ABSTRACT
USP1 deubiquitinating enzyme and its stoichiometric binding partner UAF1 play an essential role in promoting DNA homologous recombination (HR) repair in response to various types of DNA damaging agents. Deubiquitination of FANCD2 may be attributed to the key role of USP1-UAF1 complex in regulating HR repair, however whether USP1-UAF1 promotes HR repair independently of FANCD2 deubiquitination is not known. Here we show evidence that the USP1-UAF1 complex has a FANCD2-independent function in promoting HR repair. Proteomic search of UAF1-interacting proteins revealed that UAF1 associates with RAD51AP1, a RAD51-interacting protein implicated in HR repair. We show that UAF1 mediates the interaction between USP1 and RAD51AP1, and that depletion of USP1 or UAF1 led to a decreased stability of RAD51AP1. Protein interaction mapping analysis identified some key residues within RAD51AP1 required for interacting with the USP1-UAF1 complex. Cells expressing the UAF1 interaction-deficient mutant of RAD51AP1 show increased chromosomal aberrations in response to Mitomycin C treatment. Moreover, similar to the RAD51AP1 depleted cells, the cells expressing UAF1-interaction deficient RAD51AP1 display persistent RAD51 foci following DNA damage exposure, indicating that these factors regulate a later step during the HR repair. These data altogether suggest that the USP1-UAF1 complex promotes HR repair via multiple mechanisms: through FANCD2 deubiquitination, as well as by interacting with RAD51AP1.
Introduction
DNA double strand breaks (DSBs) are highly lethal LESIONS that must be repaired before cell division ensues. Homologous Recombination (HR) repair and Non-homologous end joining (NHEJ) repair represent 2 major forms of DSB repair mechanisms. The HR repair operates by duplicating genetic information from opposite sister chromatids. One of the key events in initiating HR repair is chromatin loading of RAD51, a ssDNA binding protein that facilitates homology search in the sister chromatid to copy the lost genetic material. In brief, RAD51-dependent HR pathway has a few distinct steps; a presynaptic step in which RAD51 binds the 3′end overhang of ssDNA generated at the resected DSB ends, to assemble nucleoprotein filaments, followed by strand invasion of the nucleofilament into the opposite undamaged chromatids and capture of the homology sequences, and finally DNA synthesis and resolution of the heteroduplex structures to complete the repair.Citation1,2 A number of RAD51-associated proteins support the activity of RAD51 to aid in the distinct phases during the repair process. For example, RAD51 paralogs (RAD51B, RAD51C, RAD51D, XRCC2, XRCC3) promotes the loading of RAD51 to ssDNA,Citation3 whereas RAD51AP1 (RAD51-Associated Protein 1) was suggested to function subsequent to the ssDNA-RAD51 nucleofilament formation.Citation4,5
The ubiquitin-proteasome system (UPS) is intimately implicated in the regulation of the DNA repair and DNA damage response. Deubiquitinating enzymes (DUBs) have emerged as an important class of regulators of the UPS.Citation6 By removing covalently attached ubiquitin molecules from substrates or polyubiquitinated chains, DUBs act as balancers of the ubiquitination-proteasome system. USP1, initially identified as a deubiquitinase of FANCD2,Citation7 is an essential component of the Fanconi Anemia (FA) DNA repair pathway.Citation8 Inactivation of USP1 in mouse Citation9 and chicken DT40 Citation10 cells result in increased cellular sensitivity to DNA interstrand crosslinking agents that is associated with hyper-monoubiquitination of FANCD2. The catalytic activity and stability of USP1 is promoted by its stoichiometric binding partner UAF1 (USP1-Associated Factor 1; WDR48), a WD40 repeat containing protein.Citation11 Both USP1 and UAF1 are regulators of the HR repair, as knockouts of USP1 or UAF1 in DT40 cells show reduced HR repair efficiency.Citation12 The USP1-UAF1 complex also deubiquitinates FANCI, which interacts with FANCD2,Citation13 and a replicative polymerase processive factor PCNA.Citation14 Altogether, USP1 and UAF1 are important contributors to the genome integrity at least in part by regulating the HR and TLS DNA repair pathways.
With regard to the regulation of HR repair, the current model implies that USP1 and UAF1 regulate the HR repair by facilitating the loading and unloading cycles of FANCD2 at the damaged chromatin. FANCD2 is required for efficient recruitment of CtIP,Citation15-17 an endonuclease that induces end resection at DSB sites to generate ssDNA, an important step that initiates the HR repair. Whether the role of USP1 and UAF1 in HR repair is limited to the FANCD2 and CtIP retention at the DSB sites, or whether there are other functions that directly regulate the HR repair proteins, is unknown. Intriguingly, a previous study showed that mouse Fancd2 and Usp1 are not completely epistatic, as the MEFs from double knockout of Fancd2 and Usp1 are further sensitive to Cisplatin compared to the single knockouts.Citation9 This suggests that USP1 may have other functions in DNA repair.
In an attempt to further understand the mechanistic basis for the role of UAF1 and USP1 proteins in promoting the HR repair, we performed a proteomic screen of the UAF1-interacting proteins. We reproducibly found RAD51AP1, a RAD51-binding protein involved in HR repair, to be enriched in the UAF1 immunoprecipitate. Knockdown of RAD51AP1 in human cells or knockout in DT40 cells leads to enhanced sensitivity to IR or DNA interstrand crosslinking agents.Citation4,5,18 RAD51AP1 is not essential for foci formation of RAD51,Citation4,5,18 but by directly associating with RAD51 and DNA, it stimulates the RAD51-mediated D-loop formation,Citation4,5,19 which is an intermediate structure during HR repair. Thus, RAD51AP1 is suggested to act downstream of the RAD51-ssDNA nucleoprotein filament formation. RAD51AP1 also interacts with and stimulates activity of the meiosis-specific recombinase DMC1, to enhance the HR repair in meiotic cells.Citation20,21 Whether the RAD51AP1 activity or stability is regulated by other factors is unknown. We provide evidence that RAD51AP1 interacts with USP1 through UAF1, and that stability of RAD51AP1 is promoted by USP1 and UAF1. Mapping analysis identified several residues of RAD51AP1 to be critical for interaction with UAF1. Cells expressing the UAF1-interaction deficient RAD51AP1 mutant showed increased sensitivity to DNA ICL-inducing agent, suggesting the importance of the UAF1-RAD51AP1 interaction. Our work provides a direct functional link between the deubiquitinating enzyme complex in the FA pathway and the RAD51-dependent HR repair pathway.
Results
USP1 or UAF1 depletion inhibits HR repair by primarily affecting downstream of RAD51 foci formation
USP1 and UAF1 are known to regulate HR repair,Citation12,22 and consistently, we found that USP1 or UAF1 knockdown reduces HR repair efficiency in the cell-based HR reporter assay (Fig. S1). Furthermore, we noticed that USP1 knockdown led to slightly but consistently larger reduction in the HR repair efficiency, compared to FANCD2 knockdown (). A previous study showed that cells isolated from Fancd2 and Usp1 double- knockout mice were further sensitive to DNA damaging agents compared to cells from either Fancd2 or Usp1 single knockouts, which suggested that FANCD2 and USP1 are not completely epistatic in DNA repair pathways.Citation9 Perhaps consistent with this report, co-depletion of USP1 and FANCD2 led to an additive effect on reducing the HR repair efficiency in the cell-based HR repair assay ().
Figure 1. USP1 or UAF1 depletion inhibits HR repair by primarily affecting downstream of RAD51 foci formation. A. USP1 and FANCD2 are not epistatic in the DR-GFP assay. siRNAs are transfected to the U2OS-based HR reporter cells, followed by I-Sce1 transfection. 48 hours later, cells were harvested for Flow cytometric analysis. Three independent experiments were performed (*p<0.001). B, C. UAF1 (B) or USP1 (C) knockdown impairs the disappearance of the RAD51 foci. HeLa cells were treated with UV (20 J/m2), then the cells were fixed at the corresponding time points for IF procedures. On the right are the representative images.
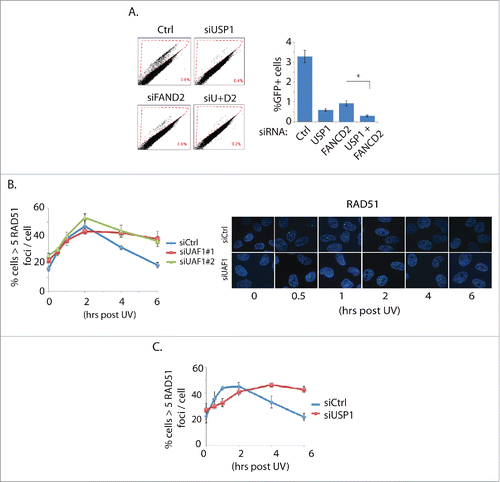
FANCD2 is required for HR repair at least in part by recruiting CtIP nuclease and thus regulating double strand DNA end resection.Citation15-17 Single-stranded stretch of DNA is exposed by end resection and bound by ssDNA binding protein RPA (replication protein A), which is in turn displaced by RAD51. The ATR-mediated phosphorylation of RPA is generally used as a surrogate marker of activated RPA and the DSB end resection status. While BRCA1 knockdown reduced the p-RPA levels, knockdown of USP1 or UAF1 did not significantly affected the p-RPA levels (. S2). This result suggests that USP1 and UAF1 do not significantly influence the DSB end-resection. The reduced HR repair efficiency of the USP1 or UAF1-depleted cells is not caused by a deficiency in the RAD51 foci formation; while initial recruitment of RAD51 foci formation was not visibly different compared to control knockdown, USP1 or UAF1 knockdown by siRNAs noticeably delayed the resolution of RAD51 foci (). The result is consistent with a previous report showing that elevated RAD51 foci are observed in the DT40 UAF1 knockout cells.Citation12 Based on these results, we concluded that USP1 or UAF1 may regulate HR repair at multiple distinct stages: they may regulate HR repair through FANCD2 deubiquitination, but also at a stage subsequent to RAD51 chromatin loading. USP1 or UAF1 did not affect the foci formation of 53BP1 or DSB-associated ubiquitin foci (Fig. S3), which suggests that USP1-UAF1 may not be involved in the canonical DSB repair signaling that can influence the HR repair. Altogether, these results support an idea that USP1 and UAF1 may have a function independent of FANCD2, and at a level downstream of RAD51 foci formation, in addition to deubiquitinating FANCD2.
The USP1-UAF1 complex interacts with RAD51AP1
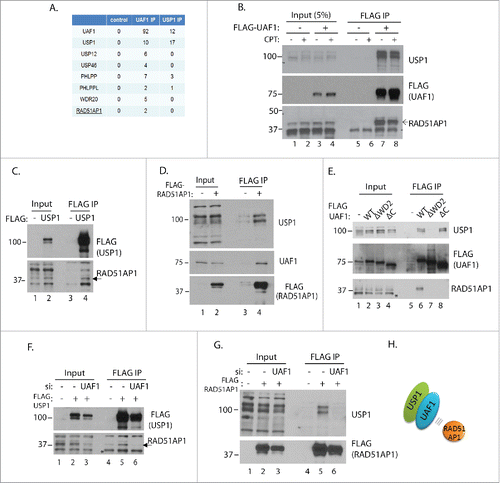
In order to search for the potential role of USP1 or UAF1 in HR repair, we undertook a proteomic approach to identify potential UAF1 or USP1 interacting proteins that regulate HR repair. Anti-FLAG immunoprecipitates of FLAG-USP1 and FLAG-UAF1 complexes isolated from HeLa S3 cells followed by LC-MS (mass spectrometry) identified previously known interacting proteins PHLPP, PHLPPL, USP1, USP12, USP46, and WDR20 (with the latter 3 only identified in UAF1 IP). In addition to these, we repeatedly identified peptides derived from RAD51AP1 protein in the UAF1 IP, in 3 independent experiments (). RAD51AP1 interacting with UAF1 was also reported previously,Citation23,24 however its functional implication remained unknown. Since RAD51AP1 was the only protein implicated in HR repair among the identified, we decided to characterize the physiological significance of the interaction further. The interaction between UAF1 and RAD51AP1 does not appear to be enhanced by exogenous DNA damage (CPT), at least in the IP analysis (). Even though we did not find RAD51AP1 peptides in the USP1 IP-mass spec, co-immunoprecipitation by anti-FLAG-USP1 showed that RAD51AP1 interacts with USP1 (). FLAG-RAD51AP1 immunoprecipitation also reversely pulled down both UAF1 and USP1, suggesting that RAD51AP1 associate with the USP1-UAF1 complex (). The WD40 repeats of UAF1 are involved in interacting with USP1, whereas the C-terminus region of UAF1 that resembles SUMO (termed SUMO-Like domain; SLD) is implicated in recognizing SUMO-interacting motifs in FANCI.Citation11,23 Our interaction analysis shows that interaction with RAD51AP1 requires both WD40 repeats and the C-terminal SLD domain of UAF1 (). To test the idea whether UAF1 serves as a substrate adaptor subunit for USP1, UAF1 was depleted by siRNA then the FLAG-USP1 or FLAG-RAD51AP1 immunoprecipitation was performed (). Indeed, UAF1 depletion significantly abrogated the association between USP1 and RAD51AP1, in both IPs, suggesting that UAF1 is a bridging factor that mediates the interaction between USP1 and RAD51AP1 (a model is shown ). Altogether, these data demonstrate that the USP1-UAF1 complex associates with RAD51AP1.
Figure 2. USP1 and UAF1 interact with RAD51AP1. A. 500 ml cultures of HeLa S3 cells expressing FLAG-UAF1 or USP1 were processed for anti-FLAG immunoprecipitation, followed by mass spec analysis. Shown is the #unique peptides identified from one experiment. B. HeLa S3 cells expressing FLAG-UAF1 were treated with or without CPT (2 uM) for ∼12 hours, then harvested for anti-FLAG IP and blotted with indicated antibodies. C. 293T cells were transfected with 3xFLAG-USP1 plasmid, followed by anti-FLAG IP and anti-RAD51AP1 western blot. D. 293T cells were transfected with 3xFLAG-RAD51AP1 plasmid, followed by anti-FLAG IP, and the western blotting with indicated antibodies. E. 293T cells were transfected with the indicated pcDNA-FLAG-UAF1 plasmids that were described previouslyCitation26,47; ΔWD2 mutant is deleted of the 2nd WD40 domain, and ΔC mutant is deleted of the C-terminal 43 amino acids) followed by anti-FLAG IP, and the anti-RAD51AP1 western blot. Note that *bands indicate a cross-reactive unknown protein. F. 293T cells were transfected with UAF1 siRNA, followed by transfecting 3xFLAG-USP1 plasmids. Anti-FLAG IP was performed using the harvested cells. G. Similar to (F), anti-FLAG-UAF1 IP was performed. H. Model: UAF1 mediates the interaction between USP1 and RAD51AP1.
Knockdown of USP1 or UAF1 destabilizes RAD51AP1
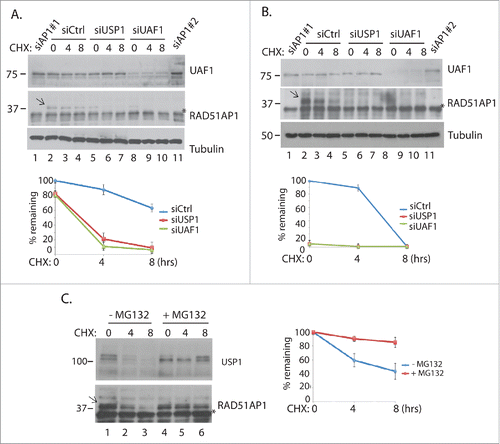
Although USP1 and UAF1 primarily deubiquitinates monoubiquitinated substrates, it has also been shown to regulate the stability ID proteins, by reversing the proteasome-targeting polyubiquitination.Citation25 Therefore we tested whether depletion of USP1 or UAF1 affects the stability of RAD51AP1, using a translation inhibitor cycloheximide (CHX), to monitor the protein half-life. Our analysis shows that RAD51AP1 is a relatively stable protein, with its half-life slightly reduced upon 4–8 hrs of CHX treatment (). Interestingly, knockdown of USP1 and UAF1 accelerated the decay rate of RAD51AP1 compared to controls in HeLa () and HCT116 cells (), suggesting that USP1 and UAF1 regulate the stability of RAD51AP1. Secondary siRNAs for each gene reproduced the similar results (Fig. S4). Treatment of proteasome inhibitor MG132 reversed the reduction of RAD51AP1 protein levels, suggesting that RAD51AP1 is degraded by the proteasome (). Knockdown of USP12 and USP46, 2 DUBs that also associate with UAF1,Citation26 did not significantly affect the RAD51AP1 stability (Fig. S5), suggesting that the effect is specific to the USP1-UAF1 complex.
Figure 3. USP1 and UAF1 promote RAD51AP1 stability. A. HeLa cells were transfected with the indicated siRNAs, then ∼60 hours later, 10 uM cycloheximide (CHX) was treated. Cells were harvested at indicated time points for western blots. Note that *bands indicate a cross-reactive unknown protein. “siAP1′ indicates “siRAD51AP1.” Below is the quantification of the band intensities using Image J software. B. The same experiment was performed using the ovarian carcinoma HEY cells. C. HeLa cells were treated with 10 uM MG132 for 4 hours, prior to treating them with cycloheximide. Cells were harvested at indicated time points for western blot. For 3A–3C, the error bars were generated from triplicate experiments.
Mapping of the UAF1-interacting region of RAD51AP1
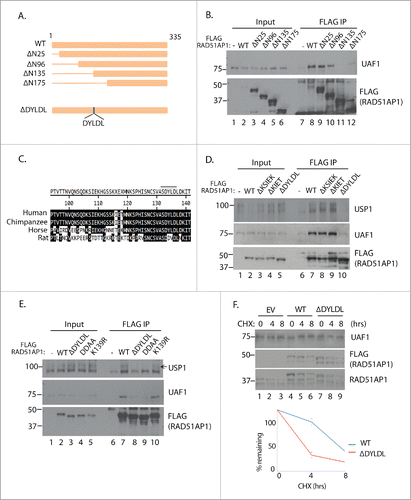
In order to specifically determine the functional significance of the UAF1-RAD51AP1 interaction, we sought to identify the residues within RAD51AP1 required for interacting with UAF1. Serial truncations of RAD51AP1 showed that the the area between 96 and 135 residues are necessary for interacting with UAF1, in the co-IP analysis (). Cross-species analysis of RAD51AP1 show that RAD51AP1 sequences are well conserved (). We randomly deleted some stretch of sequences that are particularly more conserved. After extensive mutagenesis analysis, we narrowed down to 133DYLDL137 sequence whose deletion disrupted the UAF1 interaction; while deletion of neither 106KSIEK110 nor 115KIET118 sequences had any effect on UAF1 binding, deletion of the 133DYLDL137 sequence significantly reduced the interaction capability of RAD51AP1 with both USP1 and UAF1 (). Interestingly, this area is close to the K139 residue, which was shown to be ubiquitinated in a proteomic study; the K139 residue corresponds to the K156 residue in the transcript variant 1 shown in the proteomic study.Citation27 Therefore we attempted to test whether the potential ubiquitination might regulate the UAF1 interaction, however, the K139 mutation to Arg (K139R) did not affect the interactions with UAF1 and USP1 (). However the mutation of the D133 and D136 residues within the DYLDL sequences to Ala (DDAA) abrogated the interaction, suggesting that these negatively charged residues are important in associating with UAF1. Having identified the necessary residues for interacting with UAF1, we tested whether the UAF1-interaction deficient mutant of RAD51AP1 (ΔDYLDL) is less stable than the wild type. CHX time course analysis indeed showed that the mutant degrades at a faster rate compared to the wild type (), supporting our earlier results that USP1 and UAF1 supports stabilization of RAD51AP1.
Figure 4. Mapping of the UAF1-interacton region of RAD51AP1. A. Schematic diagram of the RAD51AP1 truncates used for analysis. The DYLDL sequence whose deletion abrogated the UAF1 interaction is highlighted below. B. 293T cells were transfected with the indicated plasmids, followed by anti-FLAG IP and anti-UAF1 western blot. C. Sequence alignment of the putative interaction area of RAD51AP. D, E. 293T cells were transfected with the indicated plasmids, followed by anti-FLAG IP and anti-UAF1 western blot. F. ∼24 hours after the HeLa cells were transfected with the plasmids, cells were treated with 10 uM cycloheximide for indicated time, then harvested for western blots. Below is the quantification of the anti-FLAG bands intensity from triplicate experiments.
Phenotypes of cells expressing the UAF1 interaction-deficient mutant of RAD51AP1
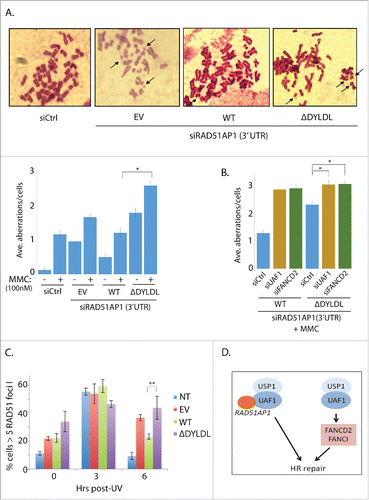
To determine the functional significance of the interaction between UAF1 and RAD51AP1, we established the complementation pair that expresses the FLAG-RAD51AP1 wild type and the interaction-deficient mutant. RAD51AP1 knockdown is known to sensitize cells to Cisplatin (or MMC).Citation4,5 Consistently, we show that cells depleted of RAD51AP1 (using the 3′UTR targeting siRNA) shows increased chromosomal aberrations in response to MMC in U2OS cells (Fig. S6). In this setting, when FLAG-RAD51AP1 wild type was expressed, it restored the chromosomal aberration (), whereas expression of the FLAG-RAD51AP1 ΔDYLDL mutant failed to do so. These results suggest that the ability of RAD51AP1 to interact with UAF1 is important in preserving the chromosomal integrity in response to the DNA ICL-inducing agent. Since UAF1 is also necessary for proper regulation of FANCD2-FANCI activation, we decided to test whether the UAF1-RAD51AP1 interaction is functionally linked to FANCD2 deubiquitination. Additional knockdown of FANCD2 in the ΔDYLDL-expressing cells further aggravated the chromosomal aberrations, suggesting that they are not in the same epistatic pathway (). Additional knockdown of UAF1 also aggravated the chromosomal aberrations () in the ΔDYLDL-expressing cells. This is not unexpected, given the multiple roles of UAF1 in the DNA repair other than the interaction with RAD51AP1.Citation11,23,28,29 Similar to the USP1 or UAF1 knockdown cells, knockdown of RAD51AP1 leads to persistent RAD51 foci retention following recovery from DNA damage exposure (Fig. S7). Importantly, the ΔDYLDL-expressing cells also display increased RAD51 foci that are not properly resolved following UV-induced DNA damage (; 6 hours post UV). These results consistently indicate that RAD51AP1 functions at a step after the RAD51-ssDNA nucleofilament formation, and that the UAF1 interaction may be necessary for this RAD51AP1 function.
Figure 5. The interaction-deficient RAD51AP1 mutant is unable to correct MMC-induced chromosomal aberrations in RAD51AP1-knockdown cells. A. U2OS cells expressing the empty vector (EV), WT, and ΔDYLDL FLAG-RAD51AP1 was treated with the RAD51AP1 3′-UTR targeting siRNAs, followed by MMC (100 nM) treatment and metaphase arrest. Representative images are only shown for MMC-treated samples. Three independent experiments were performed for the quantification in the graphs below. * p < 0.001 (n=40). B. Similar to the experimental scheme in A, U2OS cells knockdown with siRNA targeting the 3′UTR of RAD51AP1were co-transfected with siRNAs targeting UAF1 or FANCD2, followed by transfection of FLAG-RAD51AP1 WT or ΔDYLDL. After G418 selection, MMC (100 nM), colcemid (200 ng/ml) treatment, cells were fixed and stained for quantification (*p < 0.001) n=40. C. HeLa cells knockdown with RAD51AP1 and expressing EV, WT, ΔDYLDL were irradiate with UV (20 J/m2), and fixed at indicated time points for immunostainings. Three independent experiments were performed (* P < 0.001). D. Model: The USP1-UAF1 complex promotes the HR repair in multiple mechanisms.
Discussion
Here we described a functional interaction between RAD51AP1 and the USP1-UAF1 deubiquitinating enzyme complex. We have identified key residues within RAD51AP1 whose deletion impairs the interaction with UAF1. The UAF1-interaction deficient RAD51AP1 mutant is less stable than wild type, and the mutant is not able to efficiently support the cellular resistance to the DNA crosslinking agent MMC. These results suggest that reduced stability of RAD51AP1, at least partly, contributes to the HR defects in the cells depleted of USP1 or UAF1.
The USP-UAF1 complex engages in multiple interactions in DNA repair pathways, in which UAF1 was proposed as an adaptor subunit for USP1. The C-terminal SLD (SUMO-like domain) domain of UAF1 interacts with the SIM (SUMO-interacting motif) of FANCI,Citation23 as well as the SIM of ELG1.Citation28 Thus the SUMO-like delivery system was proposed as a mechanism for the USP1-UAF1 complex to recruit its substrates from these studies. Our study does not rule out the possibility that a similar mechanism plays a role for the UAF1 interaction with RAD51AP1. Clearly however, UAF1 serves as a bridging factor that mediates the interaction between USP1 and RAD51AP1 (). Since the 3D structure of RAD51AP1 is not known, it is not clear whether the DYLDL residues we identified are a part of large surface patch that interacts with UAF1, or whether it is independently engaged in interacting with UAF1. It will be interesting in future studies to see if the DYLDL residues make direct contact with UAF1.
USP1 and UAF1 have been well studied as positive regulators of the HR repair. Knockout of USP1 or UAF1 in DT40 cells are deficient in the HR repair, and the double knockout cells are similarly deficient as single knockouts, which suggested that USP1 and UAF1 are epistatic toward the HR repair.Citation12 USP1 knockout MEFs are deficient in HR repair, and the USP1 knockout DT40 cells are hypersensitive to DNA damaging agents.Citation10 UAF1 knockout mice are embryonic lethal, and the isolate MEFs are deficient in HR repair and sensitive to MMC.Citation22 These reports consistently demonstrated that USP1 and UAF1 are critical regulators of HR repair. How USP1 and UAF1 promote the HR repair is less clear however, although the deficiency in FANCD2 deubiquitination was proposed to play a major role. Thus, it is believed that USP1-mediated deubiquitination of FANCD2 is a required step for the FANCD2 activation and the functional FA pathway. USP1 and UAF1 are also implicated in regulating translesion synthesis (TLS), through regulating PCNA monoubiquitination and recruitment of TLS polymerase κ.Citation14,28,29 PCNA was shown to be required for initiation of recombination-associated DNA synthesis.Citation30 Therefore PCNA also participates in the HR repair, although whether it requires USP1 and UAF1 for the process is not known.
FANCD2 monoubiquitination (FANCD2-Ub) influences DNA repair in several ways. FANCD2-Ub recruits FAN1 nuclease to stalled replication forks,Citation31 CtIP exonuclease to damaged sites and replication forks to regulate DSB end resection,Citation15-17 and XPF-ERCC1 to repair the replication-associated ICL lesions.Citation32,33 FANCD2-Ub also interacts with the TLS polymerase eta.Citation34 Unmodified FANCD2 is also required for chromatin loading of Blm,Citation35 and functionally interacts with FANCJ Citation36-38 and PCNA.Citation39 Among these functions of FANCD2, the recruitment of CtIP to the DSB ends would have significant effects on the HR repair, as the CtIP-induced DSB end resection is the initiating event of the HR repair process. Whether USP1 and UAF1 influence DSB end resection through FANCD2 deubiquitination remains a possibility, although our analysis suggests that they may only play a minor role in the process when cells were challenged with CPT (Fig. S2). Interestingly however, USP1 or UAF1 knockdown cells show increased RAD51 foci formation, especially at the later time points of the RAD51 foci kinetic cycle (). Thus, we propose that the interaction with RAD51AP1 at the post-RAD51 foci formation step is an additional mechanism that regulates the HR repair by the USP1-UAF1 complex, independently of FANCD2 deubiquitination.
Our analysis showed that RAD51AP1 is a mildly unstable protein, and depletion of USP1 or UAF1 greatly decreases the stability. This implies that RAD51AP1 may be regulated by ubiquitination, and that USP1 deubiquitinates RAD51AP1. Indeed, RAD51AP1 was identified as a ubiquitinated protein in a proteomic study.Citation40 However, we were unable to detect the ubiquitinated species of RAD51AP1, thus we are not able to conclude whether the USP1-UAF1 complex protects RAD51AP1 from proteasomal degradation by deubiquitination.
RAD51AP1 promotes the HR repair by promoting the recombinase activities of RAD51 and its meiotic counterpart DMC1.Citation4,5,19-21 Unlike the RAD51 paralogs (e.g. RAD51C), depletion of RAD51AP1 does not affect the foci formation of RAD51.Citation4,5 Thus it was proposed that RAD51AP1 acts at a later step of the RAD51-ssDNA nucleofilament formation. Interestingly, similar to the effects of USP1 or UAF1 knockdown in RAD51 foci kinetics (), knockdown of RAD51AP1 leads to persistent foci retention at the later stage of kinetics (Fig. S7). This phenotype is consistent with previously published data using DT40 cells.Citation18 These results altogether suggests that the USP1-UAF1 complex may influence the D-loop formation mediated by RAD51 and RAD51AP1. In line with this, the ΔDYLDL-expressing cells display persistently high-level RAD51 foci formation during the UV-damage recovery kinetic assay (). It is also possible that UAF1 may affect other aspects of RAD51AP1 activity during DNA repair, such as timely recruitment of RAD51AP1 to DNA lesions during the repair process. Further studies are needed to investigate this possibility. The phenotypes of RAD51AP1 knockdown cells are similar to that of the FA-deficient cells, namely increased chromosomal breakages in response to ICL-inducing agents. Therefore it is tempting to speculate that UAF1 may serve to coordinate the interaction with the FANCD2-FANCI complex, ELG1-PCNA, and RAD51AP1, during the repair of ICL or during the replication stresses. As RAD51-driven HR reaction is necessary during the ICL repair,Citation41 the functional connection between the FA pathway and HR machineries is further supported by our work.
During the revision of our manuscript, a study from Liang et al. reported that the UAF1-RAD51AP1 complex promotes HR repair, by promoting RAD51-dependent synaptic complex.Citation42 Interestingly, this study identified the residues within the DYLDL sequence we described (which they described to be a SIM) to be also critical for interacting with UAF1. While Liang et al. found that the role of UAF1-RAD51AP1 is independent of USP1, our results indicate that USP1 and UAF1 interact with RAD51AP1 as a complex. Nonetheless, both studies revealed a critical role for the UAF1-RAD51AP1 interaction in promoting HR repair, through the DYDL sequence.
In summary, this study: 1) dissected the new function of the USP1-UAF1 complex in HR repair, 2) presents a new mode of regulation of HR repair proteins, and 3) supports the model that UAF1 is a substrate adaptation molecule for USP1. Our study also provides an additional basis for development of therapeutic inhibitors of USP1-UAF1 which may sensitize cancer cells that depend on HR repair. As RAD51AP1 amplification is associated with multiple types of cancers,Citation18,43-45 therapeutic targeting of the USP1-UAF1 complex may be beneficial in suppressing these tumors.
Materials and methods
Cell lines, plasmids and chemical reagents
HeLa, 293T, and U2OS cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM), and HEY cells (a gift from Dr. Meera Nanjundan) were grown in RPMI media. All media were supplemented with 10% bovine serum and L-glutamine. Cells were grown at 37C in 5% CO2 atmosphere. Cells were maintained in respective media containing penicillin and streptomycin. Cells were frequently tested for mycoplasma and treated with Plasmocin (Invivogen) when found positive. RAD51AP1 cDNA was obtained from mRNAs isolated from HEY cells. In brief, RAD51AP1 cDNA was reverse transcribed following First Strand Synthesis protocol from NEB. In short, 1 ug purified RNA was combined with 5 uM Oligo-dT primer and 2.5 uM dNTP. The mix is then heated for 3–5 minutes at 65C then 10 U of RNAse inhibitor, Reaction buffer and 200 U M-MuLV Reverse Transcriptase were added and incubated for 1 hr at 42C. The isolated RAD51AP1 cDNA sequence matched 100% for isoform 2 (transcript variant 2).Citation21,46 The RAD51AP1 cDNA was cloned into the p3xFLAG-CMV-10 vector (Invitrogen; gift from Dr. Mary Zhang). The p3XFLAG-CMV-10 vector was also used to ligate USP1. pcDNA-FLAG-UAF1 WT, pcDNA-FLAG-UAF1ΔWD2, and pcDNA-FLAG-UAF1ΔC were described previously.Citation11,26 Flag-tagged RAD51AP1 truncation constructs were generated by PCR. Site-directed mutagenesis was performed following the Stratagene QuikChange Site-Directed Mutagenesis protocol, using the p3xFlag-CMV-10-RAD51AP1 plasmid as a template. Etoposide, Cycloheximide, and M2 agarose were purchased from Sigma Aldrich. MG132 was purchased from Selleck Chemicals.
RNA interference
Cells were cultured in antibiotic-free medium and transfected once with 20 nM siRNA using RNAiMAX (Invitrogen) reagent following the manufacturer's protocol. The following siRNA sequences were used: UAF1#1: 5′-AAUCAGCACAAGCAAGAUCCAUAUA-3′, UAF1#2: 5′-GGACCGAGAUUAUCUUUCAAUUGAA-3′, USP1#1: 5′-AGCUUCUGAAUAUAGAGCAUCUGAA-3′, USP1#2: 5′-ACAGGCATTAATATTAGTGGA-3′ (3′UTR), RAD51AP1#1: 5′-ATGGCATATGTCTCCGATTTA-3′, RAD51AP1#2: 5′-CAGCTTTACAAGGGTGTTTAT-3′, USP12: 5′-CCGATCATGGTAGTTGATTTA-3′, USP46: 5′-TAGGGAAATGTTTGTACTATA-3′, FANCD2: 5′-UUUGGAGGCAUCUUCUGUCAGGCUC-3′. BRCA1: 5′- CAGCAGUUUAUUACUCACUAA-3′. These siRNAs were purchased from Qiagen.
Protein stability assay, western blotting, and antibodies
Cells were treated with siRNAs targeting USP1 and UAF1 for 48–72 hrs. 10 uM cycloheximide was then added for various durations of time (ex. 0, 3, 6 and 9 hrs). Cells were then prepared for western blot analysis and probed for RAD51AP1 protein levels. To test if RAD51AP1 is degraded by the proteasome, 10 uM MG132 was treated for the last 6 hrs of the 8 hr, 10 uM cycloheximide treatment. Cell extracts were run on an SDS-PAGE gel and then transferred to a PVDF membrane (Bio-Rad, Hercules, CA). Membranes were probed with primary antibodies overnight at 4C. The membranes were then washed and incubated with either mouse or rabbit secondary antibody linked to horseradish peroxidase (Cell Signaling Technologies). The bound antibodies were viewed with Pierce ECL Western Blotting Substrate (Thermo Scientific). The following antibodies were used: anti-RAD51AP1 (Abcam; cat#Ab101321), anti-FANCD2, RAD51, PCNA antibodies (Santa Cruz Biotechnology), anti-RPA, phospho-RPA32 (S4/S8), USP1, WDR20, 53BP1 rabbit polyclonal antibodies (Bethyl Laboratory), anti-ubiquitin FK2 mouse antibody (Millipore), anti-γ-H2AX (Upstate), anti-γ-Tubulin, anti-FLAG antibodies (Sigma). Anti-UAF1 antibody generated from rabbit was previously described.Citation26,47
Mass spec analysis and immunoprecipitation assays
Immunoprecipitations were carried out as follows; HEK293T cells were seeded in 6 cm dishes and transfected with 4 ug DNA of p3xFlag-RAD51AP1 or pcDNA-FLAG-UAF1 plasmids using TurboFect (Thermo Scientific). ∼24 hrs post-transfection, cells were harvested and lysed in 0.5% NP-40 (50 mM Tris, 100 mM NaCl at pH 7.0). Lysates were then incubated with the anti-FLAG M2 agarose (Sigma Aldrich) for ∼16 hrs at 4C. Beads were washed thrice with the lysis buffer, then the 2X Laemli buffer was added before boiling the samples. For the mass spec sample, the bound proteins before eluted with the addition of 4% SDS. The eluate containing total protein was processed using the FASP method, digested with trypsin-LysC and desalted using HYPERSEP C18 columns. Peptides were then concentrated by vacuum centrifugation and resuspended in 0.1% formic acid. Peptides were separated on an Acclaim PepMap C18 (75 µm × 50 cm) UPLC column (Thermo) using an EASY-nLC 1000 with gradient times of 60–90 min (2–40% acetonitrile in 0.1% formic acid). Mass spectrometric analysis was performed by a hybrid quadrupole-Orbitrap (Q Exactive Plus, Thermo) or hybrid linear ion trap-Orbitrap (Orbitrap XL) using a top 10 data-dependent acquisition method.
Immunofluorescent microscopy
48∼55 hours after siRNA transfection, the cells mounted on coverslips were irradiated with UV (15∼20J/m2), washed, pre-extracted with 0.25% Triton X-100 for 3 minutes, and then fixed with 4% paraformaldehyde for 10 min. The fixed cells incubated with primary antibody against RAD51 (SantacruzBiotechnology) at 1:500, followed by incubation with Alexa Fluor 488-anti-rabbit secondary antibody. Vectashield mounting medium with DAPI (Vector Laboratories) was used to stain nuclei. Images were collected by a Zeiss Axiovert 200 microscope equipped with a Perkin Elmer ERS spinning disk confocal imager and a 63x/1.45NA oil objective using the Velocity software (Perkin Elmer). We counted 70–120 cells from each sample for generating statistical figures for RAD51 foci.
GFP-based HR repair (DR-GFP) assays
The U2OS cell line expressing an integrated homologous recombination reporter DR-GFP (from Dr. Maria Jasin) has been described.Citation48 48 hrs post I-SceI transfection, cells were harvested and analyzed via flow cytometry for recombination efficiency using a BD Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, New Jersey). C-Flow software was used to analyze percent positive GFP cells relative to the total number of the transfected cells. Approximately 40,000 cells were counted from each sample.
Cytogenetic analysis
U2OS cells were exposed to 100 nM of MMC for 48 h. Colcemide (0.2μg/ml) was added to the medium 4 h before harvesting the cells. Cells were washed with PBS, and allow to swell in 37.5 mM KCL at room temperature for 20 min. Cells were treated with ice-cold fix solution composed with methanol : acetic acid (3:1), dropped on wet slides air dried, and stained in 3% Giemsa solution in Gurr buffer (Invitrogen) for 20 min. The samples were analyzed with Leica DM2000 microscope using a 2000x magnification. Chromosome breakage analysis was performed on 50 Giemsa-stained metaphases in MMC-treated and untreated cultures. The number and type of structural chromosomal aberration such as chromatid breaks and fragments were scored as a single break. The average number of aberrations per cell was scored for each sample.
Disclosure of potential conflicts of interest
The authors have no conflict-of-interests.
1209613_Supplemental_Material.pdf
Download PDF (328.3 KB)Acknowledgments
We thank the Stanley Stevens laboratory for helping with the mass spectrometry analysis. We thank Drs. Maria Jasin for the DR-GFP reporter cell line, and Alan D'Andrea for helpful discussions.
Funding
This work was supported by NIH 1R15HL126113 grant to Y.K.
References
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annual Rev Biochem 2008; 77:229-57; PMID:18275380; http://dx.doi.org/10.1146/annurev.biochem.77.061306.125255
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annual Rev Genetics 2010; 44:113-39; PMID:20690856; http://dx.doi.org/10.1146/annurev-genet-051710-150955
- Suwaki N, Klare K, Tarsounas M. RAD51 paralogs: roles in DNA damage signalling, recombinational repair and tumorigenesis. Seminars Cell Dev Biol 2011; 22:898-905; PMID:21821141; http://dx.doi.org/10.1016/j.semcdb.2011.07.019
- Wiese C, Dray E, Groesser T, San Filippo J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P, et al. Promotion of homologous recombination and genomic stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell 2007; 28:482-90; PMID:17996711; http://dx.doi.org/10.1016/j.molcel.2007.08.027
- Modesti M, Budzowska M, Baldeyron C, Demmers JA, Ghirlando R, Kanaar R. RAD51AP1 is a structure-specific DNA binding protein that stimulates joint molecule formation during RAD51-mediated homologous recombination. Mol Cell 2007; 28:468-81; PMID:17996710; http://dx.doi.org/10.1016/j.molcel.2007.08.025
- Kee Y, Huang TT. Role of Deubiquitinating Enzymes in DNA Repair. Mol Cell Biol 2015; 36:524-44; PMID:26644404; http://dx.doi.org/10.1128/MCB.00847-15
- Nijman SM, Huang TT, Dirac AM, Brummelkamp TR, Kerkhoven RM, D'Andrea AD, Bernards R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol Cell 2005; 17:331-9; PMID:15694335; http://dx.doi.org/10.1016/j.molcel.2005.01.008
- Kee Y, D'Andrea AD. Molecular pathogenesis and clinical management of Fanconi anemia. J Clin Invest 2012; 122:3799-806; PMID:23114602; http://dx.doi.org/10.1172/JCI58321
- Kim JM, Parmar K, Huang M, Weinstock DM, Ruit CA, Kutok JL, D'Andrea AD. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev Cell 2009; 16:314-20; PMID:19217432; http://dx.doi.org/10.1016/j.devcel.2009.01.001
- Oestergaard VH, Langevin F, Kuiken HJ, Pace P, Niedzwiedz W, Simpson LJ, Ohzeki M, Takata M, Sale JE, Patel KJ. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol Cell 2007; 28:798-809; PMID:18082605; http://dx.doi.org/10.1016/j.molcel.2007.09.020
- Cohn MA, Kowal P, Yang K, Haas W, Huang TT, Gygi SP, D'Andrea AD. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol Cell 2007; 28:786-97; PMID:18082604; http://dx.doi.org/10.1016/j.molcel.2007.09.031
- Murai J, Yang K, Dejsuphong D, Hirota K, Takeda S, D'Andrea AD. The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol Cell Biol 2011; 31:2462-9; PMID:21482670; http://dx.doi.org/10.1128/MCB.05058-11
- Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, Ballif BA, Gygi SP, Hofmann K, D'Andrea AD, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 2007; 129:289-301; PMID:17412408; http://dx.doi.org/10.1016/j.cell.2007.03.009
- Huang TT, Nijman SM, Mirchandani KD, Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R, D'Andrea AD. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol 2006; 8:339-47; PMID:16531995
- Murina O, von Aesch C, Karakus U, Ferretti LP, Bolck HA, Hanggi K, Sartori AA. FANCD2 and CtIP cooperate to repair DNA interstrand crosslinks. Cell Reports 2014; 7:1030-8; PMID:24794434; http://dx.doi.org/10.1016/j.celrep.2014.03.069
- Unno J, Itaya A, Taoka M, Sato K, Tomida J, Sakai W, Sugasawa K, Ishiai M, Ikura T, Isobe T, et al. FANCD2 binds CtIP and regulates DNA-end resection during DNA interstrand crosslink repair. Cell Reports 2014; 7:1039-47; PMID:24794430; http://dx.doi.org/10.1016/j.celrep.2014.04.005
- Yeo JE, Lee EH, Hendrickson EA, Sobeck A. CtIP mediates replication fork recovery in a FANCD2-regulated manner. Hum Mol Genetics 2014; 23:3695-705; PMID:24556218; http://dx.doi.org/10.1093/hmg/ddu078
- Parplys AC, Kratz K, Speed MC, Leung SG, Schild D, Wiese C. RAD51AP1-deficiency in vertebrate cells impairs DNA replication. DNA Repair 2014; 24:87-97; PMID:25288561; http://dx.doi.org/10.1016/j.dnarep.2014.09.007
- Dunlop MH, Dray E, Zhao W, San Filippo J, Tsai MS, Leung SG, Schild D, Wiese C, Sung P. Mechanistic insights into RAD51-associated protein 1 (RAD51AP1) action in homologous DNA repair. J Biol Chem 2012; 287:12343-7; PMID:22375013; http://dx.doi.org/10.1074/jbc.C112.352161
- Dray E, Dunlop MH, Kauppi L, San Filippo J, Wiese C, Tsai MS, Begovic S, Schild D, Jasin M, Keeney S, et al. Molecular basis for enhancement of the meiotic DMC1 recombinase by RAD51 associated protein 1 (RAD51AP1). Proc Natl Acad Sci USA 2011; 108:3560-5; PMID:21307306; http://dx.doi.org/10.1073/pnas.1016454108
- Dunlop MH, Dray E, Zhao W, Tsai MS, Wiese C, Schild D, Sung P. RAD51-associated protein 1 (RAD51AP1) interacts with the meiotic recombinase DMC1 through a conserved motif. J Biol Chem 2011; 286:37328-34; PMID:21903585; http://dx.doi.org/10.1074/jbc.M111.290015
- Park E, Kim JM, Primack B, Weinstock DM, Moreau LA, Parmar K, D'Andrea AD. Inactivation of Uaf1 causes defective homologous recombination and early embryonic lethality in mice. Mol Cell Biol 2013; 33:4360-70; PMID:24001775; http://dx.doi.org/10.1128/MCB.00870-13
- Yang K, Moldovan GL, Vinciguerra P, Murai J, Takeda S, D'Andrea AD. Regulation of the Fanconi anemia pathway by a SUMO-like delivery network. Genes Dev 2011; 25:1847-58; PMID:21896657; http://dx.doi.org/10.1101/gad.17020911
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell 2009; 138:389-403; PMID:19615732; http://dx.doi.org/10.1016/j.cell.2009.04.042
- Williams SA, Maecker HL, French DM, Liu J, Gregg A, Silverstein LB, Cao TC, Carano RA, Dixit VM. USP1 deubiquitinates ID proteins to preserve a mesenchymal stem cell program in osteosarcoma. Cell 2011; 146:918-30; PMID:21925315; http://dx.doi.org/10.1016/j.cell.2011.07.040
- Cohn MA, Kee Y, Haas W, Gygi SP, D'Andrea AD. UAF1 is a subunit of multiple deubiquitinating enzyme complexes. J Biol Chem 2009; 284:5343-51; PMID:19075014; http://dx.doi.org/10.1074/jbc.M808430200
- Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA damage response. Mol Cell 2015; 59(5):867-81
- Lee KY, Yang K, Cohn MA, Sikdar N, D'Andrea AD, Myung K. Human ELG1 regulates the level of ubiquitinated proliferating cell nuclear antigen (PCNA) through Its interactions with PCNA and USP1. J Biol Chem 2010; 285:10362-9; PMID:20147293; http://dx.doi.org/10.1074/jbc.M109.092544
- Jones MJ, Colnaghi L, Huang TT. Dysregulation of DNA polymerase kappa recruitment to replication forks results in genomic instability. EMBO J 2012; 31:908-18; PMID:22157819; http://dx.doi.org/10.1038/emboj.2011.457
- Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Mol Cell 2009; 36:704-13; PMID:19941829; http://dx.doi.org/10.1016/j.molcel.2009.09.036
- Lachaud C, Moreno A, Marchesi F, Toth R, Blow JJ, Rouse J. Ubiquitinated Fancd2 recruits Fan1 to stalled replication forks to prevent genome instability. Science 2016; 351:846-9; PMID:26797144; http://dx.doi.org/10.1126/science.aad5634
- Klein Douwel D, Boonen RA, Long DT, Szypowska AA, Raschle M, Walter JC, Knipscheer P. XPF-ERCC1 acts in Unhooking DNA interstrand crosslinks in cooperation with FANCD2 and FANCP/SLX4. Mol Cell 2014; 54:460-71; PMID:24726325; http://dx.doi.org/10.1016/j.molcel.2014.03.015
- Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, Elledge SJ, Walter JC. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Sci 2009; 326:1698-701; PMID:19965384; http://dx.doi.org/10.1126/science.1182372
- Fu D, Dudimah FD, Zhang J, Pickering A, Paneerselvam J, Palrasu M, Wang H, Fei P. Recruitment of DNA polymerase eta by FANCD2 in the early response to DNA damage. Cell Cycle 2013; 12:803-9; PMID:23388460; http://dx.doi.org/10.4161/cc.23755
- Chaudhury I, Sareen A, Raghunandan M, Sobeck A. FANCD2 regulates BLM complex functions independently of FANCI to promote replication fork recovery. Nucleic Acids Res 2013; 41:6444-59; PMID:23658231; http://dx.doi.org/10.1093/nar/gkt348
- Clark DW, Tripathi K, Dorsman JC, Palle K. FANCJ protein is important for the stability of FANCD2/FANCI proteins and protects them from proteasome and caspase-3 dependent degradation. Oncotarget 2015; 6:28816-32; PMID:26336824
- Chen X, Wilson JB, McChesney P, Williams SA, Kwon Y, Longerich S, Marriott AS, Sung P, Jones NJ, Kupfer GM. The Fanconi anemia proteins FANCD2 and FANCJ interact and regulate each other's chromatin localization. J Biol Chem 2014; 289:25774-82; PMID:25070891; http://dx.doi.org/10.1074/jbc.M114.552570
- Raghunandan M, Chaudhury I, Kelich SL, Hanenberg H, Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle 2015; 14:342-53; PMID:25659033; http://dx.doi.org/10.4161/15384101.2014.987614
- Howlett NG, Harney JA, Rego MA, Kolling FW, Glover TW. Functional interaction between the Fanconi Anemia D2 protein and proliferating cell nuclear antigen (PCNA) via a conserved putative PCNA interaction motif. J Biol Chem 2009; 284:28935-42; PMID:19704162; http://dx.doi.org/10.1074/jbc.M109.016352
- Elia AE, Boardman AP, Wang DC, Huttlin EL, Everley RA, Dephoure N, Zhou C, Koren I, Gygi SP, Elledge SJ. Quantitative Proteomic Atlas of Ubiquitination and Acetylation in the DNA damage response. Mol Cell 2015; 59:867-81; PMID:26051181; http://dx.doi.org/10.1016/j.molcel.2015.05.006
- Long DT, Raschle M, Joukov V, Walter JC. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 2011; 333:84-7; PMID:21719678; http://dx.doi.org/10.1126/science.1204258
- Liang F, Longerich S, Miller AS, Tang C, Buzovetsky O, Xiong Y, Maranon DG, Wiese C, Kupfer GM, Sung P. Promotion of RAD51-mediated homologous DNA pairing by the RAD51AP1-UAF1 complex. Cell Reports 2016; 15(10):2118-26.
- Henson SE, Tsai SC, Malone CS, Soghomonian SV, Ouyang Y, Wall R, Marahrens Y, Teitell MA. Pir51, a Rad51-interacting protein with high expression in aggressive lymphoma, controls mitomycin C sensitivity and prevents chromosomal breaks. Mutation Res 2006; 601:113-24; PMID:16920159; http://dx.doi.org/10.1016/j.mrfmmm.2006.06.016
- Schoch C, Kern W, Kohlmann A, Hiddemann W, Schnittger S, Haferlach T. Acute myeloid leukemia with a complex aberrant karyotype is a distinct biological entity characterized by genomic imbalances and a specific gene expression profile. Genes Chromosomes Cancer 2005; 43:227-38; http://dx.doi.org/10.1002/gcc.20193
- Obama K, Satoh S, Hamamoto R, Sakai Y, Nakamura Y, Furukawa Y. Enhanced expression of RAD51 associating protein-1 is involved in the growth of intrahepatic cholangiocarcinoma cells. Clin Cancer Res 2008; 14:1333-9; PMID:18316552; http://dx.doi.org/10.1158/1078-0432.CCR-07-1381
- Kovalenko OV, Golub EI, Bray-Ward P, Ward DC, Radding CM. A novel nucleic acid-binding protein that interacts with human rad51 recombinase. Nucleic Acids Res 1997; 25:4946-53; PMID:9396801; http://dx.doi.org/10.1093/nar/25.24.4946
- Kee Y, Yang K, Cohn MA, Haas W, Gygi SP, D'Andrea AD. WDR20 regulates activity of the USP12 x UAF1 deubiquitinating enzyme complex. J Biol Chem 2010; 285:11252-7; PMID:20147737; http://dx.doi.org/10.1074/jbc.M109.095141
- Pierce AJ, Johnson RD, Thompson LH, Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 1999; 13:2633-8; PMID:10541549; http://dx.doi.org/10.1101/gad.13.20.2633
