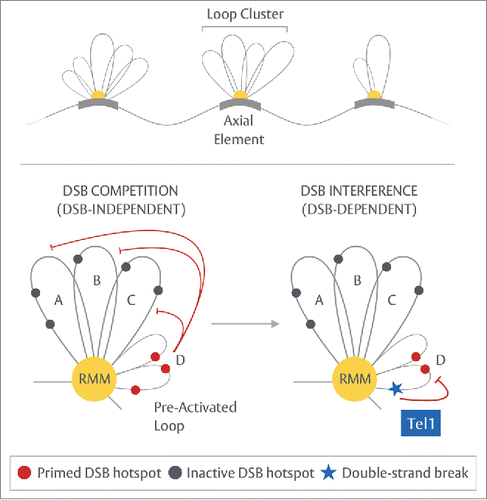
Throughout prophase I, a number of spatial surveillance mechanisms appear to sense the position of DSBs, relaying this information along and between chromatids (in-cis/trans respectively) to reactively sculpt the DSB distribution in a DSB-dependent manner. Central to the cis branch of spatial regulation within S. cerevisiae is the recently discovered phenomenon of DSB interference: a localized, suppressive effect dependent upon the DNA damage response (DDR) kinase, Tel1ATM, which operates over ∼70-100kb, reducing the frequency of coincident DSB formation below that expected by chance ().Citation46 While not explicitly investigated, the inability of Tel1ATM-dependent DSB interference to strongly manifest within the population average (R. J. Carpenter, V. Garcia and M. J. Neale unpub. obs.) suggests it is a reactive, DSB-dependent process—a hypothesis in line with known models of Tel1ATM-activation.Citation47 Abrogation of this effect by the inactivation of Tel1ATM unexpectedly results in two distinct outcomes: (i) over most distances (±20-100kb) DSBs are no longer subject to interference—forming independently of one another, with coincident DSB formation arising at frequencies similar to those expected by chance (ii) by contrast, at short range (±∼7.5kb) DSBs exhibit concerted activity—arising coincidentally at frequencies significantly greater than expected from independent behavior. Remarkably, this latter phenomenon—which results in calculated DSB interference values that are negative—is only witnessed between DSB hotspots residing within the same chromosomal loop domain (see Section on Hotspot designation and - Bottom).Citation46 Confinement of concerted activity to within the boundaries of a loop could conceivably arise if a process upstream of DSB formation “activates” the contained hotspots and where the activation of any given loop region occurs in only a subset of the population. The identity and mechanics of this hypothesized activation process remain largely unknown, however, it is pertinent, given the nature of DSB formation, to consider a state of “pre-tethering”—that is, the intimate and stable association of a loop with axial elements, prior to the induction of DSBs, which may serve to “prime” hotspots for use. The generation of localized zones of negative DSB interference may thus be a simple manifestation of a previously unconsidered and proactive consequence of the tethered-loop-axis model—one otherwise masked by Tel1, whose repressive activity ensures that only one of the primed hotspots (in any given loop) undergoes DSB formation (see ).
Figure 3. DSB interference within S. cerevisiae. Top - Meiotic chromosomes organize into linear arrays of chromatin loops bound by a proteinaceous axis. Prior to DSB formation, we propose that a sub-population of chromatin loops in any given cell exist in a pre-activated state, “priming” hotspots for usage. An attractive candidate for pre-activation may be the tethering of loop sequences to the chromosome axis as proposed within the tethered loop-axis model.Citation25–29 While pre-activation may be an unavoidable byproduct of the way in which DSB formation is setup, it may exist to actively underscore hotspot selection—the process by which the cell determines which of the available hotspots to utilize in any given round of meiosis. Middle - Within wild-type cells, a DSB at any given primed hotspot triggers a Tel1ATM- and distance-dependent suppressive effect (DSB interference), repressing DSB formation at adjacent intra-loop hotspots and within neighboring regions across ∼70-100kb in a reactive, DSB-dependent manner.Citation46 Bottom — In the absence of Tel1, DSB interference is abrogated, enabling adjacent DSBs to arise independently over mid-long range distances (>20-100kb). Over short distances (<20kb), loss of Tel1 activity unmasks the effects of pre-activation within singular loop-domains—manifesting as patches of “negative interference” (as calculated by the standard interference formula: 1-OBS/EXP) due to the concerted formation of adjacent DSBs at frequencies greater than expected from the population average [see text for further details].
![Figure 3. DSB interference within S. cerevisiae. Top - Meiotic chromosomes organize into linear arrays of chromatin loops bound by a proteinaceous axis. Prior to DSB formation, we propose that a sub-population of chromatin loops in any given cell exist in a pre-activated state, “priming” hotspots for usage. An attractive candidate for pre-activation may be the tethering of loop sequences to the chromosome axis as proposed within the tethered loop-axis model.Citation25–29 While pre-activation may be an unavoidable byproduct of the way in which DSB formation is setup, it may exist to actively underscore hotspot selection—the process by which the cell determines which of the available hotspots to utilize in any given round of meiosis. Middle - Within wild-type cells, a DSB at any given primed hotspot triggers a Tel1ATM- and distance-dependent suppressive effect (DSB interference), repressing DSB formation at adjacent intra-loop hotspots and within neighboring regions across ∼70-100kb in a reactive, DSB-dependent manner.Citation46 Bottom — In the absence of Tel1, DSB interference is abrogated, enabling adjacent DSBs to arise independently over mid-long range distances (>20-100kb). Over short distances (<20kb), loss of Tel1 activity unmasks the effects of pre-activation within singular loop-domains—manifesting as patches of “negative interference” (as calculated by the standard interference formula: 1-OBS/EXP) due to the concerted formation of adjacent DSBs at frequencies greater than expected from the population average [see text for further details].](/cms/asset/c889f877-4104-4bb4-88e3-34726ac2ebb8/kccy_a_1093709_f0003_c.gif)
Interestingly, Tel1 and its partner DDR kinase, Mec1ATR, have also been implicated in the parallel trans-branch of spatial regulation. Trans-interference describes the ability of a DSB on one chromatid to suppress formation at the corresponding locus on its sister, homolog or—in many instances—both (one-per-pair or one-per-quartet respectively).Citation45 Much less is known about the mechanics of how trans-interference is accomplished. Inactivation of Mec1 or Tel1 abolishes the occurrence of one-per-quartet constraints, indicating a loss of one specific form of trans-interference; however, whether or not the same form (inter-homolog or inter-sister) is abrogated in each mutant is not yet clear (discussed in Ref. 9). Unlike cis-interference, whose primary function may be the suppression of coincident DSBs within the same activated loop domain (see Section on DSB interference–evolution and cellular role), trans-interference likely ensures the availability of an intact repair substrate while simultaneously suppressing the potential for complex double recombination events to arise from DSB formation at the same genetic locus on both homologues. Thus, by acting in concert, cis and trans interference are likely to promote the equal spacing of recombination events across the genome—constituting a critical part of spatial regulation.
Toward the end of meiotic prophase, in order for DSB formation to be constitutively suppressed throughout the genome once sufficient inter-homolog interactions have been produced, reactive regulation must hand over to a more permanent mode of inhibition (reviewed in Ref. 2 and Ref. 9). While part of this persistent inhibitory signal may arise from global alterations in the expression and/or activity of factors required for DSB formation,Citation48,49 distinct spatial processes—such as the localized suppression of DSB formation in response to homolog synapsis (a DSB-dependent process in S. cerevisiae)Citation50–52—also appear to contribute. The multitude of reactive pathways and activities induced by DSB formation thus allow for self-regulation through multiple avenues.
DSB interference – evolution and cellular role
Substantial alterations to the DSB distribution, in the manner observed within tel1Δ backgrounds, might be expected to significantly perturb recombination and thereby reinforce a presumed importance for DSB interference within the meiotic program; yet, tel1Δ mutants display no gross, meiotic defects and exhibit only small reductions in spore viability (∼5%).Citation46,53 Thus whether it is strictly necessary for DSB interference to operate during meiosis is unclear, opening the door to an intriguing possibility: DSB interference may have emerged as an unintended byproduct of another process—persisting in meiosis by means of indirect selection for an indispensable cellular role or target of Tel1ATM. In-line with the hijacking of transcriptional markers by meiosis for hotspot designation (see Section on Hotspot designation), any process or factor altering the accessibility, presence or identity of these markers has the potential to disrupt DSB formation. Interestingly, in mitotic and vegetative states, Tel1ATM has extensive links to transcriptional regulation via the underlying epigenetic code, notably mediating the in-cis silencing of transcription in proximity to non-programmed DSBs within humans,Citation54 modulation of nucleosomal dynamics and the extensive deposition of DSB-induced histone modifications potentially spanning hundreds of kilobasesCitation55–58—a distance in S. cerevisiae similar to that of meiotic DSB interference. The availability of shared, universal substrates (e.g. histones) may thus provide a platform for the unavoidable, inadvertent acquisition—or intentional adaptation—of such Tel1ATM-dependent mitotic processes whether they are explicitly required in meiosis or not, leading to the generation of novel but potentially non-essential mechanisms (i.e. DSB interference).
While the above presents an attractive model to explain the origins of DSB interference, an ability for interference to fulfil a beneficial role is not precluded. Indeed, several important considerations remain: (i) While the impact of losing DSB interference on S. cerevisiae spore viability is relatively subtle, it may prove cumulative across generations—manifesting after successive interference-deficient meioses as a significant alteration in genetic diversity and elimination of affected lineages from the gene pool—highlighting a putative role for DSB interference in the long-term stability of the population (ii) Any potential for meiotic failure to arise from the clustering of DSB events may be suppressed or compensated for, masking an otherwise greater impact upon viability. A notable candidate for this role is crossover interference: a distinct, overlapping process regulating the placement of crossovers (COs)—the products of homologous recombination most integral to successful completion of meiosis.Citation59,60 Specifically, CO interference may have the ability to partially “correct” the faults resulting from loss of DSB interference via a second round of spatial regulation, selecting only a single DSB per cluster to enter the CO pathway. Consistent with such a role, crossover interference is notably absent within S. pombe,Citation61 an organism which, despite possessing a similar genome size, exhibits a significantly lower DSB frequency and hotspot density than S. cerevisiae (∼58 DSBs/cell and 1 hotspot/23kb vs. ∼150-200 DSBs/cell and 1 hotspot/3.4kb respectively).Citation4,7,62,63 A previously unconsidered consequence of this difference may be a lower reliance upon downstream spatial regulation to spread events along each chromatid. Instead, S. pombe may exert more stringent control at the level of DSB formation simply by placing the process in the hands of more rarely co-occurring factors. DSB and CO-interference may thus collectively guard against the risks associated with otherwise stochastic DSB deposition—preventing deleterious circumstances from arising by chance no matter how infrequently. Interestingly, and in contrast to S. cerevisiae, Atm−/− null mice develop severe meiotic complications, rendering individuals infertile,Citation64,65 a phenotype predominantly ascribed to excessive DSB formationCitation66 and thus compatible with loss of a repressive, interfering effect. Such safeguards may therefore become increasingly important for the selective fitness of mammalian organisms, which have larger chromatin loop sizesCitation22,33,67 (potentially permitting unmanageable numbers of clustered DSBs per loop), smaller populations, and greater time between sequential cycles of sexual reproduction relative to S. cerevisiae.
Proactive regulation
The introduction of a novel hotspot, either by insertion of a strong, high frequency site (e.g. HIS4::LEU2) or the tethering of Spo11/Spp1-fusion constructs to cold regions within the S. cerevisiae genome, not only induces DSB formation but also a repressive, distance-dependent effect that profoundly alters the DSB distribution over a considerable margin.Citation17,18,28,68,69 Despite the prominent similarities to DSB interference (see Section on Cis and trans interference), preliminary data from our group suggests this effect exhibits substantial Tel1-independency (R. J. Carpenter and M. J. Neale unpub. obs.), revealing a third, distinct layer of spatial regulation (see ). Furthermore, this repressive effect appears to strongly manifest itself within the population average,Citation18,28 suggesting it is a perpetually present and proactive process that does not rely upon DSB formation for activation.
This phenomenon—referred to here as “DSB competition”—may arise upstream of DSB formation out of a need for hotspots to compete over restricted and limited pools of pro-recombination factors.Citation2,18,68 Rec114, Mer2, and Mei4, which coalesce into the RMM-complex, are factors essential to Spo11-dependent DSB formation enriched on the chromatin axis.Citation26,70 Mer2, specifically, occupies a crucial role, docking to Spp1 in a manner proposed to mediate the tethering of loops for DSB formation.Citation27,28 While population-averaged binding profiles (ChIP-chip) reveal RMM to occupy ∼900 genomic loci,Citation26 an observation in line with the estimated ∼700 meiotic loops present within S. cerevisiae (per haploid genome copy),Citation35 RMM foci peak as low as ∼40-60/nucleus within individual cells,Citation49,71 identifying RMM as potential players in DSB competition. Despite this apparent limitation, S. cerevisiae is observed to form ∼150-200 DSBs/cell.Citation4,72 One way to reconcile these conflicting observations with the existence of DSB competition is to consider that, in each cell, loops aggregate around RMM foci into clustered super domains which, perhaps, nucleate at or generate those chromosomal regions that are first to assemble short, incomplete elements of the chromosomal axis during early prophase I ( - Top).Citation73,74 Within this model, we propose that DSB competition arises through successive rounds of intra-cluster but inter-loop competition for limited RMM and/or tether points, confining the repressive effect of DSB competition to the average size of a cluster ( - Bottom)—drawing considerable parallels to models previously proposed for crossover interference.Citation75 Furthermore, differences in the density of gatekeeper factors (e.g. H3K4me3) (see Section on Hotspot designation) may govern the extent to which any given loop can compete, introducing significant overlap between hotspot designation and downstream spatial regulation. Interestingly, a comparable disparity exists within mice: an estimated 10,000 loops span the genome while MEI4 foci are present at significantly lower levels (∼300/nucleus), suggesting a similar regulatory layer could operate within other species.Citation33,35,67,76 A mechanism of pre-tethering may thus underpin both the negative interference values observed within individual loops when Tel1ATM-dependent DSB interference is lost and, in part, the DSB-independent competition that arises between DSB hotspots residing in adjacent loop domains within S. cerevisiae.
Figure 4. Prospective “loop cluster” model of DSB competition. Top - During early prophase I, short stretches of axial element nucleate at scattered regions across each chromosome.Citation73,74 Upon this platform, the first meiotic loops may begin to assemble, associating together into individually acting, isolated units. Bottom – Building upon the tethered-loop axis model, we propose that within any such clustered unit, limited availability of, or access to, essential factors such as RMM (the Rec114-Mei4-Mer2 complex), coupled to a differential ability of each loop to establish a tether, could generate DSB competition by means of competitive tethering—lowering the frequency of DSB formation within the remainder of the associated loops in a proactive, DSB-independent manner. Under wild-type conditions, DSB formation subsequently induces Tel1ATM-dependent DSB interference—a process that may inhibit or dismantle cluster units thereby suppressing further DSB formation in the immediate region. As illustrated here, the apparent Tel1-independency of DSB competition suggests the strong, repressive effect observed around strong hotspots is in fact, a composite of two distinct processes.
Concluding remarks
The risk-reward tradeoff inherent in meiotic recombination places a strong demand on the cell for stringent and adaptive control at multiple stages of the process. Spatial regulation of DSB formation is rapidly emerging as a key part of this control. As proposed here, the machinery and mechanisms underlying spatial regulation within S. cerevisiae cannot be considered in isolation but rather as branches to a larger, interconnecting hierarchy within which extensive cross-talk and overlap is a possibility. That such a complex network of overlaying regulatory mechanisms exists perhaps highlights the lengths to which cells have needed to evolve in order to tolerate—and productively utilize—the DNA breaks that would otherwise be considered harmful to genome stability. Whether or not the specific mechanisms characterized (DSB interference, DSB competition and trans-interference) operate in other organisms is, however, yet to be determined and much remains to be understood within S. cerevisiae itself. Distinctions in meiotic strategy and the extent to which spatial regulation is required likely reflect far-reaching differences present in any given species at both the macro and molecular levels, and thus precisely which rules will prove to be universal and which unique, is difficult to predict. Nevertheless, we suggest that the emerging and unified view of DSB patterning presented here will help to guide future investigations.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
The authors would like to thank Scott Keeney and Rachal Allison for critical reading of this manuscript prior to publication.
Funding
Support for this research is provided by a consolidator award from the European Research Council, a project grant from the Biotechnology and Biological Research Council, and a University Research Fellowship from the Royal Society.
References
- Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 2015; 7:a016634; http://dx.doi.org/10.1101/cshperspect.a016634
- Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet 2014; 48:187-214; PMID:25421598; http://dx.doi.org/10.1146/annurev-genet-120213-092304
- Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res 2012; 22:957-65; PMID:22367190; http://dx.doi.org/10.1101/gr.130583.111
- Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, Tischfield SE, Zhu X, Neale MJ, Jasin M, Socci ND, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell 2011; 144:719-31; PMID:21376234; http://dx.doi.org/10.1016/j.cell.2011.02.009
- Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. Recombination initiation maps of individual human genomes. Science 2014; 346:1256442-2; PMID:25395542; http://dx.doi.org/10.1126/science.1256442
- Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 2011; 472:375-8; PMID:21460839; http://dx.doi.org/10.1038/nature09869
- Fowler KR, Sasaki M, Milman N, Keeney S, Smith GR. Evolutionarily diverse determinants of meiotic DNA break and recombination landscapes across the genome. Genome Res 2014; 24:1650-64; PMID:25024163; http://dx.doi.org/10.1101/gr.172122.114
- de Massy B. Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu Rev Genet 2013; 47:563-99; PMID:24050176; http://dx.doi.org/10.1146/annurev-genet-110711-155423
- Cooper TJ, Wardell K, Garcia V, Neale, MJ. Homeostatic regulation of meiotic DSB formation by ATM/ATR. Exp Cell Res 2014; 329:124-31; PMID:25116420; http://dx.doi.org/10.1016/j.yexcr.2014.07.016
- Murakami H, Nicolas A. Locally, meiotic double-strand breaks targeted by Gal4BD-Spo11 occur at discrete sites with a sequence preference. Mol Cell Biol 2009; 29:3500-16; PMID:19380488; http://dx.doi.org/10.1128/MCB.00088-09
- Prieler S, Penkner A, Borde V, Klein F. The control of Spo11's interaction with meiotic recombination hotspots. Genes Dev 2005; 19:255-69; PMID:15655113; http://dx.doi.org/10.1101/gad.321105
- Choi K, Henderson IR. Meiotic Recombination Hotspots - a Comparative View. Plant J 2015; 83:52-61; PMID:25925869; http://dx.doi.org/10.1111/tpj.12870
- Nishant KT, Rao MRS. Molecular features of meiotic recombination hot spots. Bioessays 2006; 28:45-56; PMID:16369948; http://dx.doi.org/10.1002/bies.20349
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 2009; 458 362-6; PMID:19092803; http://dx.doi.org/10.1038/nature07667
- Fan QQ, Petes TD. Relationship between nuclease-hypersensitive sites and meiotic recombination hot spot activity at the HIS4 locus of Saccharomyces cerevisiae. Mol Cell Biol 1996; 16:2037-43; PMID:8628269; http://dx.doi.org/10.1128/MCB.16.5.2037
- Kugou K, Fukuda T, Yamada S, Ito M, Sasanuma H, Mori S, Katou Y, Itoh T, Matsumoto K, Shibata T, et al. Rec8 guides canonical Spo11 distribution along yeast meiotic chromosomes. Mol Biol Cell 2009; 20:3064-76; PMID:19439448; http://dx.doi.org/10.1091/mbc.E08-12-1223
- Fukuda T, Kugou K, Sasanuma H, Shibata T, Ohta K. Targeted induction of meiotic double-strand breaks reveals chromosomal domain-dependent regulation of Spo11 and interactions among potential sites of meiotic recombination. Nucleic Acids Res 2008; 36:984-97; PMID:18096626; http://dx.doi.org/10.1093/nar/gkm1082
- Robine N, Uematsu N, Amiot F, Gidrol X, Barillot E, Nicolas A, Borde V. Genome-wide redistribution of meiotic double-strand breaks in Saccharomyces cerevisiae. Mol Cell Biol 2007; 27:1868-80; PMID:17189430; http://dx.doi.org/10.1128/MCB.02063-06
- Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. Proc Natl Acad Sci 1997; 94:5213-8; PMID:9144217; http://dx.doi.org/10.1073/pnas.94.10.5213
- Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci USA 2000; 97:11383-90; PMID:11027339; http://dx.doi.org/10.1073/pnas.97.21.11383
- Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell 2002; 111:791-802; PMID:12526806; http://dx.doi.org/10.1016/S0092-8674(02)01167-4
- Kleckner N. Chiasma formation: chromatin/axis interplay and the role(s) of the synaptonemal complex. Chromosoma 2006; 115 175-94; PMID:16555016; http://dx.doi.org/10.1007/s00412-006-0055-7
- Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev 2013; 23:147-55; PMID:23313097; http://dx.doi.org/10.1016/j.gde.2012.12.002
- Kee K, Protacio RU, Arora C, Keeney S. Spatial organization and dynamics of the association of Rec102 and Rec104 with meiotic chromosomes. EMBO J 2004; 23:1815-24; PMID:15044957; http://dx.doi.org/10.1038/sj.emboj.7600184
- Zickler D, Kleckner N. Meiotic Chromosomes: Integrating Structure and Function. Annu Rev Genet 1999; 603-754; PMID:10690419; http://dx.doi.org/10.1146/annurev.genet.33.1.603
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 2011; 146:372-83; PMID:21816273; http://dx.doi.org/10.1016/j.cell.2011.07.003
- Sommermeyer V, Béneut C, Chaplais E, Serrentino ME, Borde V. Spp1, a member of the Set1 Complex, promotes meiotic DSB formation in promoters by tethering histone H3K4 methylation sites to chromosome axes. Mol Cell 2013; 49:43-54; PMID:23246437; http://dx.doi.org/10.1016/j.molcel.2012.11.008
- Acquaviva L, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint André C, Nicolas A, Géli V. The COMPASS subunit Spp1 links histone methylation to initiation of meiotic recombination. Science 2013; 339:215-8; PMID:23160953; http://dx.doi.org/10.1126/science.1225739
- Borde V, Székvölgyi L, Dichtl B, Dichtl BS, de La Roche Saint André C, Nicolas A, Géli V. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J 2009; 28:99-111; PMID:19078966; http://dx.doi.org/10.1038/emboj.2008.257
- Tischfield SE, Keeney S. Scale matters: the spatial correlation of yeast meiotic DNA breaks with histone H3 trimethylation is driven largely by independent colocalization at promoters. Cell Cycle 2012; 11:1496-503; PMID:22433953; http://dx.doi.org/10.4161/cc.19733
- Sun X, Huang L, Markowitz TE, Blitzblau HG, Chen D, Klein F, Hochwagen A. Transcription dynamically patterns the meiotic chromosome-axis interface. Elife 2015; 4:e07424; http://dx.doi.org/10.7554/eLife.07424
- Champeimont R, Carbone A. SPoRE: a mathematical model to predict double strand breaks and axis protein sites in meiosis. BMC Bioinformatics 2014; 15:391; PMID:25495332; http://dx.doi.org/10.1186/s12859-014-0391-1
- Novak I, Wang H, Revenkova E, Jessberger R, Scherthan H, Höög C. Cohesin Smc1beta determines meiotic chromatin axis loop organization. J Cell Biol. 2008; 180:83-90; PMID:18180366; http://dx.doi.org/10.1083/jcb.200706136
- Glynn EF, Megee PC, Yu HG, Mistrot C, Unal E, Koshland DE, DeRisi JL, Gerton JL. Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol 2004; 2:E259; PMID:15309048; http://dx.doi.org/10.1371/journal.pbio.0020259
- Ito M, Kugou K, Fawcett JA, Mura S, Ikeda S, Innan H, Ohta K. Meiotic recombination cold spots in chromosomal cohesion sites. Genes Cells 2014; 19:359-73; PMID:24635992; http://dx.doi.org/10.1111/gtc.12138
- Kim KP, Weiner BM, Zhang L, Jordan A, Dekker J, Kleckner N. Sister cohesion and structural axis components mediate homolog bias of meiotic recombination. Cell 2010; 143:924-37; PMID:21145459; http://dx.doi.org/10.1016/j.cell.2010.11.015
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 2010; 327:836-40; PMID:20044539; http://dx.doi.org/10.1126/science.1183439
- Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science 2010; 327:835; PMID:20044538; http://dx.doi.org/10.1126/science.1181495
- Neale MJ. PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010; 11:104; PMID:20210982; http://dx.doi.org/10.1186/gb-2010-11-2-104
- Grey C, Barthès P, Chauveau-Le Friec G, Langa F, Baudat F, de Massy B. Mouse PRDM9 DNA-binding specificity determines sites of histone H3 lysine 4 trimethylation for initiation of meiotic recombination. PLoS Biol 2011; 9:e1001176; PMID:22028627; http://dx.doi.org/10.1371/journal.pbio.1001176
- Buard J, Barthès P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J 2009; 28:2616-24; PMID:19644444; http://dx.doi.org/10.1038/emboj.2009.207
- Berg IL, Neumann R, Lam KW, Sarbajna S, Odenthal-Hesse L, May CA, Jeffreys AJ. PRDM9 variation strongly influences recombination hot-spot activity and meiotic instability in humans. Nat Genet 2010; 42:859-63; PMID:20818382; http://dx.doi.org/10.1038/ng.658
- Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature 2012; 485:642-5; PMID:22660327; http://dx.doi.org/10.1038/nature11089
- Buard J, Rivals E, Dunoyer de Segonzac D, Garres C, Caminade P, de Massy B, Boursot P. Diversity of Prdm9 zinc finger array in wild mice unravels new facets of the evolutionary turnover of this coding minisatellite. PLoS One 2014; 9:e85021; PMID:24454780; http://dx.doi.org/10.1371/journal.pone.0085021
- Zhang L, Kim KP, Kleckner NE, Storlazzi A. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. Proc Natl Acad Sci USA 2011; 108:20036-41; PMID:22123968; http://dx.doi.org/10.1073/pnas.1117937108
- Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature 2015; 520:114-8; PMID:25539084; http://dx.doi.org/10.1038/nature13993
- Paull TT. Mechanisms of ATM Activation. Annu Rev Biochem 2015; 84:711-38; PMID:25580527; http://dx.doi.org/10.1146/annurev-biochem-060614-034335
- Gray S, Allison RM, Garcia V, Goldman ASH, Neale MJ. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR). Open Biol 2013; 3:130019; PMID:23902647; http://dx.doi.org/10.1098/rsob.130019
- Carballo JA, Panizza S, Serrentino ME, Johnson AL, Geymonat M, Borde V, Klein F, Cha RS. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet 2013; 9:e1003545; PMID:23825959; http://dx.doi.org/10.1371/journal.pgen.1003545
- Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature 2014; 510:241-6; PMID:24717437; http://dx.doi.org/10.1038/nature13120
- Kauppi L, Panizza S, Serrentino ME, Johnson AL, Geymonat M, Borde V, Klein F, Cha RS. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 2013; 27:873-86; PMID:23599345; http://dx.doi.org/10.1101/gad.213652.113
- Zickler D, Kleckner N. Recombination, Pairing, and Synapsis of Homologs during Meiosis. Cold Spring Harb Perspect Biol 2015; 7:a016626; PMID:25986558; http://dx.doi.org/10.1101/cshperspect.a016626
- Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 2008; 132:758-70; PMID:18329363; http://dx.doi.org/10.1016/j.cell.2008.01.035
- Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 2010; 141:970-81; PMID:20550933; http://dx.doi.org/10.1016/j.cell.2010.04.038
- Shiloh Y, Ziv Y. The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat Rev Mol Cell Biol 2013; 14:197-210; http://dx.doi.org/10.1038/nrm3546
- Price BD, D’Andrea AD. Chromatin remodeling at DNA double-strand breaks. Cell 2013; 152:1344-54; PMID:23498941; http://dx.doi.org/10.1016/j.cell.2013.02.011
- Lee CS, Lee K, Legube G, Haber JE. Dynamics of yeast histone H2A and H2B phosphorylation in response to a double-strand break. Nat Struct Mol Biol 2014; 21:103-9; PMID:24336221; http://dx.doi.org/10.1038/nsmb.2737
- Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Distribution and Dynamics of Chromatin Modification Induced by a Defined DNA Double-Strand Break. Curr Biol 2004; 14:1703-11; PMID:15458641; http://dx.doi.org/10.1016/j.cub.2004.09.047
- Zhang L, Wang S, Yin S, Hong S, Kim KP, Kleckner N. Topoisomerase II mediates meiotic crossover interference. Nature 2014; 511:551-6; PMID:25043020; http://dx.doi.org/10.1038/nature13442
- Jones GH, Franklin FCH. Meiotic crossing-over: obligation and interference. Cell 2006; 126:246-8; PMID:16873056; http://dx.doi.org/10.1016/j.cell.2006.07.010
- Munz P. An analysis of interference in the fission yeast Schizosaccharomyces pombe. Genetics 1994; 137:701-7; PMID:8088515
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature 2002; 415:871-80; PMID:11859360; http://dx.doi.org/10.1038/nature724
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, et al. Life with 6000 genes. Science 1996; 274, 546:563-7; http://dx.doi.org/10.1126/science.274.5287.546
- Barlow C, Liyanage M, Moens PB, Tarsounas M, Nagashima K, Brown K, Rottinghaus S, Jackson SP, Tagle D, Ried T, et al. Atm deficiency results in severe meiotic disruption as early as leptonema of prophase I. Development 1998; 125:4007-17; PMID:9735362
- Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D, et al. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell 1996; 86:159-71; PMID:8689683; http://dx.doi.org/10.1016/S0092-8674(00)80086-0
- Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature 2011; 479:237-40; PMID:22002603; http://dx.doi.org/10.1038/nature10508
- Kauppi L, Barchi M, Baudat F, Romanienko PJ, Keeney S, Jasin M. Distinct properties of the XY pseudoautosomal region crucial for male meiosis. Science 2011; 331:916-20; PMID:21330546; http://dx.doi.org/10.1126/science.1195774
- Wu TC, Lichten M. Factors that affect the location and frequency of meiosis-induced double-strand breaks in Saccharomyces cerevisiae. Genetics 1995; 140:55-66; PMID:7635308
- Fan QQ, Xu F, White MA, Petes TD. Competition between adjacent meiotic recombination hotspots in the yeast Saccharomyces cerevisiae. Genetics 1997; 145:661-70; PMID:9055076
- Maleki S, Neale MJ, Arora C, Henderson KA, Keeney S. Interactions between Mei4, Rec114, and other proteins required for meiotic DNA double-strand break formation in Saccharomyces cerevisiae. Chromosoma 2007; 116:471-86; PMID:17558514; http://dx.doi.org/10.1007/s00412-007-0111-y
- Li J, Hooker GW, Roeder GS. Saccharomyces cerevisiae Mer2, Mei4 and Rec114 form a complex required for meiotic double-strand break formation. Genetics 2006; 173:1969-81; PMID:16783010; http://dx.doi.org/10.1534/genetics.106.058768
- Martini E, Borde V, Legendre M, Audic S, Regnault B, Soubigou G, Dujon B, Llorente B. Genome-wide analysis of heteroduplex DNA in mismatch repair-deficient yeast cells reveals novel properties of meiotic recombination pathways. PLoS Genet 2011; 7:e1002305; PMID:21980306; http://dx.doi.org/10.1371/journal.pgen.1002305
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell 1991; 66:1239-56; PMID:1913808; http://dx.doi.org/10.1016/0092-8674(91)90046-2
- Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet 1998; 32:619-97; PMID:9928494; http://dx.doi.org/10.1146/annurev.genet.32.1.619
- Stahl FW, Foss HM, Young LS, Borts RH, Abdullah MF, Copenhaver GP. Does crossover interference count in Saccharomyces cerevisiae? Genetics 2004; 168:35-48; PMID:15454525; http://dx.doi.org/10.1534/genetics.104.027789
- Kumar R, Bourbon HM, de Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 2010; 24:1266-80; PMID:20551173
- Auton A, Rui Li Y, Kidd J, Oliveira K, Nadel J, Holloway JK, Hayward JJ, Cohen PE, Greally JM, Wang J, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLoS Genet 2013; 9:e1003984; PMID:24348265; http://dx.doi.org/10.1371/journal.pgen.1003984
- Muñoz-Fuentes V, Di Rienzo A, Vilà C. Prdm9, a major determinant of meiotic recombination hotspots, is not functional in dogs and their wild relatives, wolves and coyotes. PLoS One 2011; 6:e25498; http://dx.doi.org/10.1371/journal.pone.0025498
- Axelsson E, Webster MT, Ratnakumar A, Ponting CP, Lindblad-Toh K. Death of PRDM9 coincides with stabilization of the recombination landscape in the dog genome. Genome Res 2012; 22:51-63; PMID:22006216; http://dx.doi.org/10.1101/gr.124123.111
- Kaur T, Rockman MV. Crossover heterogeneity in the absence of hotspots in Caenorhabditis elegans. Genetics 2014; 196:137-48; PMID:24172135; http://dx.doi.org/10.1534/genetics.113.158857
- Barnes TM, Kohara Y, Coulson A, Hekimi S. Meiotic recombination, noncoding DNA and genomic organization in Caenorhabditis elegans. Genetics 1995; 141:159-79; PMID:8536965
- Comeron JM, Ratnappan R, Bailin S. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet 2012; 8:e1002905; PMID:23071443; http://dx.doi.org/10.1371/journal.pgen.1002905
- Steiner WW, Steiner EM, Girvin AR, Plewik LE. Novel nucleotide sequence motifs that produce hotspots of meiotic recombination in Schizosaccharomyces pombe. Genetics 2009; 182:459-69; PMID:19363124; http://dx.doi.org/10.1534/genetics.109.101253
- Yamada S, Ohta K, Yamada T. Acetylated Histone H3K9 is associated with meiotic recombination hotspots, and plays a role in recombination redundantly with other factors including the H3K4 methylase Set1 in fission yeast. Nucleic Acids Res 2013; 41:3504-17; PMID:23382177; http://dx.doi.org/10.1093/nar/gkt049
- Choi K, Zhao X, Kelly KA, Venn O, Higgins JD, Yelina NE, Hardcastle TJ, Ziolkowski PA, Copenhaver GP, Franklin FC, et al. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet 2013; 45:1327-36; PMID:24056716; http://dx.doi.org/10.1038/ng.2766
- Drouaud J, Khademian H, Giraut L, Zanni V, Bellalou S, Henderson IR, Falque M, Mézard C. Contrasted Patterns of Crossover and Non-crossover at Arabidopsis thaliana Meiotic Recombination Hotspots. PLoS Genet 2013; 9:e1003922; PMID:24244190; http://dx.doi.org/10.1371/journal.pgen.1003922
