ABSTRACT
DNA damage activated by Adriamycin (ADR) promotes ubiquitin–proteasome system-mediated proteolysis by stimulating both the activity of ubiquitylating enzymes and the proteasome. In ADR-resistant breast cancer MCF7 (MCF7ADR) cells, protein ubiquitylation is significantly reduced compared to the parental MCF7 cells. Here, we used tandem ubiquitin-binding entities (TUBEs) to analyze the ubiquitylation pattern observed in MCF7 or MCF7ADR cells. While in MCF7, the level of total ubiquitylation increased up to six-fold in response to ADR, in MCF7ADR cells only a two-fold response was found. To further explore these differences, we looked for cellular factors presenting ubiquitylation defects in MCF7ADR cells. Among them, we found the tumor suppressor p53 and its ubiquitin ligase, Mdm2. We also observed a drastic decrease of proteins known to integrate the TUBE-associated ubiquitin proteome after ADR treatment of MCF7 cells, like histone H2AX, HMGB1 or β-tubulin. Only the proteasome inhibitor MG132, but not the autophagy inhibitor chloroquine partially recovers the levels of total protein ubiquitylation in MCF7ADR cells. p53 ubiquitylation is markedly increased in MCF7ADR cells after proteasome inhibition or a short treatment with the isopeptidase inhibitor PR619, suggesting an active role of these enzymes in the regulation of this tumor suppressor. Notably, MG132 alone increases apoptosis of MCF7ADR and multidrug resistant ovarian cancer A2780DR1 and A2780DR2 cells. Altogether, our results highlight the use of ubiquitylation defects to predict resistance to ADR and underline the potential of proteasome inhibitors to treat these chemoresistant cells.
Introduction
Protein ubiquitylation regulates multiple essential cellular processes such as proteolysis, DNA repair or transcription in response to diverse stimuli.Citation1–3 Ubiquitin attachment to substrate proteins is mediated by at least three categories of enzymes known as ubiquitin-activating enzymes (E1), ubiquitin-conjugating enzymes (E2), and ubiquitin-ligases (E3). Protein substrates can be modified by one or more ubiquitin moieties and the products are known as mono or multiple monoubiquitylated proteins, respectively. Ubiquitin chains (known as polyubiquitylation) can also be formed using internal lysine residues within ubiquitin (K6, K11, K27, K29, K33, K48, K63). Furthermore, Met1-linked linear ubiquitin chains are involved in the activation of signaling events.Citation4 While K11 and K48 chains have been associated to proteasome-mediated proteolysis, K63 chains have been linked to signaling pathways, intracellular traffic, DNA repair or autophagy among other functions.Citation5–7 The cleavage of ubiquitin moieties is regulated by several families of isopeptidases known as deubiquitylating enzymes (DUBs).Citation5 Ubiquitylated proteins are recognized by multiple ubiquitin-binding domains (UBDs) that connect modified substrates with effector functions.Citation6
Point mutations and chromosomal rearrangements affecting substrates, enzymes or cofactors of the ubiquitylation/deubiquitylation cascade are known to alter ubiquitylation of multiple cellular factors that have been proposed to be at the origin of pathologies such as cancer, inflammation, immunological disorders or neurodegeneration.Citation8–10 Intracellular signaling pathways, including those activating DNA damage response, induce protein ubiquitylation changes in stimulated cells. Therapeutic doses of ADR promote UPS-mediated proteolysis by stimulating both the activity of ubiquitylating enzymes and the proteasome.Citation11,12 However, breast cancer MCF7 cells resistant to ADR show low levels of ubiquitylation and fail to accumulate ubiquitylated proteins after stimulation with ADR, compared to parental ADR sensitive cells.Citation13 The analysis of ubiquitylation defects after cell stimulation with ADR could help us to gain an improved understanding for the role of critical factors involved in the response to this drug, identify non-responding patients and ultimately, define alternative targets for intervention.
Specific anti-ubiquitin antibodies or the tandem ubiquitin-binding entities (TUBEs) are among the most popular alternatives used to analyze endogenous ubiquitylated proteins.Citation14,15 In addition to their high affinity and specificity, TUBEs show two convenient properties to improve the purification yield of ubiquitylated proteins: the protection from the action of DUBs and also from proteasome-mediated proteolysis.Citation16–19 Here, we used TUBEs to isolate and analyze ubiquitylated proteins from MCF7 cells responding or not to ADR treatment. Besides their use as affinity probes for the enrichment of ubiquitylated proteins, TUBEs were useful to quantify differences in ubiquitylation between MCF7 and MCF7ADR cells. Analysis of the obtained ubiquitylation profiles indicated that UPS is active even in the case of resistant cells, where ADR treatment failed to induce the accumulation of ubiquitylated proteins. This implies that Ubiquitin Proteasome System (UPS) inhibitors could have therapeutic value if used to treat ADR resistant cells.
Results
Analysis of protein ubiquitylation defects in adriamycin resistant cells
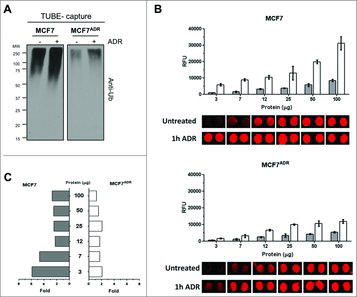
To quantify ubiquitylated proteins in response to the genotoxic agent ADR, MCF7 and MCF7ADR cells were stimulated (or not) at a time point (1 h) where we had previously observed an accumulation of protein ubiquitylation.Citation18 MCF7ADR cells showed a strong decrease of total ubiquitylated proteins at basal level and in response to ADR treatment in comparison to MCF7 cells (). We employed TUBEs-based microarrays to quantify differences in ubiquitylation profiles.Citation13 TUBEs-microarrays were incubated with known protein concentrations of MCF7 and MCF7ADR cellular extracts stimulated or not with ADR (experiment shown in ). Ubiquitylated proteins bound to TUBEs-microarrays were detected with mouse anti-ubiquitin antibody (FK2) and secondary Alexa Fluor® 647 rabbit anti-mouse antibody (). The images of individual spots are shown to demonstrate the good spot morphology and low background fluorescence obtained in the array experiments. Average fluorescence values and the SD of the mean from five replicate spots were quantified from the original images scanned at 10 μm resolution and represented as histograms above the images of individual spots for defined amounts of total protein (). Basal levels of ubiquitylated proteins in unstimulated cells showed lower relative fluorescence units (RFU) values that were proportional to the total protein content employed in the microarray assay. Protein ubiquitylation was significantly increased in response to ADR treatment only in sensitive MCF7 cells but not in MCF7ADR cells (). Interestingly, for higher protein concentrations between 50–100 μg, we observed only a modest 3-fold increase of ubiquitylation in MCF7 cells while for protein concentration below to 7 μg, ubiquitylation levels increased up to 6 fold, facilitating the comparison of ubiquitylation profiles between MCF7 and MCF7ADR cells (). This behavior might be explained by a rapid saturation of TUBEs at high concentrations of ubiquitylated proteins present in the cell extracts.Citation13 Altogether, our results indicate that TUBEs microarrays are well suited for the quantification of ubiquitylated proteins in the low microgram range.
Figure 1. Decreased of total protein ubiquitylation in MCF7ADR compared to MCF7 cells. (A) MCF7 and MCF7ADR cells were treated (+) or not for 1h with Adriamycin (1 μM). TUBE-capture fractions were analyzed by Western blot with the indicated antibodies. (B) Detection of ubiquitylated proteins on TUBEs microarrays. Analysis of ubiquitylated proteins in MCF7 or in MCF7ADR cell extracts at different concentrations of total protein (3, 7, 12, 25, 50, and 100 μg) with (white) or without (grey) ADR treatment (1h, 1 μM) is shown. Each histogram represents the average relative fluorescence units (RFU) value of five spots for each condition. (C) Fold of ubiquitylated proteins in MCF7 and MCF7ADR cell extracts at different concentrations of total protein (3, 7, 12, 25, 50, and 100 μg) comparing the level of total ubiquitylated proteins after ADR treatment to untreated cells.
Characterization of protein ubiquitylation profiles in response to adriamycin
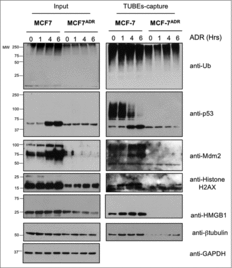
To analyze the ubiquitylation profiles of MCF7 and MCF7ADR cells in more detail, we tried to identify the key cellular factors with major ubiquitylation defects after ADR treatment. To this end, we isolated ubiquitylated proteins from the corresponding cell lines via TUBEs based affinity capture.Citation14,15 Input and captured fractions were analyzed by Western blot using a panel of selected antibodies recognizing proteins that are ubiquitylated in response to ADR in MCF7 cells Citation18 (). The most prominent ubiquitylation differences were observed for the tumor suppressor p53 and its ubiquitin E3 ligase Mdm2 (). Interestingly, p53 protein level does not increase after ADR treatment of MCF7ADR cells (, Input), explaining the low levels of expression of the p53-transcription dependent Mdm2 ligase (, input). Histone H2AX, HMGB1 or β-tubulin, that had been previously identified as part of the ADR-induced ubiquitin proteome in MCF7 cells Citation18, also showed low expression levels in resistant MCF7ADR cells, underscoring the differences observed in ubiquitylation profiles of MCF7 versus MCF7ADR cells after ADR treatment. These differences in ubiquitylation of these key cellular factors show potential for stratifying good from bad responders to ADR treatment.Citation13,20
Figure 2. Ubiquitylation defects of specific proteins in MCF7ADR cells. MCF7 and MCF7ADR cells were treated with ADR (1 μM) for the indicated times, ubiquitylated proteins were captured using TUBEs and analyzed by Western blot using the indicated antibodies. Input and TUBEs-capture fractions are shown. GAPDH was used as a loading control.
Inhibition of the ubiquitin-proteasome pathway recovers protein ubiquitylation and promote apoptosis in Adriamycin resistant cells
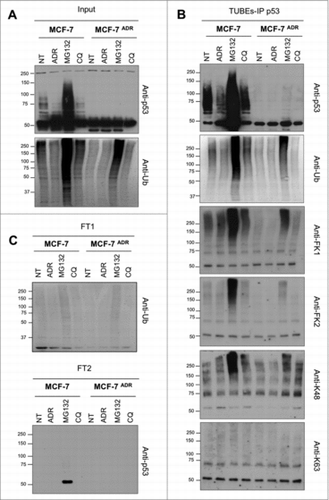
Intrigued by the low level of total protein ubiquitylation as well as the absence of p53 ubiquitylation in MCF7ADR cells, inhibitors of the proteasome and autophagy were used to accumulate and capture ubiquitylated proteins from MCF7 and MCF7ADR cells. Overnight treatment with proteasome inhibitor MG132, but not autophagy inhibitor chloroquine (CQ), partially recovered the total protein ubiquitylation in MCF7ADR cells (). The ubiquitylation of p53 was also increased after proteasome inhibition as detected using a TUBE-IP p53 procedure followed by Western blot with different ubiquitin antibodies (). These experiments were performed under conditions where most of the ubiquitylated proteins were captured, since we could hardly observe specific signal in the flow-through 1 (FT1) or flow-through 2 (FT2) (). Interestingly, the FK1 and K48 antibodies recognizing only polyubiquitylated proteins gave a stronger signal than the FK2 antibody that recognizes both mono- and polyubiquitylated species. These observations indicate that K48 polyubiquitylation of p53 is accumulated in both MCF7 and MCF7ADR cells in response to MG132 treatment. In contrast, ADR treatment modestly but consistently reduced the p53 ubiquitylation in MCF7 cells responding to this treatment.
Figure 3. Ubiquitin-chain profiles in ADR sensitive and resistant cell lines treated with proteasome and autophagy inhibitors. MCF7 and MCF7ADR cells were treated overnight with ADR (1 μM), MG132 (5 μM) or Chloroquine (CQ, 200 μM). Ubiquitylated forms were captured using TUBEs and bound material was eluted before being submitted to p53 immunoprecipitation as previously described Citation16. Input (A), TUBEs-IP p53 (B) and unbound fractions (FT1 and FT2) (C) were analyzed by Western blot using the indicated antibodies.
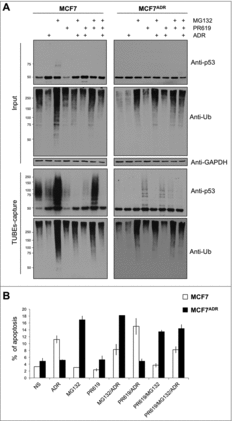
To investigate the role of isopeptidases in the defective ubiquitylation levels of MCF7ADR cells, the pan-inhibitor PR619 was used. Since this inhibitor could produce side effects in prolonged treatments, cells were treated for no longer than 15–30 min.Citation21,22 Under these experimental conditions, total ubiquitylation and p53 ubiquitylation were modestly but consistently increased after treatment of MCF7ADR cells, supporting an active role of isopeptidases in the regulation of this tumor suppressor in these chemoresistant cells ( and Supplementary ). The combination of PR619 with overnight ADR or MG132 treatments did not result in an accumulation but rather a decrease of p53 ubiquitylation, suggesting that these forms might be controlled by alternative molecular mechanisms (). Similar observations were obtained using 4h treatment with ADR and/or MG132 (Supplementary ). Interestingly, MG132 alone induced apoptosis of MCF7ADR cells, highlighting the potential use of proteasome inhibitors to treat ADR resistant cells ().
Figure 4. Proteasome inhibition promotes efficient apoptosis in MCF7ADR cells. (A) MCF7 and MCF7ADR cells were treated overnight with ADR, MG132 or PR619 (20 μM, 15 min) alone or in combination as indicated. Ubiquitylated proteins were captured using TUBEs and analyzed by Western blot. Input and TUBEs-capture fractions were shown. GAPDH was used as a loading control. (B) To track MCF7 cells undergoing apoptosis, cells were treated or not with ADR (1 μM overnight), MG132 (5 μM, overnight), or PR619 (20 μM, 15 min) alone or in combination as indicated. Data represent the mean of three independent experiments done in triplicate. A total of 10.000 events were collected and analyzed for each sample.
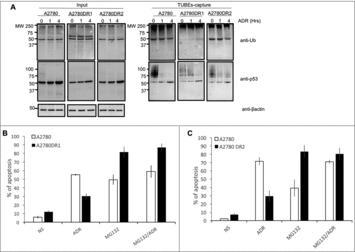
In order to investigate if all these observations could be extrapolated to other ADR resistant cells, ovarian carcinoma A2780 cells resistant to this chemotherapy agent were used. TUBE capture experiments were performed using parental A2780 and resistant A2780DR1 and A2780DR2 cells stimulated or not during 1 or 4 hours with ADR. The pattern of total ubiquitylation detected in the TUBE-captured fraction was reduced in the ADR resistant cells at basal level and after ADR stimulation (). Similarly, the pattern of ubiquitylated p53 was also reduced in ADR resistant cells at basal level but remain for longer after ADR treatment suggesting a delayed cellular response. Interestingly these differences are less obvious in the input fractions, underlining the benefit of using a TUBE-capture approach to detect these differences. We have also confirmed that MG132 alone could better induce the apoptosis of A2780DR1 and A2780DR2 cells ( and ), validating our previous observations in the breast cancer MCF7ADR cell line.
Figure 5. Proteasome inhibition promotes efficient apoptosis in A2780DR1 and A2780DR2 cells. (A) A2780, A2780DR1 and A2780DR2 cells were treated with ADR (2 μM) during 1 or 4 hours. Ubiquitylated proteins were captured using TUBEs and analyzed by Western blot with the indicated antibodies. Input and TUBEs-capture fractions were shown. β-actin was used as a loading control. (B) To track A2780DR1 and A2780DR2 cells undergoing apoptosis, these were treated or not during 24hrs with ADR (5 μM) or MG132 (2.5 μM) or in combination as indicated. Data represent the mean of three independent experiments.
Discussion
Ubiquitylation defects of critical cellular factors including those involved in the activation of the p53 pathway have been associated to distinct pathologies.Citation23,10 Strong stimuli such as DNA damage agents used in chemotherapy have the capacity to simultaneously activate multiple signaling cascades altering the protein ubiquitylation status and thus, the stability and activity of multiple proteins.Citation11 In this work, we show that ubiquitylation profiles of the totality of the proteins present in the cell can be used to distinguish between cells responding or not to the genotoxic agent ADR. Indeed, the results found by using TUBEs-microarrays revealed that total protein ubiquitylation increases up to 6 fold in ADR sensitive cells while in resistant cells the increase was less than 2-fold. Altogether, these results indicate that the analysis of protein ubiquitylation phenotypes using TUBEs-microarrays could be helpful to determine a personalized treatment option for breast cancer patients. This holds also true for specific factors such as p53 and Mdm2 that affected in their ubiquitylation levels in response to ADR. The concerted use of TUBEs with distinct antibodies could provide information on the degree and the type of accumulated protein ubiquitylation in response to an individual treatment.
The observed defects in p53 ubiquitylation in MCF7ADR cells are most probably due to the absence or high turnover of Mdm2 or other p53 ubiquitin ligases, or the hyperactivity of DUBs Citation24 but not to a general lack of p53 ubiquitylation since DUBs inhibition revealed ubiquitylated forms of this tumor suppressor. These results do not support the presence of non-tetrameric p53 monomers in MCF7ADR cells that would favor their turnover by a proteasome dependent-ubiquitin independent mechanism.Citation25,26 Thus, the fact that ubiquitylated p53 is partially accumulated in the presence of proteasome or DUBs inhibitors, indicates that ubiquitylation is mechanistically possible, however further investigations are required to better understand the origin of the reduced p53 ubiquitylation in MCF7ADR, A2780DR1 or A2780DR2 cells and its biological significance. Altogether, our characterization of ubiquitylation profiles indicated that even if ADR failed to accumulate ubiquitylated proteins in MCF7ADR, the UPS was active and could potentially be used to treat cancerous cells resistant to this chemotherapeutic agent. As matter of fact, the analysis of apoptosis using MG132 to treat ADR resistant cell lines supports this possibility ( and ).
We have observed a highly altered protein expression profile in the ADR resistant MCF7 cell lines using proteomics approaches. These changes include components of the GSH pathway that were proposed to contribute in the development of chemoresistance.Citation27,28 Interestingly, ADR significantly stimulated the formation of hydroxyl radical spin adducts [5,5-dimethyl-1-pyrroline N-oxide (DMPO)-OH] in the sensitive cells but not in the resistant cells.Citation29 The role of UPS and regulation of redox processes have emerged as essential factors to control the fate of cells upon differentiation.Citation30 This is coherent with the role of reactive oxygen species (ROS) affecting the conjugation of those ubiquitin family members that contribute to an appropriate response to chemotherapy.Citation31
The analysis of ubiquitylated proteins associated to cellular events continues to be a difficult task partly due to the highly dynamic and reversible formation of ubiquitin chains. Multiple approaches for ubiquitin analysis have been developed mainly based on the use of specific antibodies that recognize populations of ubiquitylated proteins or specific ubiquitin chains.Citation14 We show here that TUBEs combined with selected antibodies can be used to analyze total and individual ubiquitylated proteins from MCF7 and or A2780 cells in distinct protocols. This approach can be adopted to investigate ubiquitylation in response to distinct stimuli and to identify high from low responders to ADR.Citation13,20 The further development of methods to improve the analysis of protein ubiquitylation will contribute to a better understanding of the many cellular processes controlled by this post-translational modification and improve the current chemotherapeutic treatment options.
Materials and methods
Cell culture
Breast cancer MCF7 and MCF7ADR cells (gifts from Dr. Gant, MRC Toxicology Citation32,33) were grown in RPMI with 10% FBS and antibiotics. MCF7ADR cell cultures were supplemented with Adriamycin (ADR; Sigma-Aldrich; 0.5 μM) and the drug was removed 48 hours before performing experiments. Ovarian carcinoma derived cell lines A2780, A2780DR1 and A2780DR2 were grown in DMEM supplemented with 10% FBS and antibiotics.Citation34 A2780DR1 and A2780DR2 cells were grown in the presence of ADR (0.17 μM) and the drug was removed 24 hours before doing any experimental procedure.
Analysis of ubiquitylated proteins
MCF7 cells were treated or not with MG132 (Sigma-Aldrich, cat ≠ C2211, 5 μM, overnight), ADR (Sigma-Aldrich, cat ≠ 44583, as indicated), Chloroquine (Sigma-Aldrich, 200 μM, overnight) or PR619 (Merck, 20 μM, 15 min) alone or in combination. A2780 cells were treated with ADR (2 μM) during 1 or 4 hours. To capture low abundant ubiquitylated substrates and ubiquitin chains, saturating conditions were used during the TUBE-capture experiments. To proceed, the lysis buffer was supplemented with 3.5 μM of TUBEs hHR23A as previously described.Citation14,16 Lysates were clarified by cold centrifugation, and added to glutathione agarose beads (Biontex, cat ≠ R030). When indicated, glutathione beads were washed after ubiquitin capture (PBS-tween 0.05%), and the bound material was eluted before being submitted to p53 (DO.1 p53 antibody) immunoprecipitation as previously described.Citation16,35 Input, TUBEs-IP p53 and unbound fractions (FT1: obtained after the first capture with glutathione beads and FT2: obtained after TUBEs-IP p53) were analyzed using the corresponding antibodies.
Immunoblotting
Western blots were performed using the following primary antibodies: anti-p53 (clone DO1, Santa-Cruz, cat ≠ SC-126); anti-Mdm2 (Calbiochem, EMD Chemicals); anti-ubiquitin P4D1 (Santa Cruz Technology, cat ≠ SC-8017); anti-Histone H2AX (Abcam, cat ≠ Ab124781); anti-HMGMB1 (Abcam, cat ≠ Ab79823); anti-β Tubulin (Abcam, cat ≠ Ab6046); anti-GAPDH antibody (Sigma-Aldrich, cat ≠ A5441); anti-ubiquitin lys-K48 (Millipore, cat ≠ 05–1307), anti-ubiquitin lys-K63 specific (Enzo Life Sciences, cat ≠ PW0600-010), anti-ubiquitin FK1 (Enzo Life Sciences, cat ≠ BML-PW88) and anti-ubiquitin FK2 (Enzo Life Sciences, cat ≠ PW8810-0500).
Protein arrays
Detection of ubiquitylated proteins on TUBEs microarray was performed as previously described.Citation13 Cells were treated or not for 1h with ADR 1 μM and lysed for 30 min on ice. Cell lysate dilutions (100 μL) were incubated for 2 hours at room temperature on the TUBEs microarrays. Under these conditions TUBEs become saturated at high cell extract concentrations. Detection of ubiquitylated proteins was performed by incubation with anti-ubiquitin mouse monoclonal antibody (FK2) followed by Alexa Fluor®647 rabbit anti-mouse IgG H+L (Life Technologies). The slides were washed with TBS, water and dried in a slide spinner. Fluorescence measurements were performed on a microarray scanner (Agilent G2565BA, Agilent Technologies). Quantification of fluorescence was performed by ProScanArray® Express software (Perkin Elmer). Average RFU values with local background subtraction of five spots and standard deviation of the mean were reported using GraphPad Prism® software.
Apoptosis analysis
To track apoptosis, MCF7 cells were treated or not with MG132 (5 μM, overnight), ADR (1 μM, overnight), Chloroquine (200 μM, overnight) or PR619 (Merck, 20 μM, 15 min) either alone or in combination as indicated. A2780 cells were treated or not as indicated with MG132 (2.5 μM, 24 hours), ADR (5 μM, 24 hours), either alone or in combination. Co-staining with Annexin-V-DY634 (Immunostep, cat ≠ ANXVKDY-100T) and dead cell stain Sytox green (Fischer Bioblock Scientific, cat ≠ 05BR080) was performed to differentiate: early and late apoptosis as well as necrotic cells. The percentage of Annexin V+/sytox green- was analyzed by flow cytometry excluding doublets. Appropriate single staining controls were used to set voltage and compensation values. Data were collected on a FACSCanto (BD Biosciences) and were analyzed using FlowJo software (www.flowjo.com).
List of abbreviations
| ADR | = | Adriamycin |
| CQ | = | Chloroquine |
| DUBs | = | Deubiquitylating enzymes |
| TUBEs | = | Tandem ubiquitin-binding entities |
| UPS | = | Ubiquitin–proteasome system |
| UBDs | = | ubiquitin-binding domains |
Disclosure of potential conflicts of interests
The authors declare that they have no conflicts interests.
Acknowledgments
We thank Dr. Gant for providing the cell lines used in this study and laboratory members for technical support and helpful suggestions. We also thank Dr. J.E. Sarry for facilitating the access to their facilities.
Additional information
Funding
References
- Ciechanover A, Stanhill A. The complexity of recognition of ubiquitinated substrates by the 26S proteasome. Biochimica et biophysica acta. 2014;1843:86–96. doi:10.1016/j.bbamcr.2013.07.007. PMID:23872423.
- Bassermann F, Eichner R, Pagano M. The ubiquitin proteasome system – implications for cell cycle control and the targeted treatment of cancer. Biochimica et biophysica acta. 2014;1843:150–162. doi:10.1016/j.bbamcr.2013.02.028. PMID:23466868.
- Clague MJ, Heride C, Urbe S. The demographics of the ubiquitin system. Trends Cell Biol. 2015;25:417–426. doi:10.1016/j.tcb.2015.03.002. PMID:25906909.
- Walczak H, Iwai K, Dikic I. Generation and physiological roles of linear ubiquitin chains. BMC Biol. 2012;10:23. doi:10.1186/1741-7007-10-23. PMID:22420778.
- Heideker J, Wertz IE. DUBs, the regulation of cell identity and disease. Biochem J. 2015;465:1–26. doi:10.1042/BJ20140496. PMID:25631680.
- Husnjak K, Dikic I. Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annu Rev Biochem. 2012;81:291–322. doi:10.1146/annurev-biochem-051810-094654. PMID:22482907.
- Erpapazoglou Z, Walker O, Haguenauer-Tsapis R. Versatile roles of k63-linked ubiquitin chains in trafficking. Cells. 2014;3:1027–1088. doi:10.3390/cells3041027. PMID:25396681.
- Xolalpa W, Perez-Galan P, Rodriguez MS, Roue G. Targeting the ubiquitin proteasome system: beyond proteasome inhibition. Curr Pharm Des. 2013;19:4053–4093. doi:10.2174/1381612811319220014. PMID:23181575.
- Mata-Cantero L, Lobato-Gil S, Aillet F, Lang VR MS. The ubiquitin-proteasome system (UPS) as a cancer drug target: emerging mechanisms and therapeutics. Stress Response Pathways in Cancer. From Molecular Targets to Novel Therapeutics: Springer, Dordrecht,. 2015.
- Gallo LH, Ko J, Donoghue DJ. The importance of regulatory ubiquitination in cancer and metastasis. Cell Cycle. 2017;16:634–648. doi:10.1080/15384101.2017.1288326. PMID:28166483.
- Liu J, Zheng H, Tang M, Ryu YC, Wang X. A therapeutic dose of doxorubicin activates ubiquitin-proteasome system-mediated proteolysis by acting on both the ubiquitination apparatus and proteasome. Am J Physiol Heart Circ Physiol. 2008;295:H2541–H2550. doi:10.1152/ajpheart.01052.2008. PMID:18978187.
- Ranek MJ, Wang X. Activation of the ubiquitin-proteasome system in doxorubicin cardiomyopathy. Curr Hypertens Rep. 2009;11:389–395. doi:10.1007/s11906-009-0068-8. PMID:19895749.
- Serna S, Xolalpa W, Lang V, Aillet F, England P, Reichardt N, Rodriguez MS. Efficient monitoring of protein ubiquitylation levels using TUBEs-based microarrays. FEBS Lett. 2016;590:2748–2756. doi:10.1002/1873-3468.12289. PMID:27410252.
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, England P, Rodriguez MS. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Rep. 2009;10:1250–1258. doi:10.1038/embor.2009.192. PMID:19798103.
- Hjerpe R, Rodriguez MS. Efficient approaches for characterizing ubiquitinated proteins. Biochem Soc Trans. 2008;36:823–827. doi:10.1042/BST0360823. PMID:18793144.
- Aillet F, Lopitz-Otsoa F, Hjerpe R, Torres-Ramos M, Lang V, Rodriguez MS. Isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. Methods Mol Biol. 2012;832:173–183. doi:10.1007/978-1-61779-474-2_12. PMID:22350885.
- Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A data set of human endogenous protein ubiquitination sites. Molecular & cellular proteomics: MCP. 2011;10:M110 002089. doi:10.1074/mcp.M110.002089.
- Lopitz-Otsoa F, Rodriguez-Suarez E, Aillet F, Casado-Vela J, Lang V, Matthiesen R, Elortza F, Rodriguez MS. Integrative analysis of the ubiquitin proteome isolated using Tandem Ubiquitin Binding Entities (TUBEs). J Proteomics. 2012;75:2998–3014. doi:10.1016/j.jprot.2011.12.001. PMID:22178446.
- Yoshida Y, Saeki Y, Murakami A, Kawawaki J, Tsuchiya H, Yoshihara H, Shindo M, Tanaka K. A comprehensive method for detecting ubiquitinated substrates using TR-TUBE. Proc Natl Acad Sci U S A. 2015;112:4630–4635. doi:10.1073/pnas.1422313112. PMID:25827227.
- Eisenberg-Lerner A, Ciechanover A, Merbl Y. Post-translational modification profiling – A novel tool for mapping the protein modification landscape in cancer. Exp Biol Med. 2016;241:1475–1482. doi:10.1177/1535370216651732. PMID:27229346.
- Altun M, Kramer HB, Willems LI, McDermott JL, Leach CA, Goldenberg SJ, Kumar KG, Konietzny R, Fischer R, Kogan E, et al. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem Biol. 2011;18:1401–1412. doi:10.1016/j.chembiol.2011.08.018.
- Seiberlich V, Goldbaum O, Zhukareva V, Richter-Landsberg C. The small molecule inhibitor PR-619 of deubiquitinating enzymes affects the microtubule network and causes protein aggregate formation in neural cells: implications for neurodegenerative diseases. Biochimica et biophysica acta. 2012;1823:2057–2068. doi:10.1016/j.bbamcr.2012.04.011. PMID:22565157.
- Sane S, Rezvani K. Essential Roles of E3 Ubiquitin Ligases in p53 Regulation. International journal of molecular sciences. 2017;18. doi:10.3390/ijms18020442. PMID:28218667.
- Armstrong SR, Wu H, Wang B, Abuetabh Y, Sergi C, Leng RP. The Regulation of Tumor Suppressor p63 by the Ubiquitin-Proteasome System. Int J Mol Sci. 2016;17(12):2041. doi:10.3390/ijms17122041. PMID:27929429.
- Hjerpe R, Aillet F, Lopitz-Otsoa F, Lang V, Torres-Ramos M, Farras R, Hay RT, Rodriguez MS. Oligomerization conditions Mdm2-mediated efficient p53 polyubiquitylation but not its proteasomal degradation. The international journal of biochemistry & cell biology. 2010;42:725–735. doi:10.1016/j.biocel.2010.01.010.
- Lang V, Pallara C, Zabala A, Lobato-Gil S, Lopitz-Otsoa F, Farras R, Hjerpe R, Torres-Ramos M, Zabaleta L, Blattner C, et al. Tetramerization-defects of p53 result in aberrant ubiquitylation and transcriptional activity. Mol Oncol. 2014;8:1026–1042. doi:10.1016/j.molonc.2014.04.002. PMID:24816189.
- Gehrmann ML, Fenselau C, Hathout Y. Highly altered protein expression profile in the adriamycin resistant MCF-7 cell line. Journal of proteome research. 2004;3:403–409. doi:10.1021/pr0340577. PMID:15253420.
- Wang Z, Liang S, Lian X, Liu L, Zhao S, Xuan Q, Guo L, Liu H, Yang Y, Dong T, et al. Identification of proteins responsible for adriamycin resistance in breast cancer cells using proteomics analysis. Sci Rep. 2015;5:9301. doi:10.1038/srep09301. PMID:25818003.
- Sinha BK, Katki AG, Batist G, Cowan KH, Myers CE. Adriamycin-stimulated hydroxyl radical formation in human breast tumor cells. Biochemical pharmacology 1987;36:793–796. doi:10.1016/0006-2952(87)90164-X. PMID:3032195.
- Demasi M, Simoes V, Bonatto D. Cross-talk between redox regulation and the ubiquitin-proteasome system in mammalian cell differentiation. Biochimica et biophysica acta. 2015;1850:1594–1606. doi:10.1016/j.bbagen.2014.10.031. PMID:25450485.
- Bossis G, Sarry JE, Kifagi C, Ristic M, Saland E, Vergez F, Salem T, Boutzen H, Baik H, Brockly F, et al. The ROS/SUMO axis contributes to the response of acute myeloid leukemia cells to chemotherapeutic drugs. Cell Rep. 2014;7:1815–1823. doi:10.1016/j.celrep.2014.05.016. PMID:24910433.
- Davies R, Budworth J, Riley J, Snowden R, Gescher A, Gant TW. Regulation of P-glycoprotein 1 and 2 gene expression and protein activity in two MCF-7/Dox cell line subclones. British journal of cancer 1996;73:307–315. doi:10.1038/bjc.1996.54. PMID:8562335.
- Batist G, Tulpule A, Sinha BK, Katki AG, Myers CE, Cowan KH. Overexpression of a novel anionic glutathione transferase in multidrug-resistant human breast cancer cells. J Biol Chem. 1986;261:15544–15549. PMID:3782078.
- Januchowski R, Sterzynska K, Zaorska K, Sosinska P, Klejewski A, Brazert M, Nowicki M, Zabel M. Analysis of MDR genes expression and cross-resistance in eight drug resistant ovarian cancer cell lines. J Ovarian Res. 2016;9:65. doi:10.1186/s13048-016-0278-z. PMID:27756418.
- Aillet F, Lopitz-Otsoa F, Egana I, Hjerpe R, Fraser P, Hay RT, Rodriguez MS, Lang V. Heterologous SUMO-2/3-ubiquitin chains optimize IkappaBalpha degradation and NF-kappaB activity. PloS One. 2012;7:e51672. doi:10.1371/journal.pone.0051672. PMID:23284737.
