ABSTRACT
Sit4p is the catalytic subunit of a ceramide-activated PP2A-like phosphatase that regulates cell cycle, mitochondrial function, oxidative stress resistance and chronological lifespan in yeast. In this study, we show that hexokinase 2 (Hxk2p) is hyperphosphorylated in sit4Δ mutants grown in glucose medium by a Snf1p-independent mechanism and Hxk2p-S15A mutation suppresses phenotypes associated with SIT4 deletion, namely growth arrest at G1 phase, derepression of mitochondrial respiration, H2O2 resistance and lifespan extension. Consistently, the activation of Sit4p in isc1Δ mutants, which has been associated with premature aging, leads to Hxk2p hypophosphorylation, and the expression of Hxk2p-S15E increases the lifespan of isc1Δ cells. The overall results suggest that Hxk2p functions downstream of Sit4p in the control of cell cycle, mitochondrial function, oxidative stress resistance and chronological lifespan.
Introduction
Bioactive sphingolipid metabolites, such as ceramide, sphingosine and sphingosine-1-phosphate, participate actively in the regulation of signal transduction pathways and modulate a variety of cellular processes, including growth, stress responses, apoptosis and aging. Protein targets, such as ceramide-activated protein kinases and phosphatases, protein kinase C, cathepsin D and JNK, have been suggested to mediate the effects of bioactive sphingolipids.Citation1
The yeast Sit4p is the catalytic subunit of the ceramide-activated protein phosphatase (CAPP), a heterotrimeric complex that also includes Tpd3p and Cdc55p as regulatory subunits.Citation2 Sit4p is a serine-threonine protein phosphatase related to type 2A family of protein phosphatases (PP2A), and it has a high homology to other protein phosphatases, including the fission yeast PP2A and human protein phosphatase 6 that are involved in cell cycle regulation.Citation3,4 As expect for a protein phosphatase, Sit4p regulates a wide range of biological processes, including cell functions that are controlled by Pkc1p, such as cell integrity pathway, cytoskeleton organization and ribosomal gene expression, the Swi4p factor that controls the transcription of G1 cyclin genes,Citation5-7 the ubiquitin-proteasome system,Citation8 silencing in the subtelomeric region,Citation9 monovalent ion and pH homeostasis,Citation10 nutrient signaling,Citation11,12 endocytosis Citation13 and traffic from the endoplasmic reticulum to the Golgi complex.Citation14
Sit4p also plays a central role in the regulation of mitochondrial function. Sit4p deficiency prevents yeast cells to grow on respiratory substrates due to a shift of carbohydrate metabolism flux into gluconeogenesis and glycogen storage at the expense of intermediates of the Krebs cycle.Citation15,16 Wild type cells grown in glucose medium rely on fermentation for energy production and repress the transcription of genes associated with mitochondrial function and the utilization of alternative carbon sources.Citation17 Interestingly, fermentation is reduced in sit4Δ cells due to a decrease of pyruvate decarboxylase activity.Citation18 Moreover, respiration is derepressed in sit4Δ cells grown in glucose medium and mitochondrial respiration is essential for their viability since these mutants are unable to grow under anaerobic conditions.Citation16 In wild type cells grown under repressing conditions, both Mig1p and Hxk2p are dephosphorylated by the Reg1p-Glc7p protein phosphatase complex, imported into the nucleus and form a Mig1p-Hxk2p complex that represses the expression of genes associated with growth on non-fermentable carbon sources.Citation19,20 The interaction between Hxk2p and Mig1p under repressing conditions inhibits Mig1p phosphorylation at Ser311 by the Snf1p kinase and its export from the nucleus into the cytosol by Msn5p.Citation19 Under these conditions, Snf1p is dephosphorylated and inhibited by Reg1p-Glc7p and Sit4p.Citation21 The catabolite derepression in sit4Δ cells is correlated with the degradation of the Mig1p transcription factor.Citation22
Mitochondria are the main source of reactive oxygen species in cells and play a central role in oxidative stress resistance and chronological lifespan.Citation23 A recent study showed that deletion of Sit4p protects cells from defects associated with mitochondrial DNA damage, including reduced proliferation and decreased mitochondrial protein import and electrochemical potential, by mechanisms that require Snf1p.Citation24 Consistent with Sit4p being a negative regulator of mitochondrial function, sit4Δ cells show an increased chronological lifespan.Citation25 This protective effect is observed even in cells submitted to severe calorie restriction,Citation25 a regime that results in the longest survival for wild type yeast strains.Citation26 Thus, Sit4p seems to partially function through a mechanism that is not modulated by calorie restriction. Sit4p is a downstream target of Tor1p, a conserved nutrient protein kinase associated with lifespan regulation in several organisms. Sit4p deficiency also increases cellular resistance to thiol-specific oxidants and H2O2.Citation12,27 Consistent with a role for Sit4p/CAPP in the regulation of lifespan and oxidative stress resistance, the increase of dihydro-C26-ceramide and phyto-C26-ceramide levels is associated with the activation of Sit4p in isc1Δ cells, leading to mitochondrial dysfunction, H2O2 sensitivity and a shortened chronological lifespan.Citation25,28 Isc1p is an ortholog of mammalian neutral sphingomyelinase 2Citation29 and plays an important role in mitochondrial function. When aerobic respiration is induced, Isc1p is translocated from the endoplasmic reticulum into mitochondria, where it generates α-hydroxylated phytoceramides that contribute to its normal function.Citation30,31 The defective aerobic respiration in isc1Δ cells is associated with its incapacity to upregulate genes required for non-fermentable carbon source metabolism.Citation32,33
Here we provide evidence that SIT4 deletion leads to Hxk2p hyperphosphorylation and that Hxk2p-Ser15 phosphorylation is required for cellular effects such as growth arrest at G1 phase, derepression of mitochondrial respiration, H2O2 resistance and lifespan extension in sit4Δ cells. The role of Hxk2p phosphorylation on the regulation of aging is supported by data showing that Sit4p-dependent Hxk2p hypophosphorylation correlates with premature aging in isc1Δ cells and the expression of Hxk2p with a phosphomimetic glutamate residue at position 15 (Hxk2p-S15E) increases mean chronological lifespan of this mutant.
Material and methods
Yeast strains, plasmids and growth conditions
The Saccharomyces cerevisiae strains used in this study are listed in . Yeast cells were grown aerobically at 26°C in a gyratory shaker (at 140 rpm), with a ratio of flask volume / medium volume of 5:1, to early exponential phase (OD600 = 0.6) or to post-diauxic phase (OD600 = 7–8). The growth media used were YPD (1% (wt/vol) yeast extract, 2% (wt/vol) bactopeptone, 2% (wt/vol) glucose), or synthetic complete (SC) drop-out medium containing 2% (wt/vol) glucose and 0.67% (wt/vol) yeast nitrogen base without amino acids (Difco Laboratories, Detroit, USA). The deletion of ISC1 in the hxk2Δ strain was performed using a deletion fragment containing LEU2 and the flanking regions of ISC1. The deletion of SIT4 in the snf1Δ and WAY.78–1 strains was performed using a deletion fragment containing MX4HIS3 or KanMX4 and the flanking regions of SIT4, respectively. Cells were transformed by electroporation, and double mutants were selected in minimal medium lacking histidine or YPD containing 200 μg geneticin ml−1. Gene deletion was confirmed by PCR. To generate pRS316-HXK2-S15 and pRS316-HXK2-S15E plasmids, YIpHXK2-S15 and YIpHXK2-S15ECitation34 were digested with BamHI and EcoRI and the fragments containing HXK2-S15 and HXK2-S15E were cloned into the pRS316 BamHI and EcoRI sites. Cells transformed with pRS316-HXK2-S15, pRS316-HXK2-S15E or YEp352-HXK2-GFP6Citation35 were selected in minimal medium lacking uracil.
Table 1. Saccharomyces cerevisiae strains used in this study.
2D-gel electrophoresis and protein phosphorylation
Yeast cells were harvested by centrifugation, resuspended in 50 mM potassium phosphate buffer (pH 7.0) containing protease inhibitors (Complete, Mini, EDTA-free Protease Cocktail Inhibitor Tablets; Boehringer Mannhein) and phosphatase inhibitors (50 mM sodium fluoride, 5 mM sodium pyrophosphate, 1 mM sodium orthovanadate), and lysed by vigorous shaking of the cell suspension in the presence of glass beads for 5 min. Short pulses of 1 min were used, with 1 min intervals on ice. Cell debris was removed by centrifugation at 13,000 rpm for 15 min and protein content was determined by the method of Lowry, using bovine serum albumin as a standard. The proteins (200 μg) were solubilized in IEF solution (9 M Urea, 2% (wt/vol) CHAPS, 2% (vol/vol) ß-mercaptoethanol, 0.8% (vol/vol) Pharmalytes pH 3–10) and separated by 2D-gel electrophoresis, using 13 cm immobilized pH 3–10 nonlinear gradient (IPG) dry strips (GE Healthcare) in the first dimension, as previously described.Citation36 After electrophoresis, proteins were electroblotted onto a nitrocellulose membrane (Hybond-C, GE Healthcare, Little Chalfont, United Kingdom). A replica 2D-gel was silver stained and used for protein identification. For the analysis of phosphorylated proteins, the nitrocellulose membrane was incubated with the primary antibody, specific for the phosphorylated serine residues (rabbit anti-phosphoserine, Zymed Laboratories, Invitrogen, Waltham, USA), at a 1:1,000 dilution, and subsequently with the secondary antibody, goat anti-rabbit IgG-linked to horseradish peroxidase (Sigma-Aldrich, St. Louis, USA), at a 1:5,000 dilution. Immunodetection was performed by chemiluminescence, using a kit from GE Healthcare (RPN 2109). The film and the gel were scanned using a densitometer (GS-800, Bio-Rad). Images were converted to tagged image file format (TIFF) and the PDQuest v7.3 (Bio-Rad) software was used for quantification of spot intensities. Two normalization steps were performed to determine the fold changes in (phospho)protein levels. First, sampled spot intensities were divided by the intensities of all spots. Second, for each protein, normalized phosphorylation level was divided by normalized protein intensity. The relative phosphoprotein level was expressed as the ratio between the sit4Δ and BY4741 strains. Proteins that changed at least 2-fold were considered for further analysis. All values are means of the expression profiles of 3 experiments with similar results, using independent cultures grown under the same conditions.
Protein identification
Silver-stained protein spots were excised and in gel digested with trypsin (Promega, USA). Peptide extraction was performed by a 60% acetonitrile/0.1% trifluoroacetic acid solution. Protein digests were desalted and concentrated using ZipTips (Millipore, USA) and crystallized onto a MALDI plate using α-Cyano-4-hydroxycinnamic acid as a matrix. Samples were analyzed using the 4700 Proteomics Analyzer MALDI-TOF/TOF (Applied Biosystems, USA), as previously described.Citation37
Western blotting
To evaluate Snf1p phosphorylation, cells were grown to exponential phase and proteins were extracted as previously described.Citation38 For Western blotting 40 µg of proteins were separated by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. Membranes were incubated with anti-Snf1 (Santa Cruz Biotechnology, Inc., Dallas, USA) at a 1:500 dilution, or anti-AMPK (phospho T172; Cell Signaling Technology, Beverly, MA, USA) at a 1:1,000 dilution followed by incubation with appropriate secondary antibody. For analysis of Hxk2p phosphorylation, Hxk2p was immunoprecipitated by incubating protein extracts from exponential phase cells with anti-GFP (Roche, Basel, Switzerland) for 2 h at 4°C. Protein A-Sepharose beads (GE Healthcare, Little Chalfont, United Kingdom) were then added and incubated for 2 h at 4°C. After extensive washes (150 mM NaCl, 50 mM Tris, pH 7.5, 1mM EDTA, 2% (vol/vol)Triton X-100, containing protease and phosphatase inhibitors as before), immunoprecipitated samples were boiled in 2X SDS-loading buffer and the supernatant was subjected to 10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were incubated with anti-phosphoserine (Invitrogen, Waltham, USA) at a 1:10,000 dilution followed by incubation with appropriate secondary antibody. After stripping, membranes were reprobed with anti-GFP (Roche) at a 1:10,000 dilution followed by incubation with the secondary antibody. Where mentioned, stripped membranes were also reprobed with anti-phosphotyrosine (Zymed Laboratories, Invitrogen, Waltham, USA) at a 1:3,000 dilution. Immunodetection was performed by chemiluminescence using a kit from GE Healthcare. Western-blots shown are representative of at least 3 independent experiments with similar results.
Oxygen consumption
Oxygen consumption rate was measured for 7.5 × 108 cells in the culture media using an oxygen electrode (Oxygraph, Hansatech). Data was analyzed using the Oxyg32 V2.25 software (Oxygraph, Hansatech).
Oxidative stress resistance and chronological lifespan
Oxidative stress resistance was determined in yeast cells grown to exponential phase (OD600nm = 0.6) and treated with 0.5 mM H2O2 (Merck, Darmstadt, Germany) for 30 min. Chronological lifespan was assayed as previously described.Citation39 Briefly, overnight cultures were diluted to OD600nm = 0.5 and grown for 48h (stationary phase; considered t0 in the lifespan assay) and kept in culture media at 26°C for the indicated times. Cell viability was determined by standard dilution plate counts on YPD medium containing 1.5 % agar. Colonies were counted after growth at 26°C for 3 d. Viability was expressed as the percentage of the colony-forming units.
Cell cycle analysis
107 cells in exponential phase were collected and fixed with ethanol 70 % overnight at 4°C. Cells were washed twice in 1 ml of 50 mM sodium citrate pH 7.0 buffer and resuspended in the same buffer. RNase A was added to a final concentration of 0.25 mg ml−1 and incubated at 50°C for 1 h. Cells were washed in 50 mM sodium citrate pH 7.0 and sonicated for 2 min at output 2, duty cycle 20% for 2 min, incubated with propidium iodide (16 μg ml−1), incubated for 30 min at room temperature, and analyzed by flow cytometry. Fluorescence was measured on the FL-2 channel of a Becton-Dickinson FACSort flow cytometer (excitation and emission 488 and 585 nm, respectively). The data was analyzed using the FlowJo software (Tree Star).
Results
Phosphoproteomic analysis of sit4Δ cells
The reversible phosphorylation of proteins plays a major role in signal transduction, transcriptional regulation, and metabolic control by affecting protein function, activity, localization or interactions. Since Sit4p is the catalytic subunit of a type 2A ceramide-activated protein phosphatase that was previously implicated in the regulation of oxidative stress resistance and chronological lifespan,Citation25 we raised the hypothesis that changes in protein phosphorylation could mediate the phenotypes of sit4Δ cells. To identify alterations in the (phospho)proteome associated with loss of Sit4p, protein extracts prepared from parental and sit4Δ cells were separated by 2D-gel electrophoresis and silver stained or blotted into a nitrocellulose membrane. Proteins phosphorylated at serine residues were analyzed by immunodetection, using an anti-phosphoserine antibody.
The analysis of total protein patterns showed that the deficiency in Sit4p led to an increase of the levels of 6 proteins (9 spots) whereas that of 27 proteins (30 spots) decreased ( and Supplemental Table 1). Proteins differentially expressed were sorted into functional categories according to MIPS (Munich information center for protein sequences; ). The most significantly affected cell functions were related to carbohydrate metabolism and energy production. Notably, 5 out of the 6 induced proteins and 30% of repressed proteins were associated with carbohydrate metabolism. Some of these changes, such as the increase in the levels of Eno1p (expression repressed by glucose) and the decrease in the levels of Eno2p (expression induced by glucose), are consistent with the catabolite derepression in sit4Δ mutants.Citation16 Remarkably, we also observed a decrease in the levels of 5 proteins associated with stress responses, including the antioxidant defenses Sod1p and Ahp1p that catalyze the disproportionation of superoxide radicals Citation40 and the reduction of alkyl hydroperoxides,Citation41 respectively.
Figure 1. Analysis of changes in the proteome of sit4Δ cells. Yeast extracts were prepared from S. cerevisiae BY4741 and sit4Δ cells grown in YPD medium to exponential phase. Proteins were separated by 2-dimensional gel electrophoresis and visualized by silver staining (A) or blotted into a nitrocellulose membrane. Immunodetection of proteins phosphorylated in serine residues was performed using an anti-phosphoserine antibody (B), as described in Materials and Methods. The experiment was reproduced 3 times, using independent samples. A representative gel/blot is shown. Arrows indicate proteins differentially expressed in sit4Δ cells compared to BY4741 cells.
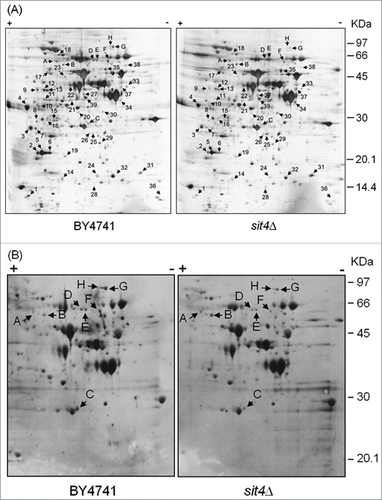
Table 2. Functional categories of proteins differentially expressed in sit4Δ cells. Proteins differentially expressed (sit4Δ vs parental cells) were sorted into functional categories according to MIPS (Munich information center for protein sequences). Proteins belonging to each category are indicated, together with their fold-change (in parentheses). Upregulated proteins are shown in bold.
The proteomic data also showed a decrease in the levels of several proteins associated with nucleotide metabolism and protein translation that may contribute to the low cellular growth rate observed in the sit4Δ strain.Citation42 One of these proteins, Tma19p, is the yeast ortholog of the translationally controlled tumor protein (TCTP) and it is translocated from cytoplasm to the outer surface of the mitochondria after induction of apoptosis by oxidative stress or replicative aging. Notably, TMA19 deletion increases the resistance to hydrogen peroxide and mother cell-specific lifespan.Citation43 The decrease in Tma19p may therefore contribute to oxidative stress resistance and increased lifespan of sit4Δ cells.
In the analysis of the phosphoproteome, 57 protein spots were detected in parental cells, 8 of them being differentially phosphorylated in sit4Δ cells (). These spots were identified by mass spectrometry and correspond to 6 proteins (). The levels of phospho-Hxk2p and phospho-Dug1p increased, as expected in a phosphatase deficient strain. The decrease in the phosphorylation status of the other proteins may result from indirect effects. Changes in the phosphorylation of Dug1p and Cys4p, 2 proteins associated with glutathione metabolism,Citation44,45 may explain the increased levels of this tripeptide in sit4Δ cells (Citation27; our results, data not shown). However, glutathione deficiency does not affect oxidative stress resistance of a sit4-110 mutant.Citation27
Table 3. Identification of proteins differentially phosphorylated in sit4Δ mutants.
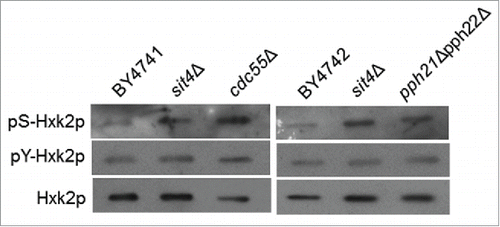
A Western blot analysis of Hxk2p immunoprecipitated from protein extracts of cells expressing Hxk2p-GFP confirmed that serine phosphorylation of Hxk2p increases in cells lacking Sit4p (). This effect seems to be specific for serine since no changes in phospho-Hxk2p levels were observed using an anti-phosphotyrosine antibody. Notably, the deletion of CDC55 gene, which encodes a regulatory subunit B of the CAPP complex,Citation2 also promoted serine phosphorylation of Hxk2p. Moreover, the levels of Hxk2p phosphorylation in cdc55Δ mutants (2.9-fold relative increase) were higher to those observed in sit4Δ cells (1.9-fold relative increase) (). This is probably due to the fact that Cdc55p is also the regulatory subunit B of type 2A protein phosphatase (PP2A), a heterotrimeric complex that contains Pph21p or Pph22p as catalytic subunit and Tpd3p as regulatory subunit A.Citation46-48 Indeed, downregulation of Pph21p and Pph22p also increased serine phosphorylation of Hxk2p (1.9-fold relative increase) to levels similar to those of sit4Δ cells (), suggesting that both CAPP and PP2A contribute to the regulation of Hxk2p phosphorylation.
Figure 2. Hxk2p is hyperphosphorylated in cells with compromised CAPP or PP2A activity. Hxk2p-GFP was immunoprecipitated from the indicated mutants and immunodetected using anti-phosphoserine, anti-phosphotyrosine or anti-GFP antibodies, as described in Material and Methods. A representative experiment is shown.
Hxk2p-S15 phosphorylation is essential for oxidative stress resistance, lifespan extension and G1 arrest in sit4Δ cells
Together with Mig1p and Snf1p, Hxk2p is part of an interconnected network that regulates both replicative and chronological lifespan in yeast.Citation49-51 The phosphorylation of Hxk2p in serine-15 was previously implicated in glucose signaling.Citation52,53 To assess the importance of Hxk2p-S15 phosphorylation in the increase of oxidative stress resistance and chronological lifespan in sit4Δ cells, SIT4 gene was deleted in S. cerevisiae WAY.78-1 cells expressing wild type hexokinase 2 (Hxk2p-S15) or a mutant protein with an alanine instead of serine at position 15 (Hxk2p-S15A; phosphoresistant mutation). This strain is isogenic to the ENY.WA parental strain (K.-D. Entian, Frankfurt, Germany). We have used a different genetic background to confirm that the phenotypes of sit4Δ cells were not specific of the BY4741 strain. Moreover, WAY.78-1 cells do not express the other 2 yeast hexokinases and one of them, Hxk1p, was also previously described as a phosphoprotein.Citation53 Therefore, the use of this strain allowed us to specifically test the role of Hxk2p phosphorylation on the phenotypes of sit4Δ cells. As observed in the BY4741 background,Citation25 SIT4 deletion in WAY.78-1 cells expressing wild type hexokinase (Hxk2p-S15) increased oxygen consumption, H2O2 resistance and chronological lifespan (). These effects were significantly decreased when SIT4 was deleted in cells expressing Hxk2p-S15A. Interestingly, the Hxk2p-S15A mutation decreased chronological lifespan even in the presence of Sit4p (), further supporting the importance of Hxk2p-S15 phosphorylation for cell survival during aging.
Figure 3. Hxk2p-S15 to Hxk2p-A15 mutation suppresses the phenotypes of sit4Δ cells. SIT4 gene was deleted in S. cerevisiae WAY.78-1 cells expressing wild type (Hxk2p-S15) or mutant (Hxk2p-S15A) hexokinase 2. (A) Oxygen consumption rates were measured as described in Materials and Methods using exponential phase cells grown in YPD medium. (B) Chronological lifespan was assessed by following the survival of cells maintained in the growth medium overtime. Cellular viability was measured at 2 to 3 d intervals and was expressed as % colony forming units (aged vs day 0). Values are mean ± SD of at least 3 independent experiments. (C) Oxidative stress resistance was assessed in cells were treated with 0.5 mM H2O2 for 30 min. Cell viability was determined by standard dilution plate counts and expressed as the percentage of the colony-forming units of non-stressed cells. Values are means ± SD of 3 independent experiments. *p < 0.05, **p < 0.01; Student's t-test. (D) Growth curves. (E) Cell cycle. Yeast cells were labeled with propidium iodide and analyzed by flow cytometry. The experiments were reproduced 3 times, using independent samples. A representative experiment is shown. (F) Quantification of the percentage of cells in the different phases of the cell cycle [from (E)].
![Figure 3. Hxk2p-S15 to Hxk2p-A15 mutation suppresses the phenotypes of sit4Δ cells. SIT4 gene was deleted in S. cerevisiae WAY.78-1 cells expressing wild type (Hxk2p-S15) or mutant (Hxk2p-S15A) hexokinase 2. (A) Oxygen consumption rates were measured as described in Materials and Methods using exponential phase cells grown in YPD medium. (B) Chronological lifespan was assessed by following the survival of cells maintained in the growth medium overtime. Cellular viability was measured at 2 to 3 d intervals and was expressed as % colony forming units (aged vs day 0). Values are mean ± SD of at least 3 independent experiments. (C) Oxidative stress resistance was assessed in cells were treated with 0.5 mM H2O2 for 30 min. Cell viability was determined by standard dilution plate counts and expressed as the percentage of the colony-forming units of non-stressed cells. Values are means ± SD of 3 independent experiments. *p < 0.05, **p < 0.01; Student's t-test. (D) Growth curves. (E) Cell cycle. Yeast cells were labeled with propidium iodide and analyzed by flow cytometry. The experiments were reproduced 3 times, using independent samples. A representative experiment is shown. (F) Quantification of the percentage of cells in the different phases of the cell cycle [from (E)].](/cms/asset/85310855-0d83-4c4e-a31f-95383cfd8673/kccy_a_1183846_f0003_b.gif)
It was previously shown that cells lacking Sit4p exhibit slow growth due to cell cycle arrest in G1 phase.Citation42 To investigate if this phenotype is associated with changes in Hxk2p phosphorylation, we measured the effect of SIT4 deletion on the growth rate of cells expressing Hxk2p-S15 or Hxk2p-S15A. As expected, sit4Δ Hxk2p-S15 cells presented a slow growth phenotype (generation time = 405min) but cell growth was not affected in sit4Δ Hxk2p-S15A cells, compared to the correspondent parental cells (Hxk2p-S15A; g = 150 min) (). To correlate these results with G1 arrest, we analyzed the cell cycle of these cells. The sit4Δ Hxk2p-S15A strain exhibited an increased number of cells in the G2/M phase, compared with that in sit4Δ Hxk2p-S15 cells (). These results show that Hxk2p functions downstream of Sit4p in the control of oxidative stress resistance, chronological lifespan and cell cycle.
Hxk2p hypophosphorylation contributes to premature aging in isc1Δ cells
As Sit4p is activated in cells lacking Isc1p,Citation25 we postulated that the phosphorylation of Hxk2p and Dug1p decrease in those mutant cells by a Sit4p-dependent mechanism. Indeed, a phosphoproteomic analysis revealed that isc1Δ cells display lower levels of phospho-Hxk2p and phospho-Dug1p that were suppressed in isc1Δsit4Δ cells (). These results further support a role of Sit4p in the regulation of Hxk2p and Dug1p.
Figure 4. Hxk2p is hypophosphorylated in isc1Δ cells by a Sit4p-dependent mechanism. (A) Yeast extracts were prepared from S. cerevisiae BY4741, isc1Δ and isc1Δsit4Δ cells and a phosphoproteome analysis was performed, as described in legend to . The region of the blots showing Hxk2p and Dug1p was selected. Spot intensities were quantified by densitometry. Values are means ± SD of 3 independent experiments. *p < 0.05. (B) Chronological lifespan of S. cerevisiae hxk2Δ and isc1Δhxk2Δ cells expressing Hxk2p-S15 or Hxk2p-S15E. Cellular viability was determined as described in legend to . Values are means ± SD of 4 independent experiments.
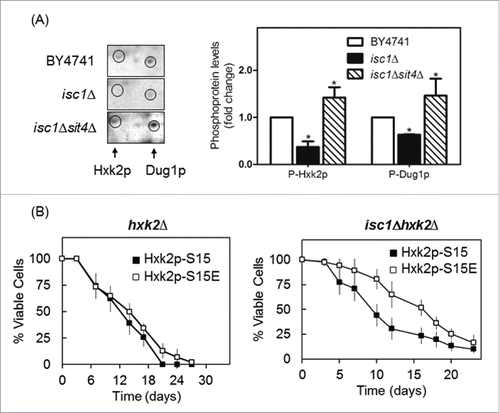
To examine whether Hxk2p hypophosphorylation contributes to the shortened lifespan of isc1Δ cells, we expressed wild type hexokinase (Hxk2p-S15) or this protein with a phosphomimetic serine to glutamate mutation at position Ser-15 (Hxk2p-S15E) in isc1Δhxk2Δ cells. As shown in , the lifespan of isc1Δhxk2Δ cells increased upon expression of Hxk2p-S15E vs Hxk2p-S15. However, the phosphomimetic Hxk2p-S15E mutation did not significantly affect the lifespan of hxk2Δ cells (). Hxk2p phosphorylation increases in wild type cells upon glucose depletion,Citation53 which also occurs before yeast cells reach stationary phase when chronological lifespan is measured. This may explain why the expression of the phosphomimetic Hxk2p-S15E did not further enhance the lifespan of parental cells. Moreover, hxk2Δ Hxk2p-S15E cells did not show the slow growth phenotype exhibited by sit4Δ mutants (data not shown). The overall results suggest that Hxk2p functions downstream of Sit4p in the control of oxidative stress resistance, chronological lifespan and cell cycle, but other unidentified protein(s) may be involved.
Hxk2p is not a direct target of Sit4p or phosphorylated by Snf1p in sit4Δ cells
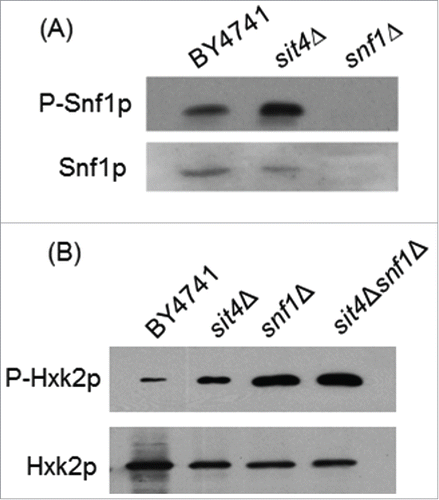
Our results led us to postulate that Sit4p may directly dephosphorylate Hxk2p. However, co-immunoprecipitation assays using Sit4p-TAP and Hxk2p-GFP constructs did not reveal a direct interaction between these proteins. In vitro assays using a Sit4p-GST construct also suggest that Hxk2p is not a direct target of Sit4p (data not shown). Sit4p is known to dephosphorylate a Thr-210 on the activation loop of Snf1p, inhibiting this kinase.Citation21 Thus, Snf1p activation in sit4Δ cells could mediate Hxk2p phosphorylation. In fact, it was previously reported that Hxk2p is a substrate of Snf1p,Citation54 a protein kinase that senses and regulates energy homeostasis.Citation17 However, other studies indicate that Snf1p is not essential for Hxk2p phosphorylation Citation34 although it seems to positively modulate Hxk2p phosphorylation.Citation55 Consistently, our results showed an increase of Snf1p phosphorylation in sit4Δ cells (). However, Hxk2p phosphorylation did not decrease (it even increased) in sit4Δsnf1Δ when comparing with sit4Δ cells (), suggesting that Snf1p activation does not contribute to Hxk2p phosphorylation in sit4Δ cells.
Figure 5. Snf1p activation does not mediate Hxk2p phosphorylation in sit4Δ cells. (A) Protein extracts from S. cerevisiae BY4741, sit4Δ and snf1Δ cells were analyzed by immunoblotting using anti-Snf1 or anti-AMPK (phospho T172) as described in Materials and Methods. (B) Hxk2p-GFP was immunoprecipitated from protein extracts of S. cerevisiae BY4741, sit4Δ, snf1Δ and sit4Δsnf1Δ cells and analyzed by immunoblotting using anti-GFP or anti-phosphoserine as described in Materials and Methods. Representative blots are shown (out of 3 independent experiments).
Discussion
The modulation and coordination of cell signaling pathways plays critical roles in oxidative stress resistance and cell longevity. Signal transduction often involves the reversible phosphorylation of proteins. Ceramide is an evolutionary conserved molecule that affects cell functions through the regulation of protein kinases or phosphatases.Citation1 We have shown that loss of Sit4p, the catalytic subunit of a ceramide-activated type 2A protein phosphatase, increases oxidative stress resistance and extends chronological lifespan.Citation25 In this study, we adopted a phosphoproteomic approach in order to define potential candidate substrates for Sit4p that could mechanistically mediate the actions of this phosphatase. Importantly, we found that hexokinase 2 (Hxk2p) and Dug1p are hyperphosphorylated in sit4Δ cells. The increase in phospho-Dug1p suggests that Sit4p has a function, direct or indirect, in the dephosphorylation of Dug1p, a protein associated with a glutathione degradation pathway.Citation44 Interestingly, the phosphorylation of Cys4p (cystathionine β-syntethase), a protein involved in the biosynthesis of cysteine that is required for glutathione production,Citation45 decreased in sit4Δ cells. Although none of these proteins have been described as phosphoproteins, changes in its phosphorylation status are probably related to the increase of glutathione levels in sit4Δ cells. However, the oxidative stress resistance of sit4Δ cells is glutathione-independent.Citation27
Hxk2p is a glycolytic enzyme involved in glucose signaling and aging.Citation49-51,56 Hxk2p phosphorylation has a regulatory function in the Mig1p-dependent glucose signaling pathway. Under derepressing conditions, Hxk2p is phosphorylated.Citation34 Under repressing conditions, Hxk2p is dephosphorylated by the Reg1p-Glc7p protein phosphatase complex and forms an active dimer that is imported into the nucleus where it interacts with the Mig1p transcription factor and represses genes associated with growth on non-fermentable carbon sources.Citation20 The phosphorylation and inhibition of Mig1p mediated by the Snf1p kinase is prevented by Snf1p dephosphorylation mediated by Reg1p-Glc7p or Sit4p.Citation21
Our results suggest that Sit4p also has an indirect function in Hxk2p dephosphorylation and Hxk2p-S15 phosphorylation is critical for mitochondrial derepression and for cell survival upon H2O2 stress and during chronological aging. Mechanistically, the mutation of S15 to A15 in Hxk2p abolished the protective effect of SIT4 deletion, thus demonstrating the critical role for this phosphorylation site in mediating the response to loss of Sit4p. In mammalian cells, the Akt-dependent phosphorylation of hexokinase 2 promotes its association with the mitochondrial outer membrane, suppressing apoptotic cell death.Citation57,58 A similar mechanism may exist in yeast, but it remains to be demonstrated.
Ceramide can be produced via de novo biosynthesis or through the hydrolysis of complex sphingolipids. We have shown that cells lacking Isc1p, an ortholog of mammalian neutral sphingomyelinase 2, present high levels of dihydro-C26-ceramide and phyto-C26-ceramide, and that the activation of Sit4p mediates mitochondrial dysfunction, oxidative stress sensitivity and premature aging of isc1Δ cells.Citation25 Notably, Hxk2p was hypophosphorylated in isc1Δ cells by a Sit4p-dependent mechanism. This result supports a role for Hxk2p phosphorylation in mitochondria function, H2O2 resistance and chronological lifespan. The regulation of Hxk2p by CAPP is further supported by our data showing that Hxk2p phosphorylation increases in cells lacking Cdc55p. Our results show that PP2A is also involved in the modulation of Hxk2p phosphorylation, probably in response to metabolic cues unrelated to sphingolipid signaling.
Our proteomic data also showed a decrease in the level of several proteins associated with purine metabolism and protein synthesis that may contribute to the slow growth phenotype of sit4Δ cells. Notably, our results show that the cell cycle arrest in G1 phase, characteristic of sit4Δ mutant cells, is also mediated by Hxk2p phosphorylation. The cell cycle arrest in sit4Δ cells may promote cell survival by providing more time to repair molecular damages and prevent mutagenesis, as previous suggested.Citation59 This probably contributes to the cellular protection observed in isc1Δsit4Δ cells since oxidized proteins and lipids accumulate to higher levels in isc1Δ mutants when stressed or aged.Citation28 DNA repair also seems to be critical for survival of isc1Δ cells.Citation60 In addition, Isc1p protects yeast from genotoxic stress by regulating Swe1p, a protein kinase that regulates the G2/M transition,Citation61 through Cdc55p, allowing cell cycle progression.Citation62,63
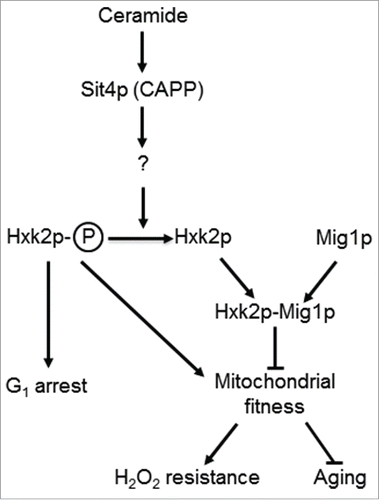
In summary, our data suggests that Hxk2p phosphorylation is required for phenotypes associated with SIT4 deletion, namely mitochondrial derepression, growth arrest in G1 phase of the cell cycle, oxidative stress resistance and lifespan extension. We propose that the activation of Sit4p in isc1Δ cells indirectly promotes Hxk2p dephosphorylation, leading to mitochondrial dysfunction, premature aging and oxidative stress sensitivity (). These results offer new insights on the regulation of mitochondrial function and aging by hexokinase 2 and sphingolipid signaling.
Figure 6. A model for the role of Sit4p in the regulation mitochondrial function. In parental cells, dihydroceramides (dh-Cer) and phytoceramide (phyto-Cer) levels are kept low, preventing the dephosphorylation of hexokinase 2 (Hxk2p) in serine-15, indirectly regulated by the ceramide-activated protein phosphatase Sit4p. In sit4Δ cells, the increase of Hxk2p phosphorylation contributes to mitochondrial fitness, increasing oxidative stress resistance and chronological lifespan. In isc1Δ cells, the higher levels of dh-Cer- and phyto-Cer activate Sit4p leading to Hxk2p dephosphorylation. This results in mitochondrial dysfunction, leading to a disturbed redox homeostasis, premature aging and oxidative stress sensitivity. Other proteins regulated by Sit4p may also contribute to these phenotypes.
Abbreviations
| CAPP | = | ceramide-activated protein phosphatase |
| PP2A | = | type 2A protein phosphatase |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KCCY_A_1183846_Supplemental.pdf
Download PDF (34.5 KB)Acknowledgments
We are grateful to Dr. Francisca Randez-Gil (CSIC, Valencia, Spain), Dr. Fernando Moreno (Universidad de Oviedo, Spain), Dr. Jeffrey E. Gerst (Weizmann Institute of Science, Israel), Dr. Manfred Schmitt (University of Saarlandes, Germany) and Dr. Johan Thevelein (KULeuven, Belgium) for generously providing yeast strains and plasmids used in this study.
Funding
This work was financed by FEDER - Fundo Europeu de Desenvolvimento Regional funds through the COMPETE 2020 - Operacional Program for Competitiveness and Internationalisation (POCI), Portugal 2020, and by Portuguese funds through FCT - Fundação para a Ciência e a Tecnologia/ Ministério da Ciência, Tecnologia e Inovação in the framework of the projects “Institute for Research and Innovation in Health Sciences” (POCI-01-0145-FEDER-007274) and FCOMP-01-0124-FEDER-028210 (PTDC/BBB-BQB/1850/2012). Clara Pereira was supported by a FCT fellowship (SFRH/BPD/100607/2014).
References
- Hannun YA, Obeid LM. Principles of bioactive lipid signalling: lessons from sphingolipids. Nat Rev Mol Cell Biol 2008; 9:139-50; PMID:18216770; http://dx.doi.org/10.1038/nrm2329
- Nickels JT, Broach JR. A ceramide-activated protein phosphatase mediates ceramide-induced G1 arrest of Saccharomyces cerevisiae. Genes Dev 1996; 10:382-94; PMID:8600023; http://dx.doi.org/10.1101/gad.10.4.382
- Bastians H, Ponstingl H. The novel human protein serine/threonine phosphatase 6 is a functional homologue of budding yeast Sit4p and fission yeast ppe1, which are involved in cell cycle regulation. J Cell Sci 1996; 109(Pt 12):2865-74; PMID:9013334
- Shimanuki M, Kinoshita N, Ohkura H, Yoshida T, Toda T, Yanagida M. Isolation and characterization of the fission yeast protein phosphatase gene ppe1+ involved in cell shape control and mitosis. Mol Biol Cell 1993; 4:303-13; PMID:8387356; http://dx.doi.org/10.1091/mbc.4.3.303
- Fernandez-Sarabia MJ, Sutton A, Zhong T, Arndt KT. SIT4 protein phosphatase is required for the normal accumulation of SWI4, CLN1, CLN2, and HCS26 RNAs during late G1. Genes Dev 1992; 6:2417-28; PMID:1334024; http://dx.doi.org/10.1101/gad.6.12a.2417
- Di Como CJ, Bose R, Arndt KT. Overexpression of SIS2, which contains an extremely acidic region, increases the expression of SWI4, CLN1 and CLN2 in sit4 mutants. Genetics 1995; 139:95-107; PMID:7705654
- Angeles de la Torre-Ruiz M, Torres J, Arino J, Herrero E. Sit4 is required for proper modulation of the biological functions mediated by Pkc1 and the cell integrity pathway in Saccharomyces cerevisiae. J Biol Chem 2002; 277:33468-76; PMID:12080055; http://dx.doi.org/10.1074/jbc.M203515200
- Singer T, Haefner S, Hoffmann M, Fischer M, Ilyina J, Hilt W. Sit4 phosphatase is functionally linked to the ubiquitin-proteasome system. Genetics 2003; 164:1305-21; PMID:12930741
- Hayashi N, Nomura T, Sakumoto N, Mukai Y, Kaneko Y, Harashima S, Murakami S. The SIT4 gene, which encodes protein phosphatase 2A, is required for telomere function in Saccharomyces cerevisiae. Curr Genet 2005; 47:359-67; PMID:15843932; http://dx.doi.org/10.1007/s00294-005-0577-1
- Masuda CA, Ramirez J, Pena A, Montero-Lomeli M. Regulation of monovalent ion homeostasis and pH by the Ser-Thr protein phosphatase SIT4 in Saccharomyces cerevisiae. J Biol Chem 2000; 275:30957-61; PMID:10921924; http://dx.doi.org/10.1074/jbc.M004869200
- Tate JJ, Feller A, Dubois E, Cooper TG. Saccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J Biol Chem 2006; 281:37980-92; PMID:17015442; http://dx.doi.org/10.1074/jbc.M606973200
- Di Como CJ, Arndt KT. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev 1996; 10:1904-16; PMID:8756348; http://dx.doi.org/10.1101/gad.10.15.1904
- McCourt PC, Morgan JM, Nickels JT, Jr. Stress-induced ceramide-activated protein phosphatase can compensate for loss of amphiphysin-like activity in Saccharomyces cerevisiae and functions to reinitiate endocytosis. J Biol Chem 2009; 284:11930-41; PMID:19254955; http://dx.doi.org/10.1074/jbc.M900857200
- Bhandari D, Zhang J, Menon S, Lord C, Chen S, Helm JR, Thorsen K, Corbett KD, Hay JC, Ferro-Novick S. Sit4p/PP6 regulates ER-to-Golgi traffic by controlling the dephosphorylation of COPII coat subunits. Mol Biol Cell 2013; 24:2727-38; PMID:23864707; http://dx.doi.org/10.1091/mbc.E13-02-0114
- Posas F, Clotet J, Arino J. Saccharomyces cerevisiae gene SIT4 is involved in the control of glycogen metabolism. FEBS Lett 1991; 279:341-5; PMID:1848194; http://dx.doi.org/10.1016/0014-5793(91)80183-4
- Jablonka W, Guzman S, Ramirez J, Montero-Lomeli M. Deviation of carbohydrate metabolism by the SIT4 phosphatase in Saccharomyces cerevisiae. Biochim Biophys Acta 2006; 1760:1281-91; PMID:16764994; http://dx.doi.org/10.1016/j.bbagen.2006.02.014
- Conrad M, Schothorst J, Kankipati HN, Van Zeebroeck G, Rubio-Texeira M, Thevelein JM. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 2014; 38:254-99; PMID:24483210; http://dx.doi.org/10.1111/1574-6976.12065
- de Assis LJ, Zingali RB, Masuda CA, Rodrigues SP, Montero-Lomeli M. Pyruvate decarboxylase activity is regulated by the Ser/Thr protein phosphatase Sit4p in the yeast Saccharomyces cerevisiae. FEMS Yeast Res 2013; 13:518-28; PMID:23692511; http://dx.doi.org/10.1111/1567-1364.12052
- Ahuatzi D, Riera A, Pelaez R, Herrero P, Moreno F. Hxk2 regulates the phosphorylation state of Mig1 and therefore its nucleocytoplasmic distribution. J Biol Chem 2007; 282:4485-93; PMID:17178716; http://dx.doi.org/10.1074/jbc.M606854200
- Moreno F, Herrero P. The hexokinase 2-dependent glucose signal transduction pathway of Saccharomyces cerevisiae. FEMS Microbiol Rev 2002; 26:83-90; PMID:12007644; http://dx.doi.org/10.1111/j.1574-6976.2002.tb00600.x
- Ruiz A, Xu X, Carlson M. Roles of two protein phosphatases, Reg1-Glc7 and Sit4, and glycogen synthesis in regulation of SNF1 protein kinase. Proc Natl Acad Sci U S A 2011; 108:6349-54; PMID: 21464305; http://dx.doi.org/10.1073/pnas.1102758108
- Jin C, Barrientos A, Epstein CB, Butow RA, Tzagoloff A. SIT4 regulation of Mig1p-mediated catabolite repression in Saccharomyces cerevisiae. FEBS Lett 2007; 581:5658-63; PMID:18022394; http://dx.doi.org/10.1016/j.febslet.2007.11.027
- Costa V, Moradas-Ferreira P. Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol Aspects Med 2001; 22:217-46; PMID:11679167; http://dx.doi.org/10.1016/S0098-2997(01)00012-7
- Garipler G, Mutlu N, Lack NA, Dunn CD. Deletion of conserved protein phosphatases reverses defects associated with mitochondrial DNA damage in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 2014; 111:1473-8; PMID:24474773; http://dx.doi.org/10.1073/pnas.1312399111
- Barbosa AD, Osorio H, Sims KJ, Almeida T, Alves M, Bielawski J, Amorim MA, Moradas-Ferreira P, Hannun YA, Costa V. Role for Sit4p-dependent mitochondrial dysfunction in mediating the shortened chronological lifespan and oxidative stress sensitivity of Isc1p-deficient cells. Mol Microbiol 2011; 81:515-27; PMID:21707788; http://dx.doi.org/10.1111/j.1365-2958.2011.07714.x
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae. Aging Cell 2003; 2:73-81; PMID:12882320; http://dx.doi.org/10.1046/j.1474-9728.2003.00033.x
- Lopez-Mirabal HR, Winther JR, Kielland-Brandt MC. Oxidant resistance in a yeast mutant deficient in the Sit4 phosphatase. Curr Genet 2008; 53:275-86; PMID:18357452; http://dx.doi.org/10.1007/s00294-008-0184-z
- Almeida T, Marques M, Mojzita D, Amorim MA, Silva RD, Almeida B, Rodrigues P, Ludovico P, Hohmann S, Moradas-Ferreira P, et al. Isc1p plays a key role in hydrogen peroxide resistance and chronological lifespan through modulation of iron levels and apoptosis. Mol Biol Cell 2008; 19:865-76; PMID:18162582; http://dx.doi.org/10.1091/mbc.E07-06-0604
- Sawai H, Okamoto Y, Luberto C, Mao C, Bielawska A, Domae N, Hannun YA. Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J Biol Chem 2000; 275:39793-8; PMID:11006294; http://dx.doi.org/10.1074/jbc.M007721200
- Vaena de Avalos S, Okamoto Y, Hannun YA. Activation and localization of inositol phosphosphingolipid phospholipase C, Isc1p, to the mitochondria during growth of Saccharomyces cerevisiae. J Biol Chem 2004; 279:11537-45; PMID:14699160; http://dx.doi.org/10.1074/jbc.M309586200
- Kitagaki H, Cowart LA, Matmati N, Vaena de Avalos S, Novgorodov SA, Zeidan YH, Bielawski J, Obeid LM, Hannun YA. Isc1 regulates sphingolipid metabolism in yeast mitochondria. Biochim Biophys Acta 2007; 1768:2849-61; PMID:17880915; http://dx.doi.org/10.1016/j.bbamem.2007.07.019
- Vaena de Avalos S, Su X, Zhang M, Okamoto Y, Dowhan W, Hannun YA. The phosphatidylglycerol/cardiolipin biosynthetic pathway is required for the activation of inositol phosphosphingolipid phospholipase C, Isc1p, during growth of Saccharomyces cerevisiae. J Biol Chem 2005; 280:7170-7; PMID:15611094; http://dx.doi.org/10.1074/jbc.M411058200
- Kitagaki H, Cowart LA, Matmati N, Montefusco D, Gandy J, de Avalos SV, Novgorodov SA, Zheng J, Obeid LM, Hannun YA. ISC1-dependent metabolic adaptation reveals an indispensable role for mitochondria in induction of nuclear genes during the diauxic shift in Saccharomyces cerevisiae. J Biol Chem 2009; 284:10818-30; PMID:19179331; http://dx.doi.org/10.1074/jbc.M805029200
- Randez-Gil F, Sanz P, Entian KD, Prieto JA. Carbon source-dependent phosphorylation of hexokinase PII and its role in the glucose-signaling response in yeast. Mol Cell Biol 1998; 18:2940-8; PMID:9566913
- Ahuatzi D, Herrero P, de la Cera T, Moreno F. The glucose-regulated nuclear localization of hexokinase 2 in Saccharomyces cerevisiae is Mig1-dependent. J Biol Chem 2004; 279:14440-6; PMID:14715653; http://dx.doi.org/10.1074/jbc.M313431200
- Costa VM, Amorim MA, Quintanilha A, Moradas-Ferreira P. Hydrogen peroxide-induced carbonylation of key metabolic enzymes in Saccharomyces cerevisiae: the involvement of the oxidative stress response regulators Yap1 and Skn7. Free Radic Biol Med 2002; 33:1507-15; PMID:12446208; http://dx.doi.org/10.1016/S0891-5849(02)01086-9
- Gomes C, Almeida A, Ferreira JA, Silva L, Santos-Sousa H, Pinto-de-Sousa J, Santos LL, Amado F, Schwientek T, Levery SB, et al. Glycoproteomic analysis of serum from patients with gastric precancerous lesions. J Proteome Res 2013; 12:1454-66; PMID:23312025; http://dx.doi.org/10.1021/pr301112x
- Dubacq C, Chevalier A, Mann C. The protein kinase Snf1 is required for tolerance to the ribonucleotide reductase inhibitor hydroxyurea. Mol Cell Biol 2004; 24:2560-72; PMID:14993292; http://dx.doi.org/10.1128/MCB.24.6.2560-2572.2004
- Teixeira V, Medeiros TC, Vilaça R, Moradas-Ferreira P, Costa V. Reduced TORC1 signaling abolishes mitochondrial dysfunctions and shortened chronological lifespan of Isc1p-deficient cells. Microbial Cell 2014; 1:21-36; http://dx.doi.org/10.15698/mic2014.01.121
- Bermingham-McDonogh O, Gralla EB, Valentine JS. The copper, zinc-superoxide dismutase gene of Saccharomyces cerevisiae: cloning, sequencing, and biological activity. Proc Natl Acad Sci U S A 1988; 85:4789-93; PMID:3290902; http://dx.doi.org/10.1073/pnas.85.13.4789
- Lee J, Spector D, Godon C, Labarre J, Toledano MB. A new antioxidant with alkyl hydroperoxide defense properties in yeast. J Biol Chem 1999; 274:4537-44; PMID:9988687; http://dx.doi.org/10.1074/jbc.274.8.4537
- Sutton A, Immanuel D, Arndt KT. The SIT4 protein phosphatase functions in late G1 for progression into S phase. Mol Cell Biol 1991; 11:2133-48; PMID:1848673; http://dx.doi.org/10.1128/MCB.11.4.2133
- Rinnerthaler M, Jarolim S, Heeren G, Palle E, Perju S, Klinger H, Bogengruber E, Madeo F, Braun RJ, Breitenbach-Koller L, et al. MMI1 (YKL056c, TMA19), the yeast orthologue of the translationally controlled tumor protein (TCTP) has apoptotic functions and interacts with both microtubules and mitochondria. Biochim Biophys Acta 2006; 1757:631-8; PMID:16806052; http://dx.doi.org/10.1016/j.bbabio.2006.05.022
- Ganguli D, Kumar C, Bachhawat AK. The alternative pathway of glutathione degradation is mediated by a novel protein complex involving three new genes in Saccharomyces cerevisiae. Genetics 2007; 175:1137-51; PMID:17179087; http://dx.doi.org/10.1534/genetics.106.066944
- Ono B, Shirahige Y, Nanjoh A, Andou N, Ohue H, Ishino-Arao Y. Cysteine biosynthesis in Saccharomyces cerevisiae: mutation that confers cystathionine beta-synthase deficiency. J Bacteriol 1988; 170:5883-9; PMID:3056921
- Healy AM, Zolnierowicz S, Stapleton AE, Goebl M, DePaoli-Roach AA, Pringle JR. CDC55, a Saccharomyces cerevisiae gene involved in cellular morphogenesis: identification, characterization, and homology to the B subunit of mammalian type 2A protein phosphatase. Mol Cell Biol 1991; 11:5767-80; PMID:1656238; http://dx.doi.org/10.1128/MCB.11.11.5767
- Sneddon AA, Cohen PT, Stark MJ. Saccharomyces cerevisiae protein phosphatase 2A performs an essential cellular function and is encoded by two genes. EMBO J 1990; 9:4339-46; PMID:2176150
- van Zyl W, Huang W, Sneddon AA, Stark M, Camier S, Werner M, Marck C, Sentenac A, Broach JR. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol 1992; 12:4946-59; PMID:1328868
- Kaeberlein M, Kirkland KT, Fields S, Kennedy BK. Genes determining yeast replicative life span in a long-lived genetic background. Mech Ageing Dev 2005; 126:491-504; PMID:15722108; http://dx.doi.org/10.1016/j.mad.2004.10.007
- Lorenz DR, Cantor CR, Collins JJ. A network biology approach to aging in yeast. Proc Natl Acad Sci U S A 2009; 106:1145-50; PMID:19164565; http://dx.doi.org/10.1073/pnas.0812551106
- Yao Y, Tsuchiyama S, Yang C, Bulteau AL, He C, Robison B, Tsuchiya M, Miller D, Briones V, Tar K, et al. Proteasomes, Sir2, and Hxk2 form an interconnected aging network that impinges on the AMPK/Snf1-regulated transcriptional repressor Mig1. PLoS Genet 2015; 11:e1004968; PMID:25629410; http://dx.doi.org/10.1371/journal.pgen.1004968
- Kriegel TM, Rush J, Vojtek AB, Clifton D, Fraenkel DG. In vivo phosphorylation site of hexokinase 2 in Saccharomyces cerevisiae. Biochemistry 1994; 33:148-52; PMID:8286332; http://dx.doi.org/10.1021/bi00167a019
- Vojtek AB, Fraenkel DG. Phosphorylation of yeast hexokinases. Eur J Biochem 1990; 190:371-5; PMID:2163841; http://dx.doi.org/10.1111/j.1432-1033.1990.tb15585.x
- Fernandez-Garcia P, Pelaez R, Herrero P, Moreno F. Phosphorylation of yeast hexokinase 2 regulates its nucleocytoplasmic shuttling. J Biol Chem 2012; 287:42151-64; PMID:23066030; http://dx.doi.org/10.1074/jbc.M112.401679
- Kaps S, Kettner K, Migotti R, Kanashova T, Krause U, Rodel G, Dittmar G, Kriegel TM. Protein kinase Ymr291w/Tda1 is essential for glucose signaling in Saccharomyces cerevisiae on the level of hexokinase isoenzyme ScHxk2 phosphorylation. J Biol Chem 2015; 290:6243-55; PMID:25593311; http://dx.doi.org/10.1074/jbc.M114.595074
- Rodriguez A, De La Cera T, Herrero P, Moreno F. The hexokinase 2 protein regulates the expression of the GLK1, HXK1 and HXK2 genes of Saccharomyces cerevisiae. Biochem J 2001; 355:625-31; PMID:11311123; http://dx.doi.org/10.1042/bj3550625
- Majewski N, Nogueira V, Robey RB, Hay N. Akt inhibits apoptosis downstream of BID cleavage via a glucose-dependent mechanism involving mitochondrial hexokinases. Mol Cell Biol 2004; 24:730-40; PMID:14701745; http://dx.doi.org/10.1128/MCB.24.2.730-740.2004
- Miyamoto S, Murphy AN, Brown JH. Akt mediates mitochondrial protection in cardiomyocytes through phosphorylation of mitochondrial hexokinase-II. Cell Death Differ 2008; 15:521-9; PMID:18064042; http://dx.doi.org/10.1038/sj.cdd.4402285
- Shackelford RE, Kaufmann WK, Paules RS. Oxidative stress and cell cycle checkpoint function. Free Radic Biol Med 2000; 28:1387-404; PMID:10924858; http://dx.doi.org/10.1016/S0891-5849(00)00224-0
- Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A 2002; 99:16934-9; PMID:12482937; http://dx.doi.org/10.1073/pnas.262669299
- Booher RN, Deshaies RJ, Kirschner MW. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J 1993; 12:3417-26; PMID:8253069
- Matmati N, Kitagaki H, Montefusco D, Mohanty BK, Hannun YA. Hydroxyurea sensitivity reveals a role for ISC1 in the regulation of G2/M. J Biol Chem 2009; 284:8241-6; PMID:19158081; http://dx.doi.org/10.1074/jbc.M900004200
- Matmati N, Metelli A, Tripathi K, Yan S, Mohanty BK, Hannun YA. Identification of C18:1-phytoceramide as the candidate lipid mediator for hydroxyurea resistance in yeast. J Biol Chem 2013; 288:17272-84; PMID:23620586; http://dx.doi.org/10.1074/jbc.M112.444802
- Castermans D, Somers I, Kriel J, Louwet W, Wera S, Versele M, Janssens V, Thevelein JM. Glucose-induced posttranslational activation of protein phosphatases PP2A and PP1 in yeast. Cell Res 2012; 22:1058-77; PMID:22290422; http://dx.doi.org/10.1038/cr.2012.20
