ABSTRACT
Mediator is considered an enhancer of RNA-Polymerase II dependent transcription but its function and regulation in pluripotent mouse embryonic stem cells (mESCs) remains unresolved. One means of controlling the function of Mediator is provided by the binding of the Cdk8 module (Med12, Cdk8, Ccnc and Med13) to the core Mediator. Here we report that Med12 operates together with PRC1 to silence key developmental genes in pluripotency. At the molecular level, while PRC1 represses genes it is also required to assemble ncRNA containing Med12-Mediator complexes. In the course of cellular differentiation the H2A ubiquitin binding protein Zrf1 abrogates PRC1-Med12 binding and facilitates the association of Cdk8 with Mediator. This remodeling of Mediator-associated protein complexes converts Mediator from a transcriptional repressor to a transcriptional enhancer, which then mediates ncRNA-dependent activation of Polycomb target genes. Altogether, our data reveal how the interplay of PRC1, ncRNA and Mediator complexes controls pluripotency and cellular differentiation.
Introduction
Polycomb group repressor complexes (PRCs) are regulators of gene expression and participate in the establishment and maintenance of embryonic stem cell fates.Citation1-3 Over the last decade the Polycomb repressive complex 1 (PRC1) and the Polycomb repressive complex 2 (PRC2)Citation4,5 have been studied thoroughly. PRC2 catalyzes the tri-methylation of histone H3 at lysine 27 (H3K27me3), which represents a hallmark of transcriptional repression and it is usually deposited at promoters of Polycomb target genes.Citation6 PRC1 generates a mono-ubiquitylation of histone H2A at lysine 119 (H2AK119ub) via its E3 ubiquitin ligase subunit Ring1A or Ring1B.Citation7 PRCs can operate successively or independently of each other to establish repression of genes. The PRC2 catalyzed H3K27me3 mark constitutes a docking site for Cbx proteins, which are subunits of PRC1 and mediate its recruitment to chromatin.Citation8,9 In mouse embryonic stem cells (mESCs) Cbx7 is the prevalent Cbx protein that controls the expression of key developmental genes during pluripotency. In the course of differentiation Cbx7 is replaced by other proteins of the Cbx family (Cbx2 and Cbx4), which play a role in later stages of development.Citation10,11 Another type of PRC1 in mESCs, which is recruited to chromatin independent of the H3K27me3 mark, comprises RYBP instead of Cbx7.Citation12 PRC1 is thought to repress transcription via direct interaction with the general transcriptional machinery and it has been proposed that Mediator subunits might facilitate this repressive function.Citation13 Further, transcriptional repression by PRC1 is likely independent of the H2AK119ub mark and rather a function of chromatin condensation.Citation14-17 PRC1 is dislocated from chromatin by Zrf1, which tethers to H2AK119ub via a specific ubiquitin-binding domain.Citation18 Hence, PRC1-mediated ubiquitylation of H2A is a prerequisite for Zrf1 recruitment and thereby PRC1 dislocation. In higher eukaryotes, the carboxy-terminus of Zrf1 harbors 2 SANT domains, which are thought to directly interact with DNA.Citation19,20 During cellular differentiation Zrf1 is recruited to promoters of Polycomb target genes where it induces the derepression of key developmental genes. Furthermore, Zrf1 is implicated in the regulation of stem cell identity and carcinogenesis.Citation21,22
A global regulator of transcription is the Mediator complex.Citation23,24 At pre-initiation complexes (PICs) Mediator conveys the activating signals from enhancers to promoters via interacting with both transcription factors and the basal transcriptional machinery.Citation25-27 Two major forms of Mediator complexes have been identified by mass spectroscopy. These consist of a core complex with or without association of the Cdk8 module,Citation28,29 which comprises the 4 proteins Cdk8, Med13, Ccnc and Med12.Citation30-33 The Cdk8 module or RNA Polymerase II (Pol II) bind mutually exclusive to Mediator, which likely constitutes a means of regulating Mediator function.Citation26,30,34,35 Still, the function of the Cdk8 module is under debate since its subunits have been implicated in both, transcriptional activation and repression.Citation29,36-40 In particular, Med12 was shown to operate as a signaling pathway hub in C. elegans suggesting a function in different cellular pathways.Citation41 Furthermore, Med12 is involved in the Polycomb-mediated repression of Hox genes in D. melanogaster contrasting a general role in transcriptional activation.Citation37 Mediator is also known to act in concert with transcriptionally activating ncRNAs.Citation42,43 These ncRNAs are transcribed in close proximity to the target gene promoter and they associate with Med12 to promote DNA looping from the ncRNA locus to the target gene enhancing transcription. However, how Mediator and Med12 operate in both transcriptional activation and silencing is still poorly understood.
Here we investigated the relationship of the Cdk8 module in PRC1-mediated gene silencing. Our data provide evidence that Med12 fulfills a dual role in gene repression and activation. During pluripotency it acts in concert with PRC1 to repress genes, which are required for differentiation and development, whereas in early stem cell differentiation Med12 activates genes in a ncRNA-dependent manner. This switch of Med12 function is facilitated by Zrf1, which dislocates PRC1 from ncRNA-Mediator complexes allowing for the incorporation of Cdk8 into the Mediator complex.
Results
PRC1 and Mediator occupy similar chromatin regions
PRC1 and Mediator constitute important regulators of transcription in mESCs. To investigate a potential interplay of PRC1 and Mediator we examined the genome-wide occupancy of PRC1 and Mediator in mESCs. To this end, we explored existing ChIP sequencing data sets.Citation10,44-47 Additionally we performed peak calling for the kinase module subunit Cdk8 (GSE44288; q value: 1e−5) since no qualitative occupancy data were available. When analyzing the genome-wide localization of Med12, Cdk8 and Ring1b we observed 2617 of Med12 target genes (26% of the Med12 targets) to be also occupied by Cdk8 ( and Table S1). Ring1b and Med12 occupy a similar number (2156) of genes (21% of the Med12 targets). Surprisingly only 486 genes (4,8% of the Med12 targets) are targeted by Cdk8, Med12 and Ring1b together. This narrow overlap of all 3 factors suggests diverging functions of Med12 with either Cdk8 or Ring1b in mESC pluripotency. Intrigued by the close relationship of Ring1b and Med12 we next investigated the genome-wide occupancy of the core Mediator subunit Med1 and other PRC subunits. Notably, at the majority of genes that are targeted by Med12 and Ring1b (80%), but not Cdk8, Med1 is also present, suggesting co-localization of Ring1b, Med12 and the core Mediator complex (). Gene Ontology (GO) term analysis predicts that genes bound by both Med12 and Ring1b are essential for development and differentiation (, S1C-E and Table S1) whereas Med12 and Cdk8 are predominantly located at metabolic genes, which are highly expressed during pluripotency (, S1F and S2C). This suggests that a subset of Mediator complexes might exert a specific function in conjunction with Ring1b in mESCs.
Figure 1. Ring1b and Med12 target key developmental genes in mouse ESCs. (A) Venn diagram and heatmap of Med12, Ring1b and Cdk8 target genes in mESCs. For the Cdk8 dataset (GSE44288) we performed peak calling with a q value of 1e-5 and annotations with the closest gene promoter (+/−5 kb around the TSS). For the other data sets the qualitative occupancy data were downloaded from the supplementary material of the respective publications. (B) Venn diagram of common Ring1b-Med12 target genes (Ring1b-Med12 only-without Cdk8-, 1670 genes) that overlap with Med1 target genes. (C) GO-enriched terms of Med12-Ring1b and (D) Med12-Cdk8 target genes in mESCs. FDR represents the corrected P-value. (E-F) Med12 but not Cdk8 localize with Cbx7-PRC1 at promoters of repressed differentiation genes in mESCs. Med12 and Cdk8, but not PRC1/PRC2, localize at promoters of expressed genes in mESCs. Screenshots from ChIP-seq profiles of Med1, Med12, Cdk8, Ring1b, Cbx7, RYBP and Suz12 for selected genes. The expression levels of the selected genes (Msx2 and Nanog) are shown. (G) Density distributions of Med12+Ring1b (red) and Med12+Cdk8 (blue) target genes expression. The majority of Med12+Cdk8 targets are expressed at high levels in mESCs whereas the Med12+Ring1b targets have low or no expression. The y-axis shows the density (probability). We omitted this axis from the graph as we only intended to focus on the shift in the x-axis.
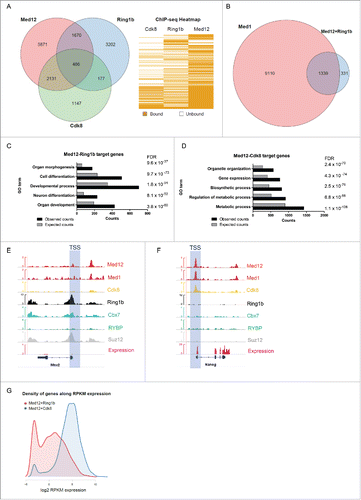
In support of this, we observed that Med12 and Med1 co-localize with both PRC2 and Cbx7-PRC1 at developmental genes, which are not occupied by Cdk8 (, S2A and Table S1). Likewise, Med12 and Med1 co-localize with RYBP-PRC1 target genes, which are not occupied by PRC2 or Cdk8 (Figure S2B and Table S1). In stark contrast, genes that are bound by both Med12 and Cdk8 are not occupied by PRC1 or PRC2 (, S2C and Table S1). Analyzing the relative expression of Med12-Ring1b targets we found that these genes are either silenced or expressed at low levels suggesting that Med12 might exert a function in Polycomb-mediated silencing (). In comparison, Med12-Cdk8 target genes are usually highly expressed (). Collectively, these data suggest that Med12 and Ring1b have a common function in the repression of developmental genes in mESCs. In conjunction with Cdk8, Med12 seems to play a different role, which is probably linked to its enhancer function in gene expression.
Med12 associates with chromatin as a function of Ring1b
Based on our previous findings we decided to study the potential role of Med12 in Polycomb-mediated silencing. To this end we generated mESC lines stably expressing either a non-specific shRNA (shNMC) or shRNAs targeting Ring1b (shRing1b) or Med12 (shMed12), respectively. First, we analyzed the relative mRNA levels (Figure S3A) and protein levels of Cdk8, Med12, and Ring1b (Figure S3B) in the aforementioned cell lines to verify the knockdown of the respective factors. We observed that depletion of one factor did not affect the expression or protein levels of the other factors tested. Next, we analyzed chromatin fractions from these cell lines either in pluripotency or after Retinoic Acid (RA) administration (). In line with previous publications, we observed in Ring1b knockdown cells dramatically reduced Ring1b and H2AK119ub levels at chromatin. Remarkably, association of both Med12 and Cdk8 with chromatin was strictly dependent on Ring1b. In contrast, in Med12 knockdown cells we found only very mild alterations of Ring1b levels at chromatin but a substantial decrease of Cdk8 recruitment during differentiation (). To further study the functional interplay of Med12, Ring1b and Mediator we performed immunoprecipitations with protein extracts from control and Ring1b knockdown cells employing Med12 antibodies (). We observed that the interaction of Med12 with the core Mediator subunit Med1 was dependent on Ring1b. To corroborate this finding we carried out immunoprecipitations from mESC nuclear extracts utilizing Ring1b antibodies (). We found a robust interaction between Ring1b and Med12 whereas neither the H2AK119ub-binding protein Zrf1 nor Cdk8 were binding to Ring1b substantiating a role for Med12 beyond its function in the Cdk8 module. To further validate our findings we performed ChIP experiments with control, Ring1b and Med12 knockdown mESCs (). We observed binding of both Med12 and Ring1b to the promoters of their target genes. At Med12-Ring1b target genes the recruitment of Med12 was impaired in Ring1b knockdown cells whereas Ring1b occupancy at the common targets was only slightly altered upon Med12 knockdown (). At Med12-Cdk8 target genes, Med12 occupancy was affected in both Ring1b and Med12 knockdown cell lines, which is in agreement with our previous findings (). Notably, we did not observe Ring1b binding to the promoters of Med12-Cdk8 target genes () suggesting that either Ring1b has an indirect effect on Med12 occupancy or that Ring1b binds only transiently to these promoters.
Figure 2. Interdependency of Ring1b and Med12 recruitment to chromatin. (A) Ring1b regulates the recruitment of subunits of the kinase module of Mediator to chromatin. Pluripotent and early differentiating mESCs (shNMC, shMed12 and shRing1b) were fractionated and the chromatin fractions were analyzed employing the respective antibodies. The arrowhead shows the corresponding band for Cdk8. The relative intensities of the protein levels were calculated as fold change to 0 timepoint shNMC samples +/− SEM, N=4, using the ponceau intensities as reference. (B) Ring1b controls the association of Med12 with the core Mediator protein Med1. Endogenous immunoprecipitations (IPs) with Med12 antibodies and protein extracts from control (shNMC) and Ring1b (shRing1b) knockdown mESCs. The precipitated material was analyzed by western blotting with the indicated antibodies. Inputs represent 7% of the material used for each IP. (C) Endogenous IPs with Ring1b antibodies and nuclear extracts from mESCs. The precipitated material was analyzed by western blotting with the indicated antibodies. Inputs represent 7% of the material used for each IP. (D) Ring1b regulates the recruitment of Med12 to promoters of Med12+Ring1b and Med12+Cdk8 target genes in mESCs. Chromatin immunoprecipitations (ChIPs) using chromatin from control (shNMC), Med12 (shMed12) and Ring1b (shRing1b) knockdown mESCs followed by qPCRs on selected promoters. The enrichments are represented as percentage (%) of recovery over input and correspond to the average of 3 independent experiments +/− SEM *** P-value ≤ 0.0003, ** P-value = 0.005, *P-value ≤ 0.03 as calculated by 2-tailed unpaired t-test. (E) Med12 does not affect Ring1b levels at the promoters of Med12+Ring1b target genes in mESCs. Ring1b is not enriched at promoters of Med12+Cdk8 target genes in mESCs. Chromatin immunoprecipitations (ChIPs) using chromatin from control (shNMC), Med12 (shMed12) and Ring1b (shRing1b) knockdown mESCs followed by qPCR on selected promoters. The enrichments are represented as percentage (%) of recovery over input and correspond to the average of 3 independent experiments +/− SEM ** P-values ≤ 0.007 as calculated by 2-tailed unpaired t test.
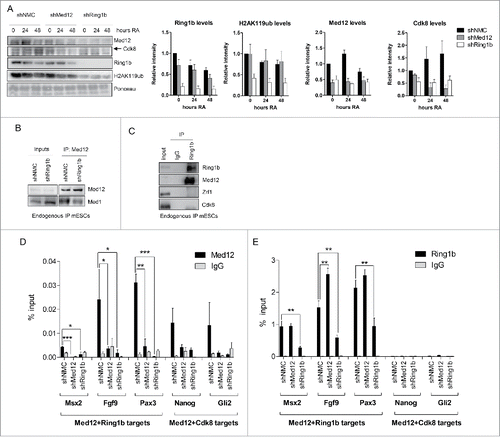
Taken together, our data suggest that Ring1b physically interacts with Med12 and that it controls both Med12 binding to Mediator and Med12 occupancy at promoters of developmental target genes.
Ring1b and Med12 repress key developmental genes during pluripotency
The physical interaction of Med12 and Ring1b and their occupancy at promoters of key developmental genes suggests a joint function of both factors in transcriptional regulation of mESCs. To analyze the impact of PRC1 and Mediator on transcription we carried out RNA-sequencing of wildtype, control, Cdk8, Ring1b and Med12 knockdown mESCs. We observed 2109 differentially expressed genes in Med12 knockdown mESCs (shMed12) and 1020 genes with altered expression levels in Ring1b knockdown mESCs (shRing1b) (, S4A and Table S2). 598 of these genes were differentially expressed in both conditions (). In comparison, Cdk8 knockdown cells exhibit a lower total number of differentially expressed genes (236 genes) suggesting that Cdk8 probably plays a minor role in transcriptional regulation in mESCs or that its function could be compensated by other kinases as for example its paralog, Cdk19.Citation48 According to GO analysis Ring1b and Med12 knockdown mESCs are essential for the transcriptional regulation of developmental genes (), which is in line with their genome-wide occupancy in mESCs (). In contrast, the 171 genes differentially expressed in Med12 and Cdk8 knockdown cells () do not exhibit significantly enriched GO terms. Interestingly, the 598 genes differentially expressed in both Ring1b and Med12 knockdown cells were only slightly affected in Cdk8 knockdown cells and the expression profile after Cdk8 knockdown contrasts sharply with the expression profile of Med12 knockdown cells (). This suggests that Cdk8 and Med12 likely play different roles in regulating these 598 genes. Next, we validated the relative expression levels of a selected number of common Ring1b-Med12 target genes by quantitative PCR (). Overall we observed a derepression of the tested genes in Ring1b and Med12 knockdown cells during pluripotency, suggesting that Med12 and Ring1b exert a common function in gene silencing. In contrast, knockdown of Cdk8 exhibited only slight alterations in the expression levels of the respective genes further substantiating diverging roles for Med12 and Cdk8 in gene regulation during pluripotency. To verify whether Ring1b and Med12 occupy the promoters of the differentially expressed genes, we compared the common 2156 Med12-Ring1b target genes () with the 598 genes differentially expressed in Ring1b and Med12 knockdown mESCs (). We found 116 differentially expressed genes directly regulated by Med12 and Ring1b (), 100 of which were derepressed in the respective knockdown cells in mESCs (). When comparing the 2131 Med12-Cdk8 targets () with the 171 differentially expressed genes in shMed12 and shCdk8, we observed only 3 genes being directly regulated by both proteins during pluripotency (Fig. S4B and S4C).
Figure 3. Ring1b and Med12 repress key developmental genes in mESCs. (A) Venn diagram of differentially expressed (DE) genes in control and knockdown mESCs. (B) GO analysis of biological functions of genes differentially expressed in both shMed12 and shRing1b mESCs. (C) Heatmap illustrating the expression profile of the genes differentially expressed in shMed12/shRing1b in control (shNMC), wild type, Cdk8 (shCdk8), Med12 (shMed12) and Ring1b (shRing1b) knockdown mESCs. (D) Knockdown of either Ring1b or Med12 causes de-repression of their common target genes in mESCs. Shown are the relative expression levels of selected Ring1b/Med12 target genes in the aforementioned cell lines. The relative mRNA levels are represented as fold change to the shNMC samples +/− SEM, n = 3. *** P-value< 0.0001, ** P-value < 0.001, * P-value < 0.02, as calculated by 2-tailed, unpaired t test. (E) Med12 and Ring1b directly control the expression of 116 genes. Venn diagram illustrating the overlap between differentially expressed genes in shMed12 and shRing1b mESCs and their common ChIP targets (). (F) Med12 and Ring1b repress the common target genes in mESCs. The plot depicts the expression Fold Change (FC) of the 116 direct target genes (). 100 out of 116 target genes (∼86%) are derepressed in both shRing1b and shMed12 mESCs.
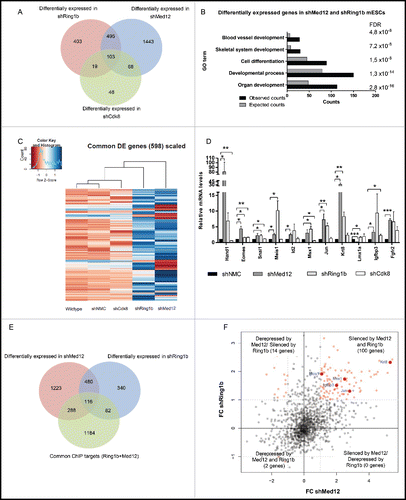
Collectively, these data suggest that both Med12 and Ring1b play an essential role in repressing key developmental genes during pluripotency. Cdk8 has a less pronounced impact on gene regulation during pluripotency likely suggesting that its function is compensated by other kinases as previously reported.
Functions of Ring1b, Med12 and Cdk8 during differentiation
Intrigued by the diverging functions of Med12-Ring1b and Med12-Cdk8 during pluripotency we addressed next whether Med12, Cdk8 and Ring1b play a role in mESC renewal. To this end we analyzed the expression of the pluripotency markers (Oct4, Sox2, Nanog, Klf4) in the respective knockdown cell lines (). The transcription levels of the pluripotency marker genes in Cdk8 depleted cells were similar to the levels observed in control mESCs. In Ring1b depleted mESCs only Sox2 levels were compromised while the expression of the other ES regulators remained unchanged in line with earlier reports.Citation49,50 Depletion of Med12 resulted in reduced levels of Oct4, Nanog and Sox2 in accordance with previous findings.Citation44 In addition we observed reduced levels of Klf4 in Med12 knockdown mESCs. Accordingly only Med12 knockdown mESCs show a tendency to spontaneously differentiate as judged by Alkaline Phosphatase staining (Figure S5A). To test whether Med12, Cdk8 and Ring1b have a function during differentiation we performed in vitro differentiation experiments. Hence, we generated embryoid bodies (EBs) from control, Ring1b, Cdk8 and Med12 knockdown cell lines and analyzed their morphology and size ( and S5B). EBs derived from shRing1b had in average smaller diameters than EBs generated from control cells ( and S5B) as previously reported.Citation10 Likewise, EBs generated from shMed12 cells exhibit smaller diameters as compared to the control EBs. In contrast, the diameters of Cdk8 knockdown and control EBs were heterogeneous in size. Taken together these data suggest that Med12 and Ring1b fulfill a critical role during in vitro differentiation. We next analyzed the relative expression of marker genes of all 3 germ layers in the aforementioned EBs throughout differentiation (). We observed that knockdown of Ring1b caused misregulation, either up- or down-regulation, of selected marker genes during EB differentiation in agreement with earlier reports.Citation10,51 In comparison, depletion of Cdk8 led only to mild alterations of the expression levels (Afp, Brachyury, Mixl1, Nestin) or caused increased expression (Gata6, VE-Cadherin, Claudin6, Gsc, Hand1, Otx2, Bmp4) of the marker genes when compared to control cells. Importantly, knockdown of Med12 caused a sustained repression of most of the developmental marker genes tested suggesting that Med12 plays a decisive role in the activation of these genes during differentiation. Similarly, when we analyzed Ring1b-Med12 target genes and other classical Polycomb target genes in RA triggered differentiation of mESCs, we noticed unchanged transcription in shCdk8 cells (). In contrast, depletion of either Med12 or Ring1b affected the expression levels of most of the tested genes significantly. Interestingly, depletion of Ring1b or Med12 showed either a similar impact on the transcription levels (shMed12=shRing1b) or opposing changes of the gene expression levels ( shMed12≠shRing1b).
Figure 4. Med12 and Ring1b knockdown mESCs do not properly differentiate. (A) shMed12 mESCs exhibit low levels of pluripotency markers. Shown are the relative mRNA levels of pluripotency markers in shNMC, shCdk8, shMed12 and shRing1b mESCs. The relative mRNA levels are depicted as fold change to the shNMC samples +/− SEM, n = 3. *** P-value< 0.0001, ** P-value < 0.001, * P-value < 0.02, as calculated by 2-tailed, unpaired t test. (B) Embryoid Bodies (EBs) derived from shMed12 and shRing1b mESCs, have smaller diameters than those from shNMC and shCdk8 mESCs. Images of EBs were taken at 4x magnification at 2, 4, 6 and 8 d after the beginning of spontaneous differentiation by LIF removal. The size of the scale bar corresponds to 500 µm. (C-E) Knockdown of Med12 causes sustained repression of differentiation marker genes from the 3 germ layers. The mRNA levels of each marker gene are represented as fold change to the levels of the Day 0 sample of each genotype. Data are represented as a mean +/− SEM, n =3. *** P-value < 0.0001, ** P-value < 0.001, * P-value < 0.02, as calculated by 2-tailed, unpaired t test. (F) Developmental genes get upregulated upon RA-induced differentiation. The relative mRNA levels of Med12-Ring1b targets (Wnt8a-Sox7) and other RA-responsive genes (Hoxb4-Hoxd11) at 24hRA are represented as fold change to the untreated samples (mESCs) +/− SEM, n = 3. ** P-value = 0.009, *P-value < 0.05, as calculated by 2-tailed, unpaired t test. (G) RA-induced differentiation genes (significantly induced from 4F) are affected in the same (shMed12 = shRing1b) or opposite ways (shMed12 ≠ shRing1b) upon Med12 or Ring1b depletion in early differentiation (24hRA). The relative mRNA levels of the tested genes are represented as fold change to the shNMC samples +/− SEM, n = 3. *** P-value =0.0004, ** P-value ≤ 0.008, * P-value ≤ 0.04, as calculated by 2-tailed, unpaired t test.
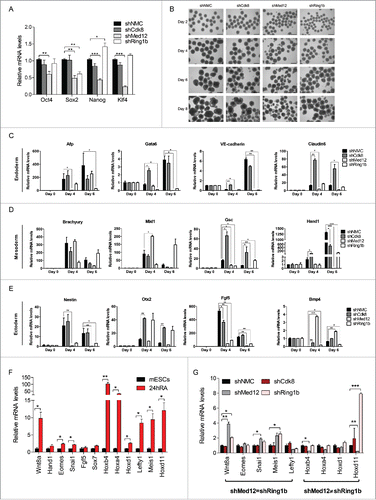
In sum, Med12 is essential for maintaining the mESC pluripotency network and both Ring1b and Med12 are important for the repression of developmental genes during pluripotency. In contrast, the activation of such genes rather relies on Med12 implying that Med12 and Ring1b play diverging roles during differentiation.
Med12 acts as a transcriptional activator of key developmental genes in retinoic acid mediated differentiation
The diverging functions of Ring1b and Med12 in transcriptional activation prompted us to study their functional interplay in differentiation further. Opposed to its silencing function in the pluripotent state, Med12 drives the transcriptional activation of Hoxd11 during RA induced differentiation (). Therefore, we focused on Hoxd11, which exhibited a striking difference regarding its expression levels in differentiating shRing1b and shMed12 cells. Of note, Hoxd11 is a classical Polycomb target gene,Citation52 which is involved in mouse forelimb development.Citation53 We verified that Med1, Med12 and Ring1b occupy the Hoxd11 locus during mESC pluripotency suggesting a common role of Cbx7-PRC1 and Med12 in silencing Hoxd11 (). Mediator and Med12 were previously shown to play a role in transcriptional activation by ncRNA-mediated mechanisms.Citation43,54 To assess a potential function of ncRNAs in the activation of Hoxd11 we analyzed a 2Mb region encompassing the HoxD locus () for the expression of ncRNAs. Interestingly, the ncRNA Dlx1as is expressed during RA induced differentiation (Fig. S6B) and progressively throughout mouse EB differentiation from a locus close to the Dlx1 gene ().Citation55 As reported for Hoxd11 mutant mice,Citation53 ablation of Dlx1as was shown to generate a skeletal phenotype.Citation56 Therefore, we asked whether Dlx1as had an impact on the transcriptional activation of Hoxd11. To this end we transfected mESCs with either control siRNA or siRNA pools targeting Dlx1as (Fig. S6C). We observed after RA induced differentiation that Hoxd11 as well as Hoxd12 and Chn1 were not activated properly, supporting a role for Dlx1as in their regulation ( and S6A). Notably, knockdown of Dlx1as did not alter the expression of Cdk8, Ring1b or Med12 during RA induced differentiation (Figure S6C) arguing that Dlx1as is probably directly involved in the activation of Hoxd11.
Figure 5. The Ring1b-Med12 target gene Hoxd11 is regulated by the ncRNA Dlx1as. (A) Med12 and Cbx7-PRC1 but not Cdk8, localize to Hoxd11 in mESCs. IGV Screenshot from RNAseq (mESCs) and ChIP-seq peaks illustrating the binding of Med12, Med1, Cdk8, Ring1b, Cbx7, RYBP and Suz12 at the Hoxd11 locus. (B) Illustration of the 2 Mb locus spanning the Dlx1as gene (green) and the HoxD cluster (red). The direction of transcription is indicated by the arrowheads. (C) The ncRNA Dlx1as regulates the expression of genes of the HoxD cluster in cis. mESCs were transfected with control siRNA or siRNA directed against Dlx1as and treated with RA. Twenty-four hours after RA administration the relative expression of genes in the 2 Mb locus were analyzed by qRT-PCR. Data are represented as a mean +/− SEM (n = 3). *** P-value<0.0001, ** P-value <0.01, * P-value < 0.02 as calculated by 2-tailed t test. (D) Med12 associates with Dlx1as in pluripotency and early differentiation. RIP experiments with Med12 antibodies and control IgGs from extracts of mESCs and differentiating cells (24h RA). The association of Dlx1as was analyzed in qRT-PCR experiments. Data are represented as a mean +/− SEM (n = 3). (E) Med12 associates with Dlx1as as a function of Ring1b. RIP experiments with Med12 antibodies and control IgGs using extracts from control and Ring1b knockdown mESCs. The association of Dlx1as, Hotairm (linc1548) and Rian was analyzed in qRT-PCR experiments. Data are represented as a mean +/− SEM (n = 3). The indicated P-value was calculated by F test to compare the differences. (F) Ring1b dissociates from Dlx1as upon differentiation. RIP experiments with Ring1b antibodies and control IgGs from extracts of mESCs and differentiating cells (24h RA). The association of the indicated ncRNAs Dlx1as, Hotairm and Rian was analyzed in qRT-PCR experiments. The figure represents an average of at least 3 independent experiments +/− SEM. The indicated P-value was calculated by F test to compare the differences. (G) The association of Ring1b and Med12 is RNA-mediated. Endogenous Ring1b IPs with nuclear extracts from wt mESCs. Precipitated material was subjected to RNaseA treatment prior to Western blotting and incubated with the indicated antibodies. Inputs represent 5% of the material used for the IPs. The relative intensity of Med12 Co-IP levels was calculated using the Ring1b levels as reference.
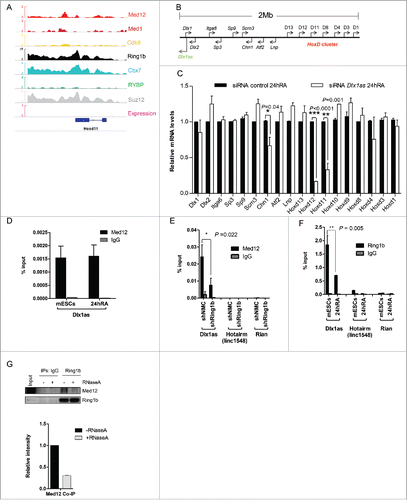
To further assess this possible direct role of Dlx1as in the transcriptional activation of Hoxd11 we performed RNA immunoprecipitation (RIP) experiments. Utilizing Med12 antibody and nuclear extracts from either mESCs or RA induced cells, we observed specific binding of Dlx1as in pluripotency and in differentiation suggesting that Dlx1as is associated to Med12 during gene repression and transcriptional activation (). Having previously demonstrated that Med12 occupancy at Med12-Ring1b target genes is dependent on Ring1b () we addressed whether the association of Med12 with Dlx1as is also mediated by Ring1b. In RIP experiments utilizing extracts from control and Ring1b knockdown mESCs we observed that during pluripotency Med12 associates with Dlx1as as a function of Ring1b (). Testing Med12 association with 2 unrelated ncRNAs (Hotairm and Rian) we observed no interaction implying specific association of Med12 with Dlx1as. Importantly, depletion of Ring1b or Cdk8 did not alter the expression levels of Dlx1as significantly (Fig. S6D). In contrast, Med12 seems to be required for the expression of Dlx1as in differentiating cells (Fig. S6D). PRC1 was recently shown to associate with ncRNAs.Citation57,58 In line with these studies we observed in RIP experiments from nuclear extracts of pluripotent mESCs Ring1b associating with Dlx1as (). Notably, in differentiating cells (24h RA) we found diminished association of Dlx1as with Ring1b. This raises the possibility that Ring1b and Med12 associate with Dlx1as in the pluripotent state. To examine the possible association of Med12 and Ring1b with ncRNAs during pluripotency, we performed immunoprecipitations with Ring1b antibodies and extracts from mESCs followed by RNase A treatment (). We observed that the interaction of both proteins is strictly dependent on RNA raising the possibility that ncRNAs stabilize Med12-PRC1 complexes during pluripotency.
Collectively our data indicate that Dlx1as is associated with PRC1 and Med12 in the pluripotent state suggesting that PRC1 likely inhibits the activating function of the ncRNA and hence expression of Hoxd11. In the course of cellular differentiation PRC1 association with Dlx1as is abrogated presumably enhancing transcription of Hoxd11.
Zrf1 associates with activating ncRNAs during differentiation
Ring1b dissociation from Dlx1as during differentiation () is reminiscent of the function of Zrf1, which displaces PRC1 from chromatin to derepress Polycomb target genes.Citation18 To verify whether Zrf1 activates developmental target genes repressed by Med12 and Ring1b, we analyzed Zrf1 knockdown cells after stimulation with RA. We found that transcriptional activation of selected Med12-Ring1b target genes was dramatically impaired in shZrf1 cells (Fig. S7A). Notably, knockdown of Zrf1 caused no alteration of both the mRNA (Fig. S7C) or the protein levels of Ring1b, Cdk8 or Med12 (Fig. S3B) supporting the idea that Zrf1 directly activates the Med12-Ring1b target genes. In line with this assumption we observed recruitment of Zrf1 to promoters of selected Ring1b-Med12 target genes (Fig. S7B) and most importantly to the Hoxd11 promoter after administration of RA (). Next we addressed whether Zrf1 was involved in ncRNA-mediated transcriptional activation of Hoxd11. Similar to siRNA-mediated depletion of Dlx1as, knockdown of Zrf1 caused diminished transcriptional activation of Hoxd11 (). The expression of additional genes within the 2Mb cluster was also affected () suggesting that Zrf1 in comparison to Dlx1as exerts a more global gene activation function. Of note, knockdown of Dlx1as did not alter Zrf1 expression further supporting a direct role of Dlx1as in the transcriptional activation of Hoxd11 (Fig. S7D). To prove a specific functional interplay of Zrf1 and Dlx1as we performed RIP experiments employing nuclear extracts from mESCs stably expressing FLAG-tagged Zrf1 (). We noted that Zrf1 associates with Dlx1as only after administration of RA suggesting a role for Zrf1 in the dissociation of PRC1 from Dlx1as (). Furthermore, we observed Zrf1 associating specifically with the ncRNA HotairmCitation59,60 after induction with RA implying that Zrf1 might act in concert with diverse ncRNAs to facilitate transcriptional activation. Having identified Zrf1 as a novel ncRNA-binding protein we next mapped its RNA binding domain. ZRF1 binds RNA with its c-terminal SANT domains as RIP experiments after expression of FLAG-tagged ZRF1 or ZRF1-SANT domain deletion mutants in HEK293T cells indicate (Fig. S7E). To further prove that the SANT domains are important for ZRF1 association with RNAs we mutated 2 amino acids (W554 and L561)Citation61 in the second SANT domain of ZRF1 (). After expression of the FLAG-tagged ZRF1 mutants in HEK293T cells we performed RIP experiments demonstrating that leucine 561 of ZRF1 is essential for RNA binding ().
Figure 6. Zrf1 and Dlx1as activate Hoxd11 during differentiation. (A) Zrf1 is recruited to the promoter of Hoxd11 during early differentiation. ChIP experiments were carried out with FLAG antibodies and chromatin from stably transfected FLAG-Zrf1 mESCs either before or after RA administration (24h RA) and subsequent qRT-PCR. Data are represented as a mean +/− SEM (n = 3). (B) Knockdown of Zrf1 causes diminished activation of Hoxd11 in RA mediated mESC differentiation. 24 hours after RA administration the relative expression of genes in the 2Mb locus () was analyzed by qRT-PCR. Data are represented as a mean +/− SEM (n=3). *** P-value<0.0001, ** P-value <0.01, * P-value < 0.02 as calculated by 2-tailed t test. The depicted p-values were calculated by 2-tailed unpaired t test. (C) Zrf1 associates with Dlx1as during RA induced differentiation. RIP experiments with FLAG antibodies and control IgGs from extracts of stably transfected FLAG-Zrf1 mESCs before or after RA administration (24h RA). The association of the indicated ncRNAs, Dlx1as, Hotairm and Rian, was analyzed by qRT-PCR. Data are represented as a mean +/− SEM (n = 3). P-value = 0.004 as calculated by the F-test. (D) Schematic representation of Zrf1 showing the DnaJ, UBD and the c-terminal SANT domains. The mutated amino acids are indicated by asterisks. (E) Leucine 561 of the human ZRF1 is essential for RNA binding. Control, FLAG-tagged ZRF1 and FLAG-tagged mutant ZRF1 plasmids were expressed in HEK293T cells. After FLAG IP the precipitated material was analyzed by Westernblot and the co-precipitated RNA was analyzed with or without RNaseA treatment.
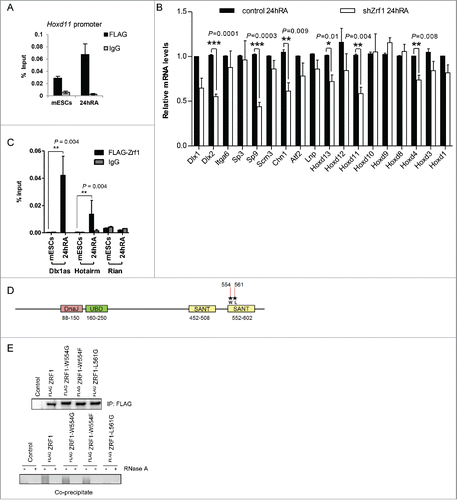
Collectively, these data suggest that Zrf1 is a novel ncRNA-binding protein that acts in concert with Dlx1as to activate the Med12-Ring1b target gene Hoxd11 in early stem cell differentiation.
Zrf1 assembles the Cdk8 module during differentiation
To further investigate the function of Zrf1 in the context of Mediator-enhanced gene activation we analyzed chromatin from control and Zrf1 knockdown cells (shZrf1) during differentiation (). In shZrf1 cells we noted stabilization of Ring1b at chromatin in agreement with our previous findings.Citation18 Med12 levels at chromatin remained unaltered but recruitment of Cdk8 was drastically impaired in Zrf1 knockdown cells. Importantly, knockdown of Zrf1 has no impact on the mRNA and the protein levels of Cdk8 (Figs. S7C and S3B) suggesting that Zrf1 and Cdk8 are functionally linked. Thus, we next analyzed the impact of Cdk8 knockdown in a similar experimental setting (). We observed no significant changes in the recruitment of Zrf1 and Med12 over time, implying that Zrf1 is essential for recruiting Cdk8 to chromatin but not vice versa. To further address the functional interplay of Zrf1 and Cdk8 we performed ChIP experiments with Cdk8 antibodies using chromatin from control and Zrf1 knockdown cells (). We observed Cdk8 binding at Hoxd11 promoters after stimulation with RA only in control but not in Zrf1 knockdown cells corroborating a role for Zrf1 in the recruitment of Cdk8 during RA induced differentiation. Similarly, purifications of FLAG-tagged H2A containing nucleosomes from HEK293T cells demonstrate that CDK8 association to chromatin is strictly dependent on ZRF1 (Fig. S8A).
Figure 7. Zrf1 mediates the recruitment of Cdk8 to the Hoxd11 promoter. (A) Zrf1 displaces Ring1b from chromatin and facilitates Cdk8 recruitment. Chromatin from control or shZrf1 knockdown cells, either pluripotent or after RA administration was isolated and subjected to Western blotting and probed with the indicated antibodies. The relative intensities of Cdk8 and Zrf1 levels were quantified by using the ponceau intensity as reference and depicted as fold change to the 0 timepoint shNMC samples, +/− SEM, n = 3 (B) Knockdown of Cdk8 has no impact on Zrf1 recruitment to chromatin. Chromatin from control or shCdk8 knockdown cells, either pluripotent or after RA administration, was isolated and subjected to Western blotting and probed with the indicated antibodies. The relative intensities of Cdk8 and Zrf1 levels were quantified by using the ponceau intensity as reference and depicted as fold change to the 0 timepoint shNMC samples, +/− SEM, n = 3 . (C) Zrf1 controls the recruitment of Cdk8 to the Hoxd11 promoter during early differentiation. ChIPs were carried out with chromatin from nuclear extracts of wt and shZrf1 pluripotent and early differentiating mESCs utilizing Cdk8 antibodies or IgGs as control. Data are represented as a mean +/− SEM (n = 3). (D) Zrf1 interacts with Cdk8. Nuclear extracts from wildtype mESCs or mESCs stably expressing FLAG-Zrf1 were used in immunoprecipitations with FLAG antibodies. The precipitated material was subjected to Western blotting and incubated with the stated antibodies. (E) Zrf1 mediates the interaction of Cdk8 with Med12 during differentiation. Nuclear extracts from control or Zrf1 knockdown cells were used in endogenous IPs with Cdk8 antibodies. The precipitated material was subjected to Western blotting and incubated with the stated antibodies. (F) Proposed model for the transcriptional activation of Med12-Ring1b repressed genes. During pluripotency (mESCs) key developmental genes are repressed by PRC1 and Med12 probably also involving ncRNAs. During differentiation Zrf1 displaces PRC1 and recruits Cdk8 probably facilitating the assembly of the kinase module of Mediator.
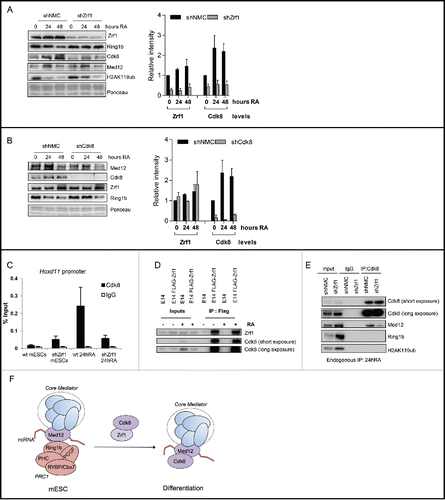
Next we performed immunoprecipitations with Zrf1 and FLAG antibodies to analyze whether Zrf1 and Cdk8 physically interact ( and S8B). We observed that Zrf1 interacts with Cdk8 in mESCs and differentiating cells further supporting a role for Zrf1 in recruiting Cdk8 to chromatin. As previously shown, at a large number of developmental genes Med12-PRC1 complexes are devoid of Cdk8 during pluripotency ( and S1). Thus we posed the question whether Zrf1 facilitates the association of Cdk8 with Med12 in the course of differentiation. First, we analyzed the interaction of Cdk8/CDK8 and Med12/MED12 in differentiating mESCs or HEK293T cells, respectively (, S8C and S8D). Knocking down Zrf1/ZRF1 in either differentiating mESCs or HEK293T cells caused diminished interaction of Med12/MED12 with Cdk8/CDK8 ( and S8D) but enhanced binding of Med12 with Ring1b/RING1b (Figs. S8F and S8G) in support of our hypothesis. Moreover, in RA stimulated differentiation of mESCs we noted stabilization of the Med12-Cdk8 interaction and reduction of the Med12-Ring1b interaction (Fig. S8C). Remarkably, the recruitment of Cdk8 had no impact on the transcriptional activation of Hoxd11 or most other Ring1b-Med12 target genes () potentially indicating that other kinases as for example Cdk19 could compensate for Cdk8 function. Still, our data show that Zrf1 recruits Cdk8 to promoters of their target genes and that it mediates the interaction between Med12 and Cdk8 in the course of differentiation. Our data further suggest that Zrf1 acts as a switch protein that remodels Mediator-associated protein complexes thereby converting repressive Mediator (Med12-Ring1b) into an active enhancer (Med12-Cdk8).
Discussion
Mediator complexes are regarded as global regulators of gene expression that act in concert with transcription factors within the PIC. The interaction with either the Cdk8 module or Pol II is thought to fine-tune Mediator function. It was demonstrated previously that subunits of the Cdk8 module represent critical regulators of developmental processes. In particular, studies in the fruit fly suggested a functional separation of Cdk8 and Med12 in regulating key developmental genes.Citation37,62 In agreement with these findings our study suggests that Med12 fulfills an important role in silencing developmental genes together with PRC1. Notably, apart from its repressive function, Med12 also keeps the ES self-renewal network active.Citation44 Hence, Med12 plays an essential role in maintaining mESC pluripotency. In contrast, Cdk8 and Med12 in conjunction with Mediator occupy highly expressed, metabolic genes during pluripotency. These findings imply that already in the pluripotent state distinct Mediator complexes exist to control different classes of genes. Mediator associated with Med12 and PRC1 is implicated in repressing developmental marker genes whereas canonical Mediator-Cdk8 complexes enhance transcription.
Furthermore, our data sheds light on the underlying molecular mechanism of Med12-PRC1 mediated gene silencing and gene activation. Med12 occupancy at repressed genes (Med12-Ring1b targets) and actively transcribed genes (Med12-Cdk8 targets) is particularly insightful. Med12-Cdk8 target genes are devoid of Ring1b (), yet Med12 occupancy at the same genes is dependent on Ring1b (). These findings support the idea that Cdk8-Med12 interaction and hence the assembly of the Cdk8 module strictly relies on Ring1b. In addition we have shown that Med12 association with the gene activating ncRNA Dlx1as relies on PRC1. Ring1b therefore operates as a repressor of developmental genes but at the same time its function is also a prerequisite for Mediator-ncRNA dependent gene activating mechanisms during early differentiation. Med12 likewise shows an ambivalent function at developmental genes. During differentiation Med12 enhances the activation of genes whereas during pluripotency it represents a transmitter of the PRC1 silencing function. Interestingly, a similar transmitter function of Mediator had been proposed in a previous report.Citation13
Our study further suggests that PRC1 and Med12 play a role in regulating differentiation. EBs derived from Ring1b knockdown cells are relatively small and do not differentiate properly, which is in agreement with an earlier study.Citation50 Although we observed a similar phenotype when analyzing EBs derived from Med12 knockdown cell lines it appears that Ring1b and Med12 have different regulatory functions at developmental genes during differentiation. Whereas knockdown of Ring1b results in misregulation of germ layer marker genes, depletion of Med12 leads to strongly diminished activation of the same genes. This suggests that the regulatory function of Med12 is converted at the onset of differentiation turning it into an enhancer of transcription. It had already been reported that Med12 acts in early mouse development,Citation63 which is in line with our findings. In stark contrast to Med12, EBs derived from Cdk8 knockdown cell lines showed no apparent phenotype despite upregulation of selected germ layer marker genes (). This would suggest that Cdk8 function is presumably not essential for the regulation of key developmental genes during EB differentiation. This is in agreement with the genome wide occupancy of Cdk8 and the transcription profile of Cdk8 knockdown mESCs. Nonetheless, Cdk8 is an important factor in early mouse development since Cdk8 knockout mice are embryonically lethal.Citation64 Importantly, the knockout phenotype is not at odds with our findings since Cdk8 seems to be required at an earlier developmental stage than the one examined in our EB analysis. Another possibility for the lack of a phenotype upon Cdk8 depletion might be attributed to redundant functions with other kinases, such as its paralog Cdk19.Citation65 How Cdk8 controls differentiation processes, how it renders Mediator function and whether this involves phosphorylation events still remains to be investigated.
It was recently reported that Mediator associates with ncRNAs to enhance transcriptional activationCitation43 and likewise ncRNAs contribute to gene repression via PRC1.Citation57,58 Given the close interplay between Med12 and PRC1 in gene regulation, we focused on one common target gene, Hoxd11, to assess ncRNA-mediated activation. The relatively similar phenotypes of mice devoid of Dlx1as and mice harboring a mutated Hoxd11 prompted us to investigate the function of Dlx1as during differentiation. We provide evidence that the ncRNA Dlx1as acts in cis to activate genes of the HoxD cluster, in particular Hoxd11. In contrast, we did not observe a reciprocal relationship between the expression of the homeobox gene Dlx1 and Dlx1as that was reported earlier ().Citation56 This suggests that Dlx1as acts rather as a gene-activating ncRNA than keeping Dlx1 mRNA levels at check. Notably, Dlx1as is associated to both PRC1 and Med12 arguing that transcriptional activation by Mediator-ncRNA complexes is likely inhibited by PRC1. In the course of differentiation Zrf1 displaces PRC1 from Med12-ncRNA complexes presumably converting the Mediator-ncRNA complex into an enhancer of transcriptional activation.
Recruitment of Zrf1 to promoters of its target genes is accompanied by its association to ncRNA, which is mediated by the c-terminal SANT domain of Zrf1. We have demonstrated that Zrf1 and Dlx1as control the transcriptional activation of Hoxd11 further underpinning a concerted function of both factors. Interestingly, Zrf1 regulates the recruitment of Cdk8 to chromatin and in particular to the promoter of the Dlx1as target gene, Hoxd11. Thus, Zrf1 not only derepresses genes by dislodging PRC1, but it also mediates the remodeling of Mediator associated protein-ncRNA complexes. During mESC differentiation Zrf1 likely mediates the assembly of the Cdk8 module at developmental genes after displacing PRC1 (). This Zrf1-dependent association of Cdk8 with Mediator might be critical for the recruitment of Pol II and potentially for Pol II-mediated elongation as discussed in a recent publication.Citation48 Further, knockdown of Cdk8 has no significant impact on the expression levels of PRC1-Med12 target genes. At any rate, ncRNA-Mediator complexes regulate Cdk8 enzymatic activity toward histone H3 serine 10,Citation43 which might constitute another layer of transcriptional control. Future research dedicated to unveiling the function of Cdk8 in transcription will certainly need to address the role of homologous cyclin-dependent kinases that could potentially play redundant roles in transcriptional activation.
Experimental procedures
Cell culture
Embryonic stem cells (E14Tg2A) were cultured in feeder-free plates, coated for at least 30 min with 0.1% Gelatin, ready to use (Millipore) or in powder form (Sigma), dissolved in MQ water after overnight stirring at 75°C. Cells were cultured in Glasgow Minimum Essential Medium (Sigma) supplemented with 15% PanSera ES Bovine Serum (PAN-Biotech), Non-Essential Amino Acids (Gibco), Sodium Pyruvate (Gibco), L-Glutamine (Gibco), Penicillin/Streptomycin (Gibco), β-mercaptoethanol (Sigma) and Leukemia Inhibitory Factor (LIF). For in vitro differentiation, cells were cultured in medium without LIF, in the presence of 10−6 M all-trans-Retinoic acid (ATRA) (Sigma).
HEK293T cells were cultured in Dulbecco's Modified Eagle Medium (Life Technologies), supplemented with 10% FBS Gold (PAA Laboratories), L-glutamine and Penicillin/Streptomycin. All transfections in HEK293T cells were carried out using calcium phosphate according to standard protocols. 48 hours post transfection cells were collected for immunoprecipitations.
Embryoid body formation
Embryoid bodies (EBs) were formed with the hanging-drop method, starting with 103 mESCs / 30 μl drop, in medium without LIF. 48 hours after the drop formation, EBs were collected and cultured from that point on in petri dishes. The medium was replaced every second day. Images were acquired with a Leica DM-IL microscope. EB size distributions were quantified with ImageJ analysis by measuring the Feret's diameter of each EB.
Extended Experimental Procedures are provided in the Supplemental information.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
1175797_Supplemental_Material.zip
Download Zip (2 MB)Acknowledgments
We thank the IMB Genomics and Bioinformatics core facilities for their expert assistance. We are grateful to Fan Lai for providing reagents. We thank the members of the Richly laboratory for comments on the manuscript.
Funding
This work was supported by grants from the Boehringer Ingelheim Foundation.
References
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol 2009; 10:697-708; PMID:19738629; http://dx.doi.org/10.1038/nrn2731
- Sauvageau M, Sauvageau G. Polycomb group proteins: multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 2010; 7:299-313; PMID:20804967; http://dx.doi.org/10.1016/j.stem.2010.08.002
- Bracken AP, Helin K. Polycomb group proteins: navigators of lineage pathways led astray in cancer. Nat Rev Cancer 2009; 9:773-84; PMID:19851313; http://dx.doi.org/10.1038/nrc2736
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell 2007; 128:735-45; PMID:17320510; http://dx.doi.org/10.1016/j.cell.2007.02.009
- Morey L, Helin K. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 2010; 35:323-32; PMID:20346678; http://dx.doi.org/10.1016/j.tibs.2010.02.009
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298:1039-43; PMID:12351676; http://dx.doi.org/10.1126/science.1076997
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004; 431:873-8; PMID:15386022; http://dx.doi.org/10.1038/nature02985
- Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 2012; 45:344-56; PMID:22325352; http://dx.doi.org/10.1016/j.molcel.2012.01.002
- Luis NM, Morey L, Di Croce L, Benitah SA. Polycomb in stem cells: PRC1 branches out. Cell Stem Cell 2012; 11:16-21; PMID:22770239; http://dx.doi.org/10.1016/j.stem.2012.06.005
- Morey L, Pascual G, Cozzuto L, Roma G, Wutz A, Benitah SA, Di Croce L. Nonoverlapping functions of the Polycomb group Cbx family of proteins in embryonic stem cells. Cell Stem Cell 2012; 10:47-62; PMID:22226355; http://dx.doi.org/10.1016/j.stem.2011.12.006
- O'Loghlen A, Munoz-Cabello AM, Gaspar-Maia A, Wu HA, Banito A, Kunowska N, Racek T, Pemberton HN, Beolchi P, Lavial F, et al. MicroRNA regulation of Cbx7 mediates a switch of Polycomb orthologs during ESC differentiation. Cell Stem Cell 2012; 10:33-46; PMID:22226354; http://dx.doi.org/10.1016/j.stem.2011.12.004
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012; 148:664-78; PMID:22325148; http://dx.doi.org/10.1016/j.cell.2011.12.029
- Simon JA, Kingston RE. Occupying chromatin: Polycomb mechanisms for getting to genomic targets, stopping transcriptional traffic, and staying put. Mol Cell 2013; 49:808-24; PMID:23473600; http://dx.doi.org/10.1016/j.molcel.2013.02.013
- Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science 2004; 306:1574-7; PMID:15567868; http://dx.doi.org/10.1126/science.1100576
- Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, et al. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 2010; 38:452-64; PMID:20471950; http://dx.doi.org/10.1016/j.molcel.2010.02.032
- Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell 2005; 20:845-54; PMID:16359901; http://dx.doi.org/10.1016/j.molcel.2005.12.002
- Illingworth RS, Moffat M, Mann AR, Read D, Hunter CJ, Pradeepa MM, Adams IR, Bickmore WA. The E3 ubiquitin ligase activity of RING1B is not essential for early mouse development. Genes Dev 2015; 29:1897-902; PMID:26385961; http://dx.doi.org/10.1101/gad.268151.115
- Richly H, Rocha-Viegas L, Ribeiro JD, Demajo S, Gundem G, Lopez-Bigas N, Nakagawa T, Rospert S, Ito T, Di Croce L. Transcriptional activation of polycomb-repressed genes by ZRF1. Nature 2010; 468:1124-8; PMID:21179169; http://dx.doi.org/10.1038/nature09574
- Inoue T, Shoji W, Obinata M. MIDA1 is a sequence specific DNA binding protein with novel DNA binding properties. Genes Cells 2000; 5:699-709; http://dx.doi.org/10.1046/j.1365-2443.2000.00362.x
- Inoue T, Shoji W, Obinata M. MIDA1, an Id-associating protein, has two distinct DNA binding activities that are converted by the association with Id1: a novel function of Id protein. Biochem Biophys Res Commun 1999; 266:147-51; PMID:10581180; http://dx.doi.org/10.1006/bbrc.1999.1779
- Demajo S, Uribesalgo I, Gutierrez A, Ballare C, Capdevila S, Roth M, Zuber J, Martin-Caballero J, Di Croce L. ZRF1 controls the retinoic acid pathway and regulates leukemogenic potential in acute myeloid leukemia. Oncogene 2014; 33:5501-10; PMID:24292673; http://dx.doi.org/10.1038/onc.2013.501
- Aloia L, Demajo S, Di Croce L. ZRF1: a novel epigenetic regulator of stem cell identity and cancer. Cell Cycle 2015; 14:510-5; PMID:25665097; http://dx.doi.org/10.4161/15384101.2014.988022
- Ansari SA, He Q, Morse RH. Mediator complex association with constitutively transcribed genes in yeast. Proc Natl Acad Sci U S A 2009; 106:16734-9; PMID:19805365; http://dx.doi.org/10.1073/pnas.0905103106
- Takagi Y, Kornberg RD. Mediator as a general transcription factor. J Biol Chem 2006; 281:80-9; PMID:16263706; http://dx.doi.org/10.1074/jbc.M508253200
- Borggrefe T, Yue X. Interactions between subunits of the Mediator complex with gene-specific transcription factors. Semin Cell Dev Biol 2011; 22:759-68; PMID:21839847; http://dx.doi.org/10.1016/j.semcdb.2011.07.022
- Naar AM, Taatjes DJ, Zhai W, Nogales E, Tjian R. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev 2002; 16:1339-44; PMID:12050112; http://dx.doi.org/10.1101/gad.987602
- Boyer TG, Martin ME, Lees E, Ricciardi RP, Berk AJ. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 1999; 399:276-9; PMID:10353252; http://dx.doi.org/10.1038/20466
- Liu Y, Ranish JA, Aebersold R, Hahn S. Yeast nuclear extract contains two major forms of RNA polymerase II mediator complexes. J Biol Chem 2001; 276:7169-75; PMID:11383511
- Belakavadi M, Fondell JD. Cyclin-dependent kinase 8 positively cooperates with Mediator to promote thyroid hormone receptor-dependent transcriptional activation. Mol Cell Biol 2010; 30:2437-48; PMID:20231357; http://dx.doi.org/10.1128/MCB.01541-09
- Elmlund H, Baraznenok V, Lindahl M, Samuelsen CO, Koeck PJ, Holmberg S, Hebert H, Gustafsson CM. The cyclin-dependent kinase 8 module sterically blocks Mediator interactions with RNA polymerase II. Proc Natl Acad Sci U S A 2006; 103:15788-93; PMID:17043218; http://dx.doi.org/10.1073/pnas.0607483103
- Knuesel MT, Meyer KD, Donner AJ, Espinosa JM, Taatjes DJ. The human CDK8 subcomplex is a histone kinase that requires Med12 for activity and can function independently of mediator. Mol Cell Biol 2009; 29:650-61; PMID:19047373; http://dx.doi.org/10.1128/MCB.00993-08
- Sato S, Tomomori-Sato C, Banks CA, Sorokina I, Parmely TJ, Kong SE, Jin J, Cai Y, Lane WS, Brower CS, et al. Identification of mammalian Mediator subunits with similarities to yeast Mediator subunits Srb5, Srb6, Med11, and Rox3. J Biol Chem 2003; 278:15123-7; PMID:12584197; http://dx.doi.org/10.1074/jbc.C300054200
- Sato S, Tomomori-Sato C, Parmely TJ, Florens L, Zybailov B, Swanson SK, Banks CA, Jin J, Cai Y, Washburn MP, et al. A set of consensus mammalian mediator subunits identified by multidimensional protein identification technology. Mol Cell 2004; 14:685-91; PMID:15175163; http://dx.doi.org/10.1016/j.molcel.2004.05.006
- Myers LC, Gustafsson CM, Bushnell DA, Lui M, Erdjument-Bromage H, Tempst P, Kornberg RD. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev 1998; 12:45-54; PMID:9420330; http://dx.doi.org/10.1101/gad.12.1.45
- Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ. The human CDK8 subcomplex is a molecular switch that controls Mediator coactivator function. Genes Dev 2009; 23:439-51; PMID:19240132; http://dx.doi.org/10.1101/gad.1767009
- Carrera I, Janody F, Leeds N, Duveau F, Treisman JE. Pygopus activates Wingless target gene transcription through the mediator complex subunits Med12 and Med13. Proc Natl Acad Sci U S A 2008; 105:6644-9; PMID:18451032; http://dx.doi.org/10.1073/pnas.0709749105
- Gaytan de Ayala Alonso A, Gutierrez L, Fritsch C, Papp B, Beuchle D, Muller J. A genetic screen identifies novel polycomb group genes in Drosophila. Genetics 2007; 176:2099-108; PMID:17717194; http://dx.doi.org/10.1534/genetics.107.075739
- Wang JC, Walker A, Blackwell TK, Yamamoto KR. The Caenorhabditis elegans ortholog of TRAP240, CeTRAP240/let-19, selectively modulates gene expression and is essential for embryogenesis. J Biol Chem 2004; 279:29270-7; PMID:15073178; http://dx.doi.org/10.1074/jbc.M401242200
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol Cell 2007; 27:121-33; PMID:17612495; http://dx.doi.org/10.1016/j.molcel.2007.05.026
- Donner AJ, Ebmeier CC, Taatjes DJ, Espinosa JM. CDK8 is a positive regulator of transcriptional elongation within the serum response network. Nat Struct Mol Biol 2010; 17:194-201; PMID:20098423; http://dx.doi.org/10.1038/nsmb.1752
- Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet 2006; 38:896-903; PMID:16845399; http://dx.doi.org/10.1038/ng1844
- Orom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010; 143:46-58; PMID:20887892; http://dx.doi.org/10.1016/j.cell.2010.09.001
- Lai F, Orom UA, Cesaroni M, Beringer M, Taatjes DJ, Blobel GA, Shiekhattar R. Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 2013; 494:497-501; PMID:23417068; http://dx.doi.org/10.1038/nature11884
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 2010; 467:430-5; PMID:20720539; http://dx.doi.org/10.1038/nature09380
- Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep 2013; 3:60-9; PMID:23273917; http://dx.doi.org/10.1016/j.celrep.2012.11.026
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006; 441:349-53; PMID:16625203; http://dx.doi.org/10.1038/nature04733
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 2013; 153:307-19; PMID:23582322; http://dx.doi.org/10.1016/j.cell.2013.03.035
- Allen BL, Taatjes DJ. The Mediator complex: a central integrator of transcription. Nat Rev Mol Cell Biol 2015; 16:155-66; PMID:25693131; http://dx.doi.org/10.1038/nrm3951
- van der Stoop P, Boutsma EA, Hulsman D, Noback S, Heimerikx M, Kerkhoven RM, Voncken JW, Wessels LF, van Lohuizen M. Ubiquitin E3 ligase Ring1b/Rnf2 of polycomb repressive complex 1 contributes to stable maintenance of mouse embryonic stem cells. PLoS One 2008; 3:e2235; PMID:18493325; http://dx.doi.org/10.1371/journal.pone.0002235
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev 2010; 24:265-76; PMID:20123906; http://dx.doi.org/10.1101/gad.544410
- Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol 2007; 178:219-29; PMID:17620408; http://dx.doi.org/10.1083/jcb.200612127
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell 2010; 140:99-110; PMID:20085705; http://dx.doi.org/10.1016/j.cell.2009.12.022
- Favier B, Le Meur M, Chambon P, Dolle P. Axial skeleton homeosis and forelimb malformations in Hoxd-11 mutant mice. Proc Natl Acad Sci U S A 1995; 92:310-4; PMID:7816839; http://dx.doi.org/10.1073/pnas.92.1.310
- Orom UA, Derrien T, Guigo R, Shiekhattar R. Long noncoding RNAs as enhancers of gene expression. Cold Spring Harb Symp Quant Biol 2010; 75:325-31; PMID:21502407; http://dx.doi.org/10.1101/sqb.2010.75.058
- Dinger ME, Amaral PP, Mercer TR, Pang KC, Bruce SJ, Gardiner BB, Askarian-Amiri ME, Ru K, Solda G, Simons C, et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res 2008; 18:1433-45; PMID:18562676; http://dx.doi.org/10.1101/gr.078378.108
- Kraus P, Sivakamasundari V, Lim SL, Xing X, Lipovich L, Lufkin T. Making sense of Dlx1 antisense RNA. Dev Biol 2013; 376:224-35; PMID:23415800; http://dx.doi.org/10.1016/j.ydbio.2013.01.035
- Bonasio R, Lecona E, Narendra V, Voigt P, Parisi F, Kluger Y, Reinberg D. Interactions with RNA direct the Polycomb group protein SCML2 to chromatin where it represses target genes. eLife 2014; 3:e02637; PMID:24986859; http://dx.doi.org/10.7554/eLife.02637
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011; 477:295-300; PMID:21874018; http://dx.doi.org/10.1038/nature10398
- Zhang X, Lian Z, Padden C, Gerstein MB, Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM, Newburger PE. A myelopoiesis-associated regulatory intergenic noncoding RNA transcript within the human HOXA cluster. Blood 2009; 113:2526-34; PMID:19144990; http://dx.doi.org/10.1182/blood-2008-06-162164
- Diaz-Beya M, Brunet S, Nomdedeu J, Pratcorona M, Cordeiro A, Gallardo D, Escoda L, Tormo M, Heras I, Ribera JM, et al. The lincRNA HOTAIRM1, located in the HOXA genomic region, is expressed in acute myeloid leukemia, impacts prognosis in patients in the intermediate-risk cytogenetic category, and is associated with a distinctive microRNA signature. Oncotarget 2015; 6:31613-27; PMID:26436590
- Saikumar P, Murali R, Reddy EP. Role of tryptophan repeats and flanking amino acids in Myb-DNA interactions. Proc Natl Acad Sci U S A 1990; 87:8452-6; PMID:2236054; http://dx.doi.org/10.1073/pnas.87.21.8452
- Gobert V, Osman D, Bras S, Auge B, Boube M, Bourbon HM, Horn T, Boutros M, Haenlin M, Waltzer L. A genome-wide RNA interference screen identifies a differential role of the mediator CDK8 module subunits for GATA/ RUNX-activated transcription in Drosophila. Mol Cell Biol 2010; 30:2837-48; PMID:20368357; http://dx.doi.org/10.1128/MCB.01625-09
- Rocha PP, Scholze M, Bleiss W, Schrewe H. Med12 is essential for early mouse development and for canonical Wnt and Wnt/PCP signaling. Development 2010; 137:2723-31; PMID:20630950; http://dx.doi.org/10.1242/dev.053660
- Westerling T, Kuuluvainen E, Makela TP. Cdk8 is essential for preimplantation mouse development. Mol Cell Biol 2007; 27:6177-82; PMID:17620419; http://dx.doi.org/10.1128/MCB.01302-06
- Conaway RC, Conaway JW. The Mediator complex and transcription elongation. Biochim Biophys Acta 2013; 1829:69-75; PMID:22983086; http://dx.doi.org/10.1016/j.bbagrm.2012.08.017
