ABSTRACT
Bloom Syndrome (BS) is a rare genetic disease characterized by high levels of chromosomal instability and an increase in cancer risk. Cytidine deaminase (CDA) expression is downregulated in BS cells, leading to an excess of cellular dC and dCTP that reduces basal PARP-1 activity, compromising optimal Chk1 activation and reducing the efficiency of downstream checkpoints. This process leads to the accumulation of unreplicated DNA during mitosis and, ultimately, ultrafine anaphase bridge (UFB) formation. BS cells also display incomplete sister chromatid disjunction when depleted of cohesin. Using a combination of fluorescence in situ hybridization and chromosome spreads, we investigated the possible role of CDA deficiency in the incomplete sister chromatid disjunction in cohesin-depleted BS cells. The decrease in basal PARP-1 activity in CDA-deficient cells compromised sister chromatid disjunction in cohesin-depleted cells, regardless of BLM expression status. The observed incomplete sister chromatid disjunction may be due to the accumulation of unreplicated DNA during mitosis in CDA-deficient cells, as reflected in the changes in centromeric DNA structure associated with the decrease in basal PARP-1 activity. Our findings reveal a new function of PARP-1 in sister chromatid disjunction during mitosis.
Introduction
Chromosome segregation is crucial for faithful genome transmission. Errors leading to abnormal chromosome segregation are a major source of genetic instability, promoting aneuploidy and, thus, carcinogenesis.Citation1,2 Correct chromosome segregation requires the sister chromatids to be held together until the onset of anaphase. Cohesin is a protein complex involved in the maintenance of sister chromatid cohesion. Cohesin is loaded onto chromatin in early G1 phase and is necessary to establish sister chromatid cohesion during S phase. Cohesin remains bound to the chromosome during G2 and is released during mitosis. In yeast, cohesin is released in a single step at metaphase-anaphase transition.Citation3,4 In vertebrates, it is released in 2 steps. In the first step, cohesin is dissociated from the chromosome arms at the onset of mitosis,Citation5,6 but a small pool of cohesin complex located at the centromeres is protected by Shugoshin (Sgo1) and remains in place until the start of anaphase, thereby preventing the precocious separation of sister chromatids.Citation6 At the metaphase-anaphase transition, Rad21, the cleavable subunit of the cohesin complex,Citation5,7 is cleaved by separase. This leads to a loss of cohesion between sister chromatids, resulting in chromosome segregation.
Mutation of both copies of the BLM gene, encoding BLM RecQ helicase, leads to Bloom Syndrome (BS), a rare genetic disease displaying one of the strongest known correlations between chromosomal instability and an increase in cancer risk.Citation8 We previously showed that BLM deficiency is associated with changes in centromeric DNA structure and an increase in the percentage of cohesin-depleted prometaphase cells with incomplete sister chromatid disjunction.Citation9 We also reported that the genetic instability associated with BS phenotype results at least partly from disequilibrium of the pyrimidine pool due to a cytidine deaminase (CDA) defect.Citation10,11,12 CDA is an enzyme of the pyrimidine salvage pathway catalyzing the hydrolytic deamination of cytidine and deoxycytidine into uridine and deoxyuridine, respectively.Citation13 CDA deficiency leads to intracellular dC and dCTP accumulation, lowering the basal activity of poly(ADP-ribose) polymerase 1 (PARP-1), a multifunctional enzyme involved in many cellular processes, including the response to DNA damage.Citation14 The decrease in basal PARP-1 activity disturbs Chk1 activation and decreases the efficiency of downstream checkpoints, leading to the accumulation, during mitosis, of unreplicated DNA at “difficult-to-replicate” loci, such as centromeres and fragile sites, resulting in ultrafine anaphase bridge (UFB) formation.Citation11,12
Based on these observations, we investigated whether the incomplete sister chromatid disjunctions and the changes in centromeric DNA structure we reported in BS cells were the consequence of BLM deficiency per se, or of the nucleotide pool disequilibrium and the subsequent decrease in basal PARP-1 activity resulting from CDA defect in BS cells.Citation9
We found – in cohesin-depleted cells –, that CDA deficiency, but not BLM deficiency, leads to an increase in the frequency of cells presenting an incomplete sister chromatid disjunction in mitosis. This “incomplete disjunction” phenotype results from the nucleotide pool disequilibrium induced by CDA defect. Moreover, the excess of dC and dCTP resulting from CDA deficiency also affects centromere structure. Finally, we report that both centromere structural changes and incomplete sister chromatid disjunctions result from the reduction of basal PARP-1 activity due to the pyrimidine pool disequilibrium in CDA-deficient cells. Our data highlight a new function for PARP-1 in the regulation of sister chromatid disjunction during mitosis.
Results
CDA deficiency leads to an increase in the frequency of Rad21-depleted prometaphase cells with incomplete sister chromatid disjunction
We have previously shown that BS cells display an increase in the percentage of prometaphase cells with incomplete sister chromatid disjunction when cohesin is depleted.Citation9 Given that BLM deficiency greatly decreases CDA expression, contributing to the genetic instability associated with Bloom Syndrome,Citation10,11,12 we investigated whether the high percentage of prometaphase cells with incomplete sister chromatid disjunction in BLM-deficient cells was due to CDA deficiency. We analyzed chromosome spreads from the following cell lines with Rad21 depletion: BS cells expressing neither BLM nor CDA (BS-Ctrl, BLM−/CDA−); BS cells stably expressing exogenous BLM, restoring CDA expression (BS-BLM, BLM+/CDA+), and BS cells expressing exogenous CDA but not BLM (BS-CDA, BLM−/CDA+) ().Citation10
Table 1. Characterisitics of cell lines.
Rad21, the cleavable subunit of cohesin,Citation5 was depleted from all cell lines with a specific siRNA (siRad21). As previously reported,Citation9 siRNA-mediated Rad21 depletion was associated with 3 distinct metaphase phenotypes: 1) chromosomes present the classical X-shaped structure, probably corresponding to cells not transfected with the Rad21 siRNA (X-shapes), 2) chromosomes are fully disjoined due to cohesin depletion (complete disjunction), 3) separated sister chromatids are still physically linked, reflecting incomplete sister chromatid disjunction (incomplete disjunction) (Supplementary Fig. 1).
Figure 1. CDA deficiency leads to an increase in the frequency of Rad21-depleted prometaphase cells with incomplete sister chromatid disjunction. (A) Percentage of BS prometaphase cells expressing BLM and/or CDA, transfected with siRNA specific for Rad21 and sorted on the basis of their phenotype: X-shapes, incomplete disjunction or complete disjunction (n = 3, > 275 prometaphase cells analyzed). (B) Representative immunoblot of BS-Ctrl(BLM), BS-BLM, BS-Ctrl(CDA) and BS-CDA cells transfected with the indicated siRNA. (C) Percentage of HeLa prometaphase cells with and without CDA expression, transfected with siRNA specific for Rad21 and sorted on the basis of their phenotype: X-shapes, incomplete disjunction or complete disjunction (n = 3, > 285 prometaphase cells analyzed). (D) Representative immunoblot of HeLa-Ctrl(CDA) and HeLa-shCDA cells transfected with the indicated siRNA. (E) Percentage of HeLa Rad21-depleted prometaphase cells with and without CDA expression, left untreated or treated with 1 mM dC for 16 hours and sorted on the basis of their phenotype: X-shapes, incomplete disjunction or complete disjunction (n = 3, > 150 prometaphase cells analyzed). (F) Representative immunoblot of HeLa-Ctrl(CDA) and HeLa-shCDA cells left untreated or treated with 1 mM dC for 16 hours and transfected with the indicated siRNA. Error bars represent the mean ± SD. Statistical significance was assessed with Student's t-test.
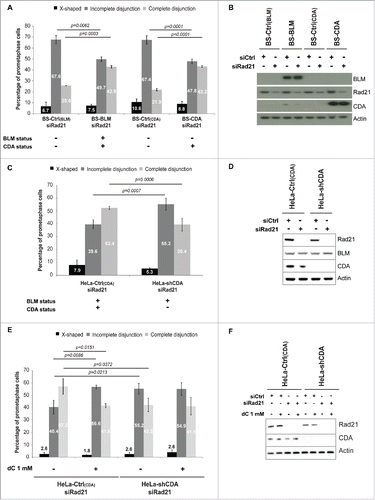
We determined the frequency of each metaphase phenotype in the 4 cell lines. As expected, BLM expression in BS cells (BS-BLM cell line, BLM+/CDA+) significantly reduced the frequency of Rad21-depleted prometaphase cells with incomplete sister chromatid disjunction (67.6% vs 49.7%, p = 0.0062) ( and ) and increased the frequency of Rad21-depleted prometaphase cells displaying complete sister chromatid disjunction (25.6% vs 42.8%, p = 0.0003) ( and ). Similar results were obtained for Rad21-depleted BS cells expressing CDA (BLM−/CDA+), in which the frequency of cells presenting an “incomplete disjunction” phenotype decreased (67.4% vs 47.8%, p = 0.0001), and the frequency of cells with a “complete disjunction” phenotype increased (21.9% vs 43.2%, p<0.0001). The frequency of cells presenting the “incomplete disjunction” phenotype was significantly reduced in Rad21-depleted BS-CDA cells (BLM-/CDA+) to levels similar to that of BS cells expressing BLM and, thus, CDA (BS-BLM cells; BLM+/CDA+). These results suggest that CDA expression was sufficient to prevent incomplete sister chromatid disjunction in BS cells, independently of BLM expression ( and ).
We investigated whether CDA deficiency alone was sufficient to increase the frequency of cells presenting an “incomplete disjunction” phenotype in Rad21-depleted cells, by using HeLa cells stably depleted of CDA with an adenoviral short hairpin RNA (shRNA) specific for CDA (HeLa-shCDA, BLM+/CDA−).Citation11,12 CDA depletion in BLM-expressing cells led to a significant increase in the frequency of Rad21-depleted prometaphase cells with incomplete sister chromatid disjunction (39.6% vs 59.3%, p = 0.0007) ( and ), and a significant decrease in the percentage of Rad21-depleted prometaphase cells displaying complete sister chromatid disjunction (52.4% vs 39.4%, p = 0.0006) ( and ). Thus, CDA depletion is sufficient to induce an increase in the frequency of incomplete sister chromatid disjunction in Rad21-depleted cells, regardless of their BLM expression status.
CDA deficiency leads to an excess of intracellular dC and dCTP, resulting in genetic instability.Citation10,11,12 The addition of dC to the cell culture medium of CDA-proficient cells leads to an excess of intracellular dCTP, mimicking CDA deficiency.Citation10,11,12 We treated Rad21-depleted CDA-proficient and -deficient cells with 1 mM dC for 10 hours. We observed no effect of dC treatment on CDA-deficient cells. However, the treatment of CDA-proficient cells with dC led to a significant increase in the frequency of cells presenting incomplete sister chromatid disjunction (40.4% vs 56.6%, p = 0.0086), to levels similar to those observed in CDA-depleted cells left untreated or treated with dC (55.2%) ( and ). Thus, the culture of CDA-expressing cells in the presence of dC is sufficient to promote the “incomplete sister chromatid disjunction” phenotype associated with CDA deficiency, indicating that this phenotype results from the nucleotide pool disequilibrium induced by CDA defect.
CDA deficiency is associated with structural changes to the centromere
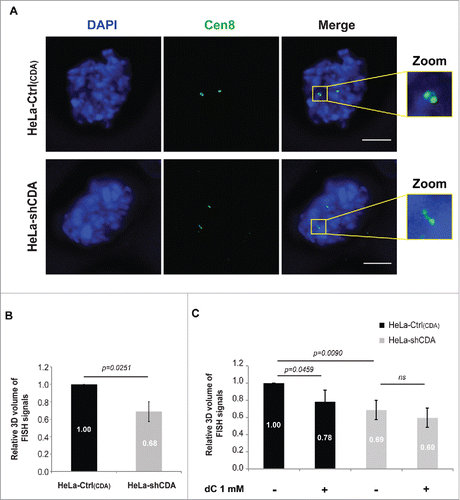
We previously reported the presence of structural defects at the centromeres of BLM-deficient cells associated with incomplete sister chromatid disjunction.Citation9 We investigated possible structural changes to the centromeres of CDA-deficient cells, by using a FISH (fluorescence in situ hybridization)-based assay to measure the volume of centromeric DNA for chromosome 8 (CEN-8 probe)Citation9,15 during prometaphase and metaphase (). The CEN-8 signal volume in CDA-depleted HeLa cells was significantly smaller than that in CDA-expressing HeLa cells (), indicating that CDA depletion is itself associated with an abnormal structure of centromeric DNA. We then investigated whether the pyrimidine pool disequilibrium resulting from CDA deficiency was responsible for the abnormal centromeric DNA structure. We found that the addition of 1 mM dC to the culture medium of CDA-proficient cells reduced the volume of the CEN-8 signal to levels similar to those in CDA-deficient cells, whereas it had no effect when added to the medium of CDA-depleted cells (). Thus, the excess of dC and dCTP affects centromere structure in CDA-proficient cells, establishing a link between pyrimidine pool disequilibrium and the structural changes to the centromere observed in the absence of CDA. These findings suggest that the changes in centromeric DNA structure in CDA-deficient cells may result from the pyrimidine pool disequilibrium.
Figure 2. CDA deficiency is associated with structural changes to the centromere. (A) Representative deconvoluted z-projection immunofluorescence images of prometaphase HeLa cells with and without CDA expression. DNA was visualized by staining with DAPI (in blue). The centromeres of chromosome 8 were stained with a Cen8 probe (in green). Scale bar: 5 µm. (B) Relative 3D volume of FISH signals in HeLa-Ctrl(CDA) (black bars) and HeLa-shCDA (gray bars) cell lines (n = 3, > 90 prometaphase cells analyzed). (C) Relative 3D volume of FISH signals in HeLa-Ctrl(CDA) (black bars) and HeLa-shCDA (gray bars) cells left untreated or treated with 1 mM dC for 16 hours (n = 3, > 285 prometaphase cells analyzed). Error bars represent the mean ± SD. Statistical significance was evaluated with Student's t-test; ns: not significant.
Optimal PARP-1 activity is required to prevent abnormal centromeric DNA structure and incomplete sister chromatid disjunction
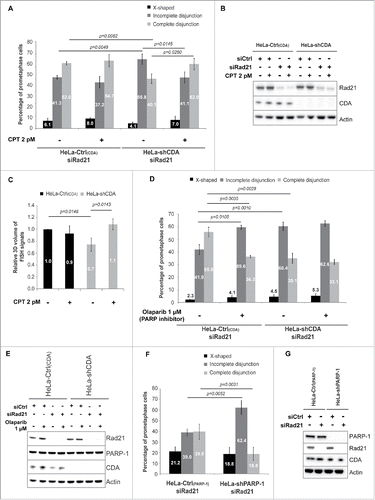
Some phenotypes associated with CDA deficiency are due to the decrease in basal PARP-1 activity resulting from the pyrimidine pool disequilibrium. Optimal PARP-1 activity can be restored with very low doses of camptothecin (CPT).Citation11,12 We thus investigated whether the low basal levels of PARP-1 activity in CDA-deficient cells were involved in the changes in centromeric DNA structure and the incomplete sister chromatid disjunction phenotype. The treatment of Rad21-depleted CDA-deficient cells with 2 pM CPT restored both centromeric volume and the frequency of cells with incomplete sister chromatid disjunction to levels similar to those in Rad21-depleted CDA-proficient cells (). The importance of PARP-1 activity for correct sister chromatid disjunction was confirmed by analyses of chromosome spreads from Rad21-depleted cells treated with olaparib, a chemical PARP inhibitor.Citation16 Indeed, treatment with 1 µM olaparib increased the frequency of Rad21-depleted prometaphase cells displaying incomplete disjunction in CDA-proficient cells (41.9% vs 59.6%, p = 0.0105) to levels similar to those in CDA-deficient cells (60.4%) ( and ). Consistent with these findings, we found that shRNA-mediated PARP-1 depletion in HeLa cells led to an increase in the percentage of Rad21-depleted prometaphase cells with incomplete disjunction (39.0% vs 62.4%, p = 0.0052), and a decrease in the percentage of Rad21-depleted prometaphase cells displaying complete disjunction (39.8% vs 18.8%, p = 0.0031) ( and ). These results highlight the importance of PARP-1 activity for preventing changes in centromeric DNA structure and incomplete sister chromatid disjunction during mitosis.
Figure 3. Optimal PARP-1 activity is required to prevent incomplete sister chromatid disjunction and abnormal centromeric DNA structure. (A) Percentage of HeLa Rad21-depleted prometaphase cells with and without CDA expression, left untreated or treated with 2 pM CPT for 16 hours and sorted on the basis of their phenotype: X-shapes, incomplete disjunction or complete disjunction (n = 3, > 230 prometaphase cells analyzed). (B) Representative immunoblot of HeLa-Ctrl(CDA) and HeLa-shCDA cells left untreated or treated with 2 pM CPT for 16 hours and transfected with the indicated siRNA. (C) Relative 3D volume of FISH signals in HeLa-Ctrl(CDA) (black bars) and HeLa-shCDA (gray bars) cells left untreated or treated with 2 pM CPT for 16 hours (n = 3, > 290 prometaphase cells analyzed). (D) Percentage of HeLa Rad21-depleted prometaphase cells with and without CDA expression, left untreated or treated with 1 µM olaparib for 16 hours and sorted on the basis of their phenotype: X-shapes, incomplete disjunction or complete disjunction (n = 3, > 110 prometaphase cells analyzed). (E) Representative immunoblot of HeLa-Ctrl(CDA) and HeLa-shCDA cells left untreated or treated with 1 µM olaparib for 16 hours and transfected with the indicated siRNA. (F) Percentage of HeLa Rad21-depleted prometaphase cells with and without PARP-1 expression, sorted on the basis of phenotype: X-shapes, incomplete disjunction or complete disjunction (n = 3, > 230 prometaphase cells analyzed). (G) Representative immunoblot of HeLa-Ctrl(PARP-1) and HeLa-shPARP-1 cell lines transfected with the indicated siRNAs. Error bars represent the mean ± SD. Statistical significance was assessed with Student's t-test.
Discussion
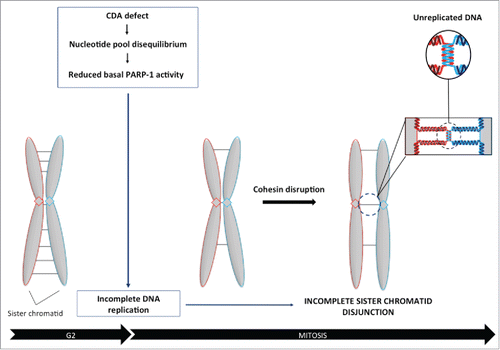
We report here a new mechanism by which an unbalanced nucleotide pool can affect sister chromatid disjunction. An excess of cellular dC and dCTP due to CDA deficiency, decreases basal PARP-1 activity, reducing centromeric volume, probably due to the accumulation of unreplicated DNA. As a consequence, sister chromatids remain physically linked in cohesin-depleted cells, leading to a defect in chromosome segregation ().
Figure 4. Balanced pyrimidine pool is required to promote optimal sister chromatid disjunction. Excess cellular dCTP, due to CDA deficiency, compromises basal PARP-1 activity, leading to the accumulation of unreplicated DNA sequences during mitosis, as revealed by the decrease in centromeric volume. As a consequence, sister chromatids remain physically linked in cohesin-depleted cells, revealing a defect in chromosome segregation.
Our results demonstrate that the structural changes at centromeres and the incomplete sister chromatid disjunction associated with BLM deficiencyCitation9 are entirely due to the nucleotide pool disequilibrium resulting from the lack of CDA associated with BLM deficiency, rather than BLM deficiency per se. Indeed, CDA expression in BS cells is sufficient to prevent these phenotypes, regardless of BLM expression status.
We also observed a correlation between structural changes at centromeres and incomplete sister chromatid disjunction, suggesting that these 2 phenotypes are linked. Both result from the lower levels of basal PARP-1 activity in CDA-deficient cells, highlighting the major role of PARP-1 in preventing structural changes to the centromere and incomplete sister chromatid disjunction.
Incomplete sister chromatid disjunction has previously been reported to be prevented by mitotic processes involving either ChiR1 or PICH DNA helicases.Citation17,18 Our data suggest that completion of replication is also of major importance to prevent incomplete sister chromatid disjunction during mitosis. Indeed, our data establish a new link between PARP-1 activity, centromeric structure and sister chromatid disjunction. We previously reported that CDA deficiency leads to a decrease in basal PARP-1 activity, promoting the accumulation of unreplicated DNA during mitosis. In particular, we observed that some centromeres were not fully replicated.Citation11,12 The presence of unreplicated DNA sequences at centromeres may decrease the amount of DNA available for FISH probe hybridization, resulting in the detection of a structural change. The incomplete sister chromatid disjunction phenotype in CDA-deficient cells may result from unreplicated DNA sequences physically linking 2 sister chromatids, impairing their segregation ().
In conclusion, our findings highlight a new role for CDA in the regulation of chromosome segregation: by maintaining a balanced nucleotide pool, CDA promotes optimal PARP-1 activity, which, in turn, ensures faithful sister chromatid disjunction by preventing the persistence of unreplicated DNA until mitosis.
Materials and methods
Cell culture and treatments
Cell lines were cultured in DMEM supplemented with 10% FCS. BS-Ctrl(BLM), BS-BLM, BS-Ctrl(CDA) and BS-CDA cells were obtained and cultured as described previously.Citation10 HeLa-Ctrl(CDA) and HeLa-shCDA cells were obtained and cultured as described previously.Citation11 HeLa-Ctrl(PARP-1) and HeLa-shPARP-1 cells were cultured as described previouslyCitation11
Camptothecin (CPT) was provided by Sigma Aldrich (C9911) and was added to the cell culture medium at a final concentration of 2 pM. Olaparib was provided by SelleckChem (S1060) and was added to the cell culture medium at a final concentration of 1 µM. dC was provided by Sigma Aldrich (D0779) and was added to the cell culture medium at a final concentration of 1 mM.
All cells were routinely checked for mycoplasma infection.
Western blot analysis and antibodies
Cells were lysed in 8 M urea, 50 mM Tris HCl, pH 7.5 and 150 mM β-mercaptoethanol, sonicated and heated at 75°C for 10 minutes. Samples (equivalent of 2 × 105 cells) were subjected to electrophoresis in NuPAGE Novex 4–12% Bis-Tris pre-cast gels (Life Technologies). The procedures used for gel electrophoresis and immunoblotting have been described elsewhere.Citation11 Primary and secondary antibodies were used at the following dilutions: rabbit anti-BLM antibody (1:5,000; ab2179 from Abcam); rabbit anti-CDA antibody (1:500; ab56053 from Abcam); rabbit anti-β-actin antibody (1:10,000; from Sigma); rabbit anti-PARP-1 antibody (1:4,000; ALX-210–302 from Enzo Life Sciences); mouse anti-Rad21 (1:1,000; 53A03 from Upstate); horseradish peroxidase-conjugated goat anti-rabbit or goat anti-mouse IgG (1:5,000; Santa Cruz Biotechnology).
Chromosome spread
Cells were left untreated or were transfected as indicated. After 48 h of transfection with an siRNA specific for Rad21, cells were transferred to slides in 6-well plates. After 24 h at 37°C, colchicine was added to a final concentration of 0.1 µg/ml and the cells were incubated for 1 hour. The cells were then incubated in hypotonic solution (1:5 (vol/vol) FCS-distilled water) and fixed by incubation with a 3:1 (vol/vol) mixture of methanol and acetic acid. Cells were then stained by incubation with 2% Giemsa solution (VWR) for 16 minutes, rinsed in distilled water, dried, and mounted. Chromosomes were observed with a Leica DMRB microscope at 100x magnification. Metaphases were captured with a SONY DXC 930 P camera.
FISH analysis
FISH was performed with the CEP-8 probe (Cen-8) from Abbott Vysis, according to the manufacturer's instructions and as described previously.Citation9
Statistical analysis
At least 3 independent experiments were performed to generate each data set and the statistical significance of differences was calculated with Student's t-test, as indicated in the figure legends.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Author contributions
SG performed the experiments, participated in the design of the experiments and data analysis, generated the figures and cowrote the manuscript. GBL and ROD performed experiments. CJ contributed to data analysis and preparation of the manuscript. MA-G supervised the study, analyzed the data and cowrote the manuscript.
1317413_Supplemental_Material.docx
Download MS Word (28.3 MB)Funding
This work was supported by grants from the Institut Curie (PICSysBio), the Center National de la recherche Scientifique (CNRS), the Ligue contre le Cancer (Comité de l'Essonne), the Association pour la Recherche sur le Cancer (ARC, SFI20121205645), the Agence Nationale de la Recherche (ANR-14-CE14–0004–01) and by a fellowship awarded to S.G by the Ministère de l'Education, de l'Enseignement Supérieur et de la Recherche and the ARC (DOC20140601310), and Institut Curie (PIC SysBio). C.J. is an investigator at the INSERM.
References
- Gelot C, Magdalou I, Lopez BS. Replication stress in Mammalian cells and its consequences for mitosis. Genes (Basel) 2015; 6:267-98; PMID:26010955; https://doi.org/10.3390/genes6020267
- Baxter J. “Breaking up is hard to do:” the formation and resolution of sister chromatid intertwines. J Mol Biol 2015; 427:590-607; PMID:25194916; https://doi.org/10.1016/j.jmb.2014.08.022
- Ciosk R, Zachariae W, Michaelis C, Shevchenko A, Mann M, Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell 1998; 3:1067-76; https://doi.org/10.1016/S0092-8674(00)81211-8
- Funabiki H, Kumada K, Yanagida M. Fission yeast Cut1 and Cut2 are essential for sister chromatid separation, concentrate along the metaphase spindle and form large complexes. Embo J 1996; 15:6617-28; PMID:8978688
- Losada A. Cohesin regulation: Fashionable ways to wear a ring. Chromosoma 2007; 116:321-9; PMID:17333234; https://doi.org/10.1007/s00412-007-0104-x
- Waizenegger IC, Hauf S, Meinke A, Peters JM. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell 2000; 103:399-410; PMID:11081627; https://doi.org/10.1016/S0092-8674(00)00132-X
- Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, Peters JM, et al. Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell 2001; 1:759-70; PMID:11740938; https://doi.org/10.1016/S1534-5807(01)00088-0
- German J. Bloom's syndrome. XX. The first 100 cancers. Cancer Genet Cytogenet 1997; 93:100-6; PMID:9062585; https://doi.org/10.1016/S0165-4608(96)00336-6
- Rouzeau S, Cordelières FP, Buhagiar-Labarchède G, Hurbain I, Onclercq-Delic R, Gemble S, Magnaghi-Jaulin L, Jaulin C, Amor-Guéret M. Bloom's syndrome and PICH helicases cooperate with topoisomerase IIalpha in centromere disjunction before anaphase. PLoS One 2012; 7:e33905; PMID:22563370; https://doi.org/10.1371/journal.pone.0033905
- Chabosseau P, Buhagiar-Labarchède G, Onclercq-Delic R, Lambert S, Debatisse M, Brison O, Amor-Guéret M. Pyrimidine pool imbalance induced by BLM helicase deficiency contributes to genetic instability in Bloom syndrome. Nat Commun 2011; 2:368; PMID:21712816; https://doi.org/10.1038/ncomms1363
- Gemble S, Ahuja A, Buhagiar-Labarchède G, Onclercq-Delic R, Dairou J, Biard DS, Lambert S, Lopes M, Amor-Guéret M. Pyrimidine pool disequilibrium induced by a cytidine deaminase deficiency inhibits PARP-1 activity, leading to the under replication of DNA. PLoS Genet 2015; 11:e1005384; PMID:26181065; https://doi.org/10.1371/journal.pgen.1005384
- Gemble S, Buhagiar-Labarchède G, Onclercq-Delic R, Biard D, Lambert S, Amor-Guéret M. A balanced pyrimidine pool is required for optimal Chk1 activation to prevent ultrafine anaphase bridge formation. J Cell Sci 2016; 129:3167-77; PMID:27383768; https://doi.org/10.1242/jcs.187781
- Nygaard P. On the role of cytidine deaminase in cellular metabolism. Adv Exp Med Biol 1986; 195 Pt B:415-20; PMID:3532704
- Tallis M, Morra R, Barkauskaite E, Ahel I. Poly(ADP-ribosyl)ation in regulation of chromatin structure and the DNA damage response. Chromosoma 2013; 123:79-90; PMID:24162931; https://doi.org/10.1007/s00412-013-0442-9
- Batram C, Baddeley D, Kreth G, Cremer C. High precision size measurement of centromere 8 and the 8q24/c-myc gene region in metaphase and interphase human fibroblasts indicate differential condensation. J Struct Biol 2008; 164:293-303; PMID:18835450; https://doi.org/10.1016/j.jsb.2008.09.002
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med 2009; 361:123-34; PMID:19553641; https://doi.org/10.1056/NEJMoa0900212
- Parish JL, Rosa J, Wang X, Lahti JM, Doxsey SJ, Androphy EJ. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J Cell Sci 2006; 119:4857-65; PMID:17105772; https://doi.org/10.1242/jcs.03262
- Nielsen CF, Huttner D, Bizard AH, Hirano S, Li TN, Palmai-Pallag T, Bjerregaard VA, Liu Y, Nigg EA, Wang LH, et al. PICH promotes sister chromatid disjunction and co-operates with topoisomerase II in mitosis. Nat Commun 2015; 6:8962; PMID:26643143; https://doi.org/10.1038/ncomms9962
