Abstract
Glioblastoma (GBM) is an incurable cancer, with survival rates of just 14-16 months after diagnosis.Citation1 Functional genomics have identified numerous genetic events involved in GBM development. One of these, the deregulation of microRNAs (miRNAs), has been attracting increasing attention due to the multiple biologic processes that individual miRNAs influence. Our group has been studying the role of miR-182 in GBM progression, therapy resistance, and its potential as GBM therapeutic. Oncogenomic analyses revealed that miR-182 is the only miRNA, out of 470 miRNAs profiled by The Cancer Genome Atlas (TCGA) program, which is associated with favorable patient prognosis, neuro-developmental context, temozolomide (TMZ) susceptibility, and most significantly expressed in the least aggressive oligoneural subclass of GBM. miR-182 sensitized glioma cells to TMZ-induced apoptosis, promoted glioma initiating cell (GIC) differentiation, and reduced tumor cell proliferation via knockdown of Bcl2L12, c-Met and HIF2A.Citation2 To deliver miR-182 to intracranial gliomas, we have characterized Spherical Nucleic Acids covalently functionalized with miR-182 sequences (182-SNAs). Upon systemic administration, 182-SNAs crossed the blood-brain/blood-tumor barrier (BBB/BTB), reduced tumor burden, and increased animal subject survival.Citation2-4 Thus, miR-182-based SNAs represent a tool for systemic delivery of miRNAs and a novel approach for the precision treatment of malignant brain cancers.
Introduction
miRNAs are evolutionary conserved short non-coding RNAs, which upon binding to cis-regulatory elements within the 3’untranslated region (UTR) repress gene expression. Gene silencing occurs by translational repression or mRNA degradation.Citation5 The extent of complementarity between miRNA and target mRNA determines the silencing mechanism. In case of perfect complementarity, mRNA cleavage is initiated by the RNA-Induced Silencing (RISC) complex. When there is partial complementarity, translational repression of the target mRNA is predominant, leading to reduction in protein levels without affecting transcript abundance.Citation6 As the majority of miRNAs have only partial complementarity to their target 3’UTR, and many genes are regulated by multiple miRNAs engaging distinct 3-UTR-localized binding sites, miRNAs fine-tune the expression and activity of hundreds of target genes and their associated signaling networks. miRNA-mediated gene silencing is further complicated by the fact that miRNAs can have diverse functions in different tissues and different developmental stages, and in many instances, can concomitantly regulate diametrically opposed signaling pathways.
Multiple studies have identified miRNAs with important roles in normal and cancerous growth. miRNAs impact animal and plant development, including lineage differentation processes, which guide tissue morphogenesis. They also play fundamental roles in regulating tumor development, further underscoring the general observation that tumor cells deregulate developmental factors to reaquire proliferative, pro-migratory and pro-invasive capabilities.Citation7 In this perspective, we will highlight the importance of miRNA biology for regulating normal and cancerous processes in the central nervous system (CNS). We will focus on GBM-associated miR-182, and will describe results from our functional oncogenomic investigations to characterize the impact of miRNA-182 on GBM cell growth, differentiation and therapy susceptibility. We will describe how miRNA discovery can inform the rational design of miRNA-based therapies, in particular Spherical Nucleic Acids (SNAs) conjugated with miRNA sequences, a highly prominent and potent gene regulating platform designed to systemically deliver RNA interference to intracranial brain tumors.
miRNAs regulate the GBM phenotype
miRNAs show defined expression in the developing CNS, and play important roles in brain pattern formation.Citation8,9 During normal development, miRNAs regulate the transition of neural progenitors into differentiated neurons that contribute to brain morphogenesis. Supporting a role of miRNAs in maintaining a differentiated cell state, neuron-specific ablation of Dicer, the key miRNA-generating enzyme, causes neuronal cell death, cerebellar degeneration and ataxia.Citation10 Developmental miRNA candidates include miR-9 and miR-124, which drive neuronal differentiation of embryonic stem cells by targeting members of the REST transcriptional complex that inhibits the expression of genes implicated in neuronal cell fate decisions.Citation11-14 In particular, developmental induction of miR-124 expression within the subventricular zone of the mouse brain cortex promotes adult neurogenesis. miR-124 regulate Dlx2 and Sox9 expression, and in so doing, promotes the transition of the transit amplifying cell to neuroblast stage.Citation15 Together with miR-9, miR-124 inhibits the differentiation of progenitor cells into glial cells and contributes to increased neuron to glia ratio.Citation11 In addition, miR-134 is abundantly expressed in the brain, in particular in the hippocampus, localizes to dendrites at the synaptic sites, and regulates dendrite spine development by modulating Lim-domain containing kinase (LIMK)-1 activity.Citation16,17 Other miRNAs that are found in the synaptosome and are important for synaptic transmission are miR-9 and miR-138.Citation18 miR-138 regulates the size of dendritic spines, without affecting spine density, by targeting the adenine phosphoribosyltransferase APT1, which regulates the depalmitoylation of proteins. Furhtermore, miR-125b and miR-24a have been implicated in regulating apoptosis during neural development by regulating p53 and other apoptosis effector proteins.Citation19,20
Reflecting their roles in CNS development, deregulated miRNA expression drives or contributes to the development of malignant brain cancers, in particular GBM. Multiple miRNAs are differentially expressed in GBM vs. normal brain tissue, and target key molecules with central functions in angiogenesis, glioma cell proliferation, invasion and susceptibility toward extant therapies.Citation21 GBM is the most common and lethal type of brain cancer in adults with a median patient survival of only 15 months after diagnosis. Conventional (i.e., TMZ and radiation) and molecularly targeted therapies, foremost inhibitors of receptor tyrosine kinase (RTK) activity, failed to demonstrate survival benefits for GBM patients. Here, the existence of complex anti-apoptotic signaling mechanisms,Citation22 substantial inter- and intratumoral heterogeneity, together with a therapy/apoptosis-resistant glioma stem cell (GSC) population Citation23 conspired to make GBM a highly enigmatic and incurable disease. GSCs, a sub-population of cells embedded within GBM tissues, form tumors in orthotopic transplants in vivo and generate a diversified neuron-like and glia-like postmitotic progeny, and are critical drivers of tumor recurrence and therapy resistance.Citation23,24
Similiarities between GSC and neural stem cell (NSC) biology have motivated investigations to identify miRNAs with neurodevelopmental annotations. In particular, GBM tumors have recently been classified into 5 groups according to miRNA clustering of the TCGA dataset, with each group having distinct molecular, biological, and clinical characteristics.Citation25 In contrast to mRNA-based hierachical clustering, miRNA-based classification yielded robust survival differences between subclasses, each corresponding to a different neural stem cell differentiation stage, e.g. oligoneural, radial glial, neural, neuromesenchymal and astrocytic. Oligoneural GBM, a subclass of proneural tumors, are known for longer patient survival than other GBM subtypes, and show repression of neural and embryonic stem cell-related mRNA signatures.Citation25 To identify high-priority miRNAs with critical functions in GBM, Johnson and colleagues prioritized miRNAs based on highly variable expression levels [median absolute deviation (MAD) >1], correlation with patient survival (significance in univariate Cox model < 0.1), and implication in neurodevelopment. Seven miRNAs fulfilled all of these criteria (miR-146b, miR-182, miR-21, miR-210, miR-221, miR-9/miR-9*). Among these, miR-9 was identified as inhibitor of oncogenic JAK-STAT3-CEBP-β signaling, with potent growth-suppressive and dedifferentiation activities in glioma-initiating cells (GICs).Citation11,25 Importantly, analyses of miRNA-mRNA expression correlations revealed that miR-9 showed the highest level of connectivity, suggesting that miR-9 may represent a core regulator of gene expression in GBM.
In addition to miR-9, miR-21 was one of the first miRNAs found to be highly expressed in GBM. miR-21 has oncogenic functions and promotes cell proliferation by targeting the phosphatase and tensin homolog (PTEN), sprouty RTK signaling antagonist 2 (SPRY2) tumor suppressor pathways, and by activating the nuclear factor of kappa light polypeptide gene enhancer in B-cells (NF-kB) and rat sarcoma viral oncogene (Ras)-signaling.Citation26,27 Further molecular analyses revealed that miR-21 promoted cell invasion by targeting the matrix metaloproteinase regulators reversion-inducing cysteine-rich protein with kazal motifs (RECK) and tissue inhibitor of metallopeptidase 3 (TIMP3),Citation28 and inhibited apoptosis via repression of heterogeneous nuclear ribonucleoprotein K (HNRPK) and programmed cell death 4 (PDCD4).Citation29 miR-10b and -26a represent additional tumor-promoting miRNAs. miR-10b expression is correlated with GBM grade. By targeting the pro-apoptotic Bcl-2 family protein Bim, the urokinase plasminogen activator receptor (uPAR) and the Ras homolog gene family member (RhoC) as well as cell cycle regulators and genes implicated in glioma cell migration, miR-10b is a potent regulator of apoptosis, cell cycle and invasive processes.Citation30-32 miR-26a promotes transformation by targeting PTEN and retinoblastoma 1 (RB1), 2 of the most frequently deleted tumor suppressors in GBM, and by modulating JNK activation.Citation33,34 Finally, by regulating Notch, RTK and Smad4 signaling, miR-34a was identified as a tumor suppressor in proneural tumors.Citation35-37 In particular, applying a Context Likelihood of Relatedness (CLR) algorithm, Chin and collegues sought to identify high-priority miRNA-mRNA relationships in the TCGA data set. Relationships were prioritized based on negative correlation between miRNA and mRNA expression, the presence of miRNA binding sites within 3’UTR target gene sequences, and copy-number-driven expression of miRNA and mRNA. The latter criteria are based on the assumption that biologically relevant miRNAs or mRNAs are likely deregulated on multiple levels. These multidimensional oncogenomic investigations pointed to the miR-34a-platelet-derived growth factor receptor α polypeptide (PDGFRA) axis as a top-priority regulatory relationship with strong discriminatory potential between proneural and mesenchymal GBM subtypes.Citation37
miR-182 expression influences multiple tumor biological properties
Similar to miR-9, miR-182 emerged as a high priority miRNA in GBM, characterized by highly variable expression, functions in neurodevelopment, and association with the oligoneural GBM subclass (). Unlike miR-9, our studies suggest that miR-182 also influences the apoptotic response of glioma cells to a variety of anti-cancer agents, including TMZ and RTK inhibitors, by downregulating the caspase and p53 inhibitor Bcl2L12.Citation2 In addition, we demonstrated that miR-182 expression provoked a more differentiated state in multipotent GICs, which is evidenced by decreased expression of stem cell markers Oct4, CD44, Nanog and Sox2, and by elevated levels of microtubule-associated protein 2 (MAP2) and glial fibrillary acidic protein (GFAP) expression. Furthermore, miR-182 is expressed at lower levels in CD133-positive, GSC-enriched GICs compared to CD133-negative non-stem cells.Citation2 To define miR-182-dependent transcriptional effects, we transduced patient-derived GICs, of the mesenchymal subtype, harboring low-level expression of endogenous miR-182, with miR-182 or control (Co)-miR lentivirus. Cultures were subjected to whole genome expression profiling. The expression profiling data were used for sequence-based prediction of specific interactions with miR-182 (TargetScan algorithm). A comparative analysis of these targets with mRNA signatures upregulated in different stages of neural cell differentiation [datasets described in ref.Citation25] revealed that the miR-182 repressed target sequences that are enriched for stem cell-related genes. To molecularly define the miR-182 modus operandi, we used Ingenuity Pathway analysis to integrate miR-182 target gene functions. The top scoring signaling pathway that emerged contained a signature that in addition to Bcl2L12 included c-Met and HIF2A.Citation2 c-Met is hyperactivated in GBM Citation38-40 and represents a critical driver of GBM growth, invasion, hypoxia-induced dedifferentiation and tumor progression.Citation41,42 HIF2A is an important effector of hypoxia-driven stem cell maintenance that promotes GIC self-renewal, growth, and tumorigenicity.Citation43 miR-182 recognizes an evolutionary conserved site within the 3’UTR of HIF2A and c-Met, and in so doing, reduces cell proliferation as well as stem cell marker expression, to promote a more differentiated cell state. The miR-182 target gene c-Met is a member of the RTK family and is overexpressed in GBM. It is well appreciated that c-Met regulates GBM growth, invasion as well as hypoxia-induced dedifferentiation.Citation41,44 c-Met is also involved in stem cell reprogramming and transitioning from a quiescent to an active self proliferating state.Citation45 Similarly, HIF2A is a hypoxia regulated transcription factor and effector of stem cell maintenance that drives GIC self-renewal, growth, and tumorigenicity, and its expression negatively correlates with glioma patient survival.Citation43 With respect to the regulation of differentiation processes, the miR-182-repressed transcriptome was enriched for a stem cell specific signature, mirrored phenotypically by reduced sphere size and expansion, and enhanced differentiation along neuronal and astroglial axes.Citation2 In further support of a tumor suppressive role of miR-182 in GBM, our studies in orthotopic xenograft models using transformed glioma cells and mesenchymal GICs harboring low levels of endogenous miR-182 showed that ectopic expression of miR-182 increases animal subject survival, and slows tumor growth, as determined by measurement of tumor weights, and by bioluminescence imaging of luciferase-modified tumors.Citation2 Thus, our studies point to miR-182 as a critical tumor suppressor and prognostic factor in GBM, which integrates cell death, growth and differentiation processes in GBM.
miR-182 has been the topic of extensive research in developmental and cancer biology. The miR-183/96/182 cluster regulates gene expression in neurosensory tissues. The cluster is transcribed in the same direction, the individual miRNAs have similar sequence and can be co-expressed. miR-182 together with its family members, miR-96 and miR-183, were first described as developmental regulators in the mouse inner ear, where a spatiotemporal expressing profiling indicated a very restricted and dynamic expression. miR-182 and miR-183 are co-expressed, whereas miR-96, which has almost identical seed sequence and gene targets as miR-182, is expressed at different developmental stages. Similarly, miR-182 is abundantly expressed in the mouse retina and dorsal root gangliaCitation46,47 During embryonic development, miR-182 levels are low, but significantly increase post-natally, to drive terminal differentiation of retinal progenitor cells and to maintain mature retinal neuron function. Mechanistically, miR-182 promotes cellular differentiation and mesenchymal to epithelial transition, by regulating the expression of the transcription factor SNAI2.Citation48 In summary, these and many additional studies identified miR-182 as a critcal factor that controls cell fate specification and cell differentiation during development through targeting of a diverse network of developmental genes.Citation49,50
In vivo miRNA delivery for GBM therapy
With the advent of functional genomics, precision cancer medicine has begun to enter clinical practice. Personalized therapeutic regimens target specific genetic aberrations that drive disease progression. The majority of cancer genes, however, represent unprecedented, non-enzymatic targets with unknown function that cooperate to promote tumor growth and therapy resistance. To target the complex, undruggable oncogenome, RNA interference technologies have been developed. Delivery of these small, negatively charged RNAs is particularly challenging, due to the inherent instability of RNA molecules, limited bioavailability, and endosomsal entrapment.Citation49 Thus, siRNA and miRNA delivery remains a significant challenge and an unmet clinical need. For the systemic treatment of GBM, the presence of the BBB/BTBs represents a major physiological barrier. Tight endothelial cell junctions do not permit extensive drug trespassing, and thus, reduce therapy efficacy.Citation51 miRNA-based therapeutics are an emerging class of cancer treatment, as they can target multiple genes and pathways, to restrain unabated expansion of genomically and genetically high complex cancers, such as GBM.
miRNAs have already been used as therapies in clinical trials. Miravirsen is a miR-122 antagonist used for the treatment of Hepatitis C virus infectionCitation52 and TargoMiR, a miR-16 mimic, has been used in patients with recurrent malignant pleural mesothelioma (MPM) and advanced non-small cell lung cancer (NSCLC), delivered intavenously with epidermal growth factor receptor (EGFR)-targeted minicells.Citation53-55 A variety of nanoparticle-based delivery methods have been developed for the robust and safe delivery of siRNA and miRNA payloads, including inorganic, polymer or lipid-based nanoparticles.Citation2,56 A silica-based nanoparticle conjugated with a disialoganglioside GD2 antibody as targeting moiety was used to express miR-34a in neuroblastoma, which reduced tumor growth and vascularization by decreasing the levels of N-myc and increasing tissue inhibitor of metallopeptidase 2 (TIMP2).Citation57 Polymer-based particles, such as poly(lactic-co-glycolic acid) (PLGA) and polyethyleneimine (PEI) based materials were used to transfer miR-21 and -10b antagonists, and miR-145 into colon carcinomas,Citation58 where they regulated the avian myelocytomatosis viral oncogene (c-Myc) and ERK5 (see for nano-based miRNA therapeutics Citation59-62).
Table 1. Summary of nanomaterial-based carriers for miRNA delivery.
We have developed miR-182-based Spherical Nucleic Acids (182-SNAs), which consist of 13 nm gold nanoparticles densely functionalized with highly oriented miR-182 sequences. Robust cell penetration and activity of these SNAs were supported by confocal microscopy and western blot results, showing intracellular accumulation and significantly decreased Bcl2L12 and c-Met protein levels, as compared to Co-SNA-treated cultures. In addition, 182-SNAs amplified apoptosis responses, and diminished proliferation of glioma cells. In vivo, when administered intravenously into various orthotopic GBM xenograft models, 182-SNAs crossed an intact BBB in non-glioma bearing mice (∼1010 particles per gram of brain tissue as determined by ICP-MS), and via enhanced permeability and retention of the tumor-associated vasculature, showed even higher accumulation in intracranial gliomas (∼1.5 × 1011 particles per gram of tissue). Pharmacokinetic and biodistribution analyses provided evidence of significant brain tumor retention, with brain clearance of 85% of SNAs within 72 hours post injection.Citation2,3 Importantly, we found highly significant reduction in tumor burden of 182-SNA-treated orthotopic xenograft-bearing mice as assessed by analyses of tumor weight and bioluminescence imaging of a luciferase-labeled mesenchymal GIC model, and increased animal subject survival in the absence of adverse side effects and immunogenicity. Hence, our results indicate that 182-SNAs represent a novel strategy for the precision treatment of GBM, which can overcome some of major challenges of drug delivery to the CNS.
Future studies
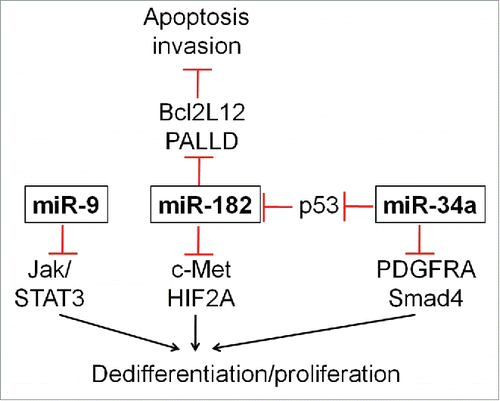
In future studies, we will develop combinatorial treatment strategies to further improve anti-tumor activity of 182-SNA monotherapy. We envision using 182-SNAs as adjuvant for TMZ. As TMZ increases the glioma stem cell compartment,Citation63 we will test the hypothesis that due to the pro-differentiation effects of miR182, 182-SNAs, in addition to its ability to regulate Bcl2L12, will increase TMZ effectiveness by promoting a more differentiated tumor phenotype. In addition, we will evaluate whether miR-9 and miR-34a, 2 additional high-priority miRNAs show enhanced anti-tumor effects when combined with miR-182 (). As pointed out above, miR-34a suppresses p53, a negative regulator of miR-182 and downregulates PDGFRA and a Smad4 transcriptional network. miR-9 is a core regulator of GBM differentiation, inhibits oncogenic JAK-STAT3-CEBP-β and suppresses growth and dedifferentiation of neural progenitor cells.Citation25 Finally, to simultaneously deliver miR-182 and miR-9 or miR-34a, bi- or trimodal SNA conjugates will be designed to target multiple oncogenic pathways. These conjugates are expected to display greater anti-tumor effect when compared to SNAs conjugated with a single miRNA.
Figure 1. Identification of miR-182 as a high-priority miRNA in GBM. To identify high-priority miRNAs, 470 miRNAs were filtered based on variable expression [green box, low expressing miRNAs, MAD ≤ 0.1; red box, highly variable miRNAs, MAD ≥ 1] correlation with patient survival, neuro-developmental annotation, ability to sensitize cells toward temozolomide-induced apoptosis (miR-182 increases, anti-miR-182 decreases caspase-3/7 activation), and association with oligoneural tumors. miR-182 emerged as the only miRNA that fulfills all criteria. Of note, a positive correlation between miR-182 expression and patient survival is most prominent in GBM with proneural/oligoneural characteristics.
![Figure 1. Identification of miR-182 as a high-priority miRNA in GBM. To identify high-priority miRNAs, 470 miRNAs were filtered based on variable expression [green box, low expressing miRNAs, MAD ≤ 0.1; red box, highly variable miRNAs, MAD ≥ 1] correlation with patient survival, neuro-developmental annotation, ability to sensitize cells toward temozolomide-induced apoptosis (miR-182 increases, anti-miR-182 decreases caspase-3/7 activation), and association with oligoneural tumors. miR-182 emerged as the only miRNA that fulfills all criteria. Of note, a positive correlation between miR-182 expression and patient survival is most prominent in GBM with proneural/oligoneural characteristics.](/cms/asset/da32291c-6220-4e95-be5f-69b2177e02d8/kccy_a_1093711_f0001_c.gif)
Disclosure of Potential Conflicts of Interest
AHS is an advisory board member of Exicure.
References
- Furnari FB, Fenton T, Bachoo RM, Mukasa A, Stommel JM, Stegh A, Hahn WC, Ligon KL, Louis DN, Brennan C, et al., Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 2007; 21(21): p. 2683-710; PMID:17974913; http://dx.doi.org/10.1101/gad.1596707
- Kouri FM, Hurley LA, Daniel WL, Day ES, Hua Y, Hao L, Peng CY, Merkel TJ, Queisser MA, Ritner C, et al., miR-182 integrates apoptosis, growth, and differentiation programs in glioblastoma. Genes Dev 2015; 29(7): p. 732-45; PMID:25838542; http://dx.doi.org/10.1101/gad.257394.114
- Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et al., Spherical Nucleic Acid Nanoparticle Conjugates as an RNAi-Based Therapy for Glioblastoma. Sci Transl Med 2013; 5(209): p. 209ra152; PMID:24174328; http://dx.doi.org/10.1126/scitranslmed.3006839
- Mirkin CA, Stegh AH. Spherical nucleic acids for precision medicine. Oncotarget 2014; 5(1): p. 9-10; PMID:24398537; http://dx.doi.org/10.18632/oncotarget.1757
- Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 2009; 11(3): p. 228-34; PMID:19255566; http://dx.doi.org/10.1038/ncb0309-228
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004; 116(2): p. 281-97; PMID:14744438; http://dx.doi.org/10.1016/S0092-8674(04)00045-5
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011 12(12): p. 861-74; PMID:22094949; http://dx.doi.org/10.1038/nrg3074
- Coolen M, Bally-Cuif L. MicroRNAs in brain development and physiology. Curr Opin Neurobiol 2009; 19(5): p. 461-70; PMID:19846291; http://dx.doi.org/10.1016/j.conb.2009.09.006
- Wang W, Kwon EJ, Tsai LH. MicroRNAs in learning, memory, and neurological diseases. Learn Mem 2012 19(9): p. 359-68; PMID:22904366; http://dx.doi.org/10.1101/lm.026492.112
- Schaefer A, O'Carroll D, Tan CL, Hillman D, Sugimori M, Llinas R, Greengard P. Cerebellar neurodegeneration in the absence of microRNAs. J Exp Med 2007; 204(7): p. 1553-8; PMID:17606634; http://dx.doi.org/10.1084/jem.20070823
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells 2006; 24(4): p. 857-64; PMID:16357340; http://dx.doi.org/10.1634/stemcells.2005-0441
- Conaco C, Otto S, Han JJ, Mandel G. Reciprocal actions of REST and a microRNA promote neuronal identity. Proc Natl Acad Sci U S A 2006; 103(7): p. 2422-7; PMID:16461918; http://dx.doi.org/10.1073/pnas.0511041103
- Packer AN, Xing Y, Harper SQ, Jones L, Davidson BL. The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J Neurosci 2008; 28(53): p. 14341-6; PMID:19118166; http://dx.doi.org/10.1523/JNEUROSCI.2390-08.2008
- Makeyev EV, Zhang J, Carrasco MA, Maniatis T. The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol Cell 2007; 27(3): p. 435-48; PMID:17679093; http://dx.doi.org/10.1016/j.molcel.2007.07.015
- Cheng LC, Pastrana E, Tavazoie M, Doetsch F. miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat Neurosci 2009; 12(4): p. 399-408; PMID:19287386; http://dx.doi.org/10.1038/nn.2294
- Meng Y, Zhang Y, Tregoubov V, Janus C, Cruz L, Jackson M, Lu WY, MacDonald JF, Wang JY, Falls DL, et al., Abnormal spine morphology and enhanced LTP in LIMK-1 knockout mice. Neuron 2002; 35(1): p. 121-33; PMID:12123613; http://dx.doi.org/10.1016/S0896-6273(02)00758-4
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature 2006; 439(7074): p. 283-9; PMID:16421561; http://dx.doi.org/10.1038/nature04367
- Siegel G, Obernosterer G, Fiore R, Oehmen M, Bicker S, Christensen M, Khudayberdiev S, Leuschner PF, Busch CJ, Kane C, et al., A functional screen implicates microRNA-138-dependent regulation of the depalmitoylation enzyme APT1 in dendritic spine morphogenesis. Nat Cell Biol 2009; 11(6): p. 705-16; PMID:19465924; http://dx.doi.org/10.1038/ncb1876
- Le MT, Teh C, Shyh-Chang N, Xie H, Zhou B, Korzh V, Lodish HF, Lim B. MicroRNA-125b is a novel negative regulator of p53. Genes Dev 2009; 23(7): p. 862-76; PMID:19293287; http://dx.doi.org/10.1101/gad.1767609
- Walker JC, Harl RM. microRNA-24a is required to repress apoptosis in the developing neural retina. Genes Dev 2009; 23(9): p. 1046-51; PMID:19372388; http://dx.doi.org/10.1101/gad.1777709
- Silber J, James CD, Hodgson JG. microRNAs in gliomas: small regulators of a big problem. Neuromolecular Med 2009; 11(3): p. 208-22; PMID:19731102; http://dx.doi.org/10.1007/s12017-009-8087-9
- Nagane M, Levitzki A, Gazit A, Cavenee WK, Huang HJ. Drug resistance of human glioblastoma cells conferred by a tumor-specific mutant epidermal growth factor receptor through modulation of Bcl-XL and caspase-3-like proteases. Proc Natl Acad Sci U S A 1998; 95(10): p. 5724-9; PMID:9576951; http://dx.doi.org/10.1073/pnas.95.10.5724
- Stiles CD, Rowitch DH. Glioma stem cells: a midterm exam. Neuron 2008; 58(6): p. 832-46; PMID:18579075; http://dx.doi.org/10.1016/j.neuron.2008.05.031
- Gilbertson RJ, Rich JN. Making a tumour's bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer 2007; 7(10): p. 733-6; PMID:17882276; http://dx.doi.org/10.1038/nrc2246
- Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 2011; 71(9): p. 3387-99; PMID:21385897; http://dx.doi.org/10.1158/0008-5472.CAN-10-4117
- Zhou X, Ren Y, Moore L, Mei M, You Y, Xu P, Wang B, Wang G, Jia Z, Pu P, et al., Downregulation of miR-21 inhibits EGFR pathway and suppresses the growth of human glioblastoma cells independent of PTEN status. Lab Invest 2010; 90(2): p. 144-55; PMID:20048743; http://dx.doi.org/10.1038/labinvest.2009.126
- Kwak HJ, Kim YJ, Chun KR, Woo YM, Park SJ, Jeong JA, Jo SH, Kim TH, Min HS, Chae JS, et al., Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene 2011; 30(21): p. 2433-42; PMID:21278789; http://dx.doi.org/10.1038/onc.2010.620
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, Krichevsky AM. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 2008; 28(17): p. 5369-80; PMID:18591254; http://dx.doi.org/10.1128/MCB.00479-08
- Zhou X, Zhang J, Jia Q, Ren Y, Wang Y, Shi L, Liu N, Wang G, Pu P, You Y, et al., Reduction of miR-21 induces glioma cell apoptosis via activating caspase 9 and 3. Oncol Rep 2010; 24(1): p. 195-201; PMID:20514462
- Gabriely G, Teplyuk NM, Krichevsky AM, Context effect: microRNA-10b in cancer cell proliferation, spread and death. Autophagy 2011; 7(11): p. 1384-6; PMID:21795860; http://dx.doi.org/10.4161/auto.7.11.17371
- Gabriely G, Yi M, Narayan RS, Niers JM, Wurdinger T, Imitola J, Ligon KL, Kesari S, Esau C, Stephens RM, et al., Human glioma growth is controlled by microRNA-10b. Cancer Res 2011; 71(10): p. 3563-72; PMID:21471404; http://dx.doi.org/10.1158/0008-5472.CAN-10-3568
- Guessous F, Alvarado-Velez M, Marcinkiewicz L, Zhang Y, Kim J, Heister S, Kefas B, Godlewski J, Schiff D, Purow B , et al., Oncogenic effects of miR-10b in glioblastoma stem cells. J Neurooncol 2013; 112(2): p. 153-63; PMID:23307328; http://dx.doi.org/10.1007/s11060-013-1047-0
- Huse JT, Brennan C, Hambardzumyan D, Wee B, Pena J, Rouhanifard SH, Sohn-Lee C, le Sage C, Agami R, Tuschl T, et al., The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev 2009; 23(11): p. 1327-37; PMID:19487573; http://dx.doi.org/10.1101/gad.1777409
- Kim H, Huang W, Jiang X, Pennicooke B, Park PJ, Johnson MD. Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci U S A 2010; 107(5): p. 2183-8; PMID:20080666; http://dx.doi.org/10.1073/pnas.0909896107
- Li Y, Guessous F, Zhang Y, Dipierro C, Kefas B, Johnson E, Marcinkiewicz L, Jiang J, Yang Y, Schmittgen TD, et al., MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 2009; 69(19): p. 7569-76; PMID:19773441; http://dx.doi.org/10.1158/0008-5472.CAN-09-0529
- Silber J, Jacobsen A, Ozawa T, Harinath G, Pedraza A, Sander C, Holland EC, Huse JT. miR-34a repression in proneural malignant gliomas upregulates expression of its target PDGFRA and promotes tumorigenesis. PLoS One 2012; 7(3): p. e33844; PMID:22479456; http://dx.doi.org/10.1371/journal.pone.0033844
- Genovese G, Ergun A, Shukla SA, Campos B, Hanna J, Ghosh P, Quayle SN, Rai K, Colla S, Ying H, et al., microRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-beta signaling in glioblastoma. Cancer Discov 2012; 2(8): p. 736-49; PMID:22750848; http://dx.doi.org/10.1158/2159-8290.CD-12-0111
- Cancer Genome Atlas Research, N., Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008; 455(7216): p. 1061-8; PMID:18772890; http://dx.doi.org/10.1038/nature07385
- Huang PH, Mukasa A, Bonavia R, Flynn RA, Brewer ZE, Cavenee WK, Furnari FB, White FM. Quantitative analysis of EGFRvIII cellular signaling networks reveals a combinatorial therapeutic strategy for glioblastoma. Proc Natl Acad Sci U S A 2007; 104(31): p. 12867-72; PMID:17646646; http://dx.doi.org/10.1073/pnas.0705158104
- Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C, et al., Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science 2007; 318(5848): p. 287-90; PMID:17872411; http://dx.doi.org/10.1126/science.1142946
- Boccaccio C, Comoglio PM. Invasive growth: a MET-driven genetic programme for cancer and stem cells. Nat Rev Cancer 2006; 6(8): p. 637-45; PMID:16862193; http://dx.doi.org/10.1038/nrc1912
- Boccaccio C, Comoglio PM. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Res 2013; 73(11): p. 3193-9; PMID:23695554; http://dx.doi.org/10.1158/0008-5472.CAN-12-4039
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al., Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell 2009; 15(6): p. 501-13; PMID:19477429; http://dx.doi.org/10.1016/j.ccr.2009.03.018
- Boccaccio C, Comoglio PM. The MET oncogene in glioblastoma stem cells: implications as a diagnostic marker and a therapeutic target. Cancer Res 2013; 73(11): p. 3193-9; PMID:23695554; http://dx.doi.org/10.1158/0008-5472.CAN-12-4039
- Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai CR, et al., mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 2014; 510(7505): p. 393-6; PMID:24870234
- Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell 2006; 11(4): p. 441-50; PMID:17011485; http://dx.doi.org/10.1016/j.devcel.2006.09.009
- Weston MD, Pierce ML, Jensen-Smith HC, Fritzsch B, Rocha-Sanchez S, Beisel KW, Soukup GA. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev Dyn 2011; 240(4): p. 808-19; PMID:21360794; http://dx.doi.org/10.1002/dvdy.22591
- Qu Y, Li WC, Hellem MR, Rostad K, Popa M, McCormack E, Oyan AM, Kalland KH, Ke XS. MiR-182 and miR-203 induce mesenchymal to epithelial transition and self-sufficiency of growth signals via repressing SNAI2 in prostate cells. Int J Cancer 2013; 133(3): p. 544-55; PMID:23354685; http://dx.doi.org/10.1002/ijc.28056
- Kiernan AE, Pelling AL, Leung KK, Tang AS, Bell DM, Tease C, Lovell-Badge R, Steel KP, Cheah KS. Sox2 is required for sensory organ development in the mammalian inner ear. Nature 2005; 434(7036): p. 1031-5; PMID:15846349; http://dx.doi.org/10.1038/nature03487
- Brooker R, Hozumi K, Lewis J. Notch ligands with contrasting functions: Jagged1 and Delta1 in the mouse inner ear. Development 2006; 133(7): p. 1277-86; PMID:16495313; http://dx.doi.org/10.1242/dev.02284
- Kim SS, Harford JB, Pirollo KF, Chang EH. Effective treatment of glioblastoma requires crossing the blood-brain barrier and targeting tumors including cancer stem cells: the promise of nanomedicine. Biochem Biophys Res Commun 2015; http://dx.doi.org/10.1016/j.bbrc.2015.06.137
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, et al., Treatment of HCV infection by targeting microRNA. N Engl J Med 2013; 368(18): p. 1685-94; PMID:23534542; http://dx.doi.org/10.1056/NEJMoa1209026
- Reid G, Pel ME, Kirschner MB, Cheng YY, Mugridge N, Weiss J, Williams M, Wright C, Edelman JJ, Vallely MP, et al., Restoring expression of miR-16: a novel approach to therapy for malignant pleural mesothelioma. Ann Oncol 2013; 24(12): p. 3128-35; PMID:24148817; http://dx.doi.org/10.1093/annonc/mdt412
- MacDiarmid JA, Mugridge NB, Weiss JC, Phillips L, Burn AL, Paulin RP, Haasdyk JE, Dickson KA, Brahmbhatt VN, Pattison ST, et al., Bacterially derived 400 nm particles for encapsulation and cancer cell targeting of chemotherapeutics. Cancer Cell 2007; 11(5): p. 431-45; PMID:17482133; http://dx.doi.org/10.1016/j.ccr.2007.03.012
- MacDiarmid JA, Amaro-Mugridge NB, Madrid-Weiss J, Sedliarou I, Wetzel S, Kochar K, Brahmbhatt VN, Phillips L, Pattison ST, Petti C, et al., Sequential treatment of drug-resistant tumors with targeted minicells containing siRNA or a cytotoxic drug. Nat Biotechnol 2009; 27(7): p. 643-51; PMID:19561595; http://dx.doi.org/10.1038/nbt.1547
- Jensen SA, Day ES, Ko CH, Hurley LA, Luciano JP, Kouri FM, Merkel TJ, Luthi AJ, Patel PC, Cutler JI, et al., Spherical nucleic acid nanoparticle conjugates as an RNAi-based therapy for glioblastoma. Sci Transl Med 2013; 5(209): p. 209ra152; PMID:24174328; http://dx.doi.org/10.1126/scitranslmed.3006839
- Tivnan A, Orr WS, Gubala V, Nooney R, Williams DE, McDonagh C, Prenter S, Harvey H, Domingo-Fernández R, Bray IM, et al., Inhibition of neuroblastoma tumor growth by targeted delivery of microRNA-34a using anti-disialoganglioside GD2 coated nanoparticles. PLoS One 2012; 7(5): p. e38129; PMID:22662276; http://dx.doi.org/10.1371/journal.pone.0038129
- Chiou GY, Cherng JY, Hsu HS, Wang ML, Tsai CM, Lu KH, Chien Y, Hung SC, Chen YW, Wong CI, et al., Cationic polyurethanes-short branch PEI-mediated delivery of Mir145 inhibited epithelial-mesenchymal transdifferentiation and cancer stem-like properties and in lung adenocarcinoma. J Control Release 2012; 159(2): p. 240-50; PMID:22285547; http://dx.doi.org/10.1016/j.jconrel.2012.01.014
- Muthiah M, Islam MA, Lee HJ, Moon MJ, Cho CS, Park IK. MicroRNA delivery with osmotic polysorbitol-based transporter suppresses breast cancer cell proliferation. Int J Biol Macromol 2015; 72: p. 1237-43; PMID:25450545; http://dx.doi.org/10.1016/j.ijbiomac.2014.10.041
- Qureshi AT, Doyle A, Chen C, Coulon D, Dasa V, Del Piero F, Levi B, Monroe WT, Gimble JM, Hayes DJ. Photoactivated miR-148b-nanoparticle conjugates improve closure of critical size mouse calvarial defects. Acta Biomater 2015; 12: p. 166-73; PMID:25462528; http://dx.doi.org/10.1016/j.actbio.2014.10.010
- Chen Y, Gao DY, Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv Drug Deliv Rev 2015; 81: p. 128-41; PMID:24859533; http://dx.doi.org/10.1016/j.addr.2014.05.009
- Wang H, Jiang Y, Peng H, Chen Y, Zhu P, Huang Y. Recent progress in microRNA delivery for cancer therapy by non-viral synthetic vectors. Adv Drug Deliv Rev 2015; 81: p. 142-60; PMID:25450259; http://dx.doi.org/10.1016/j.addr.2014.10.031
- Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU, Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ 2014; 21(7): p. 1119-31; PMID:24608791; http://dx.doi.org/10.1038/cdd.2014.31