ABSTRACT
MicroRNA let-7 has been reported to be down-regulated in several human cancers and is now characterized as a tumor suppressor. IGF1R is over-expressed in many cancers and IGF1/IGF1R pathway is attractive target for anticancer therapy. However, the crosstalk between let-7 and IGF1/IGF1R are largely unknown. The present study showed IGF1R were significantly over-expressed in colorectal cancer tissues compared with adjacent normal tissues through immunohistochemical analysis. qRT-PCR results showed that let-7a, let-7b and let-7e were down-regulated in colorectal cancer tissues. Bioinformatics analysis revealed that both IGF1 and IGF1R mRNA are potential targets for let-7 miRNA family. Ectopic transfection of let-7e led to a significant reduction in IGF1R at protein level and their downstream Akt inhibition, as well as a reduction in cell proliferation, migration and invasion in colorectal cancer cells, while inhibition of let-7e enhanced the expression of IGF1R. On the other hand, IGF1 stimulation can significantly down-regulate the expression of let-7e in colorectal cancer cells. Taken together, our findings identify a negative feedback regulation between let-7e and IGF1/IGF1R, and suggest that let-7e could be used in IGF1R-targeted therapeutics in anticancer therapy.
Abbreviations: IGF1: insulin-like growth factor 1; IGF1R: IGF1 receptor; miRNA: microRNA; CRC: colorectal cancer; EGFR: epidermal growth factor receptor; HRP: horseradish peroxidase; MTT: 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide; p-Akt: phospho-Akt; PI3K: phosphoinositide 3-kinase; qRT–PCR: quantitative reverse transcription–PCR; IHC: immunohistochemical; siRNA: small interfering RNA; 3′-UTR: 3′-untranslated region
Introduction
Colorectal cancer (CRC) is among the five most common cancers in the world in terms of incidence for both men and women.[Citation1] According to the American Cancer Society, the 3 most prevalent cancers among males are prostate (43%), colorectal (9%), and melanoma of the skin (7%), and those among females are breast (41%), uterine corpus (8%), and colorectal (8%) in the United States. It is estimated that there are nearly 1.2 million men and women living in the United States with a previous diagnosis of colorectal cancer, and an additional 143,460 will be diagnosed in 2012.[Citation2]
The type I insulin-like growth factor receptor (IGF1R) is a member of a family of transmembrane tyrosine kinases. Accumulating evidence implicated the IGF-1R and its ligands, IGF-1 and IGF-2, in the development and progression of cancer including colorectal cancer.[Citation3,Citation4] The principal pathways for transduction of the IGF signal are the mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase/Akt (PI3K/Akt) pathways.[Citation5] Amplified IGF-1/IGF-1R signaling is not only associated with an increased relative risk for development of CRC, but also contributes to CRC cell survival, invasion, metastasis and resistance to chemotherapeutic drugs.[Citation6,Citation7] Although up-regulation of IGF1R has been found in many types of cancers including CRC, the mechanisms of the increase remains elusive.
Post-transcriptional gene regulation by miRNAs is an increasingly well-appreciated regulatory mechanism involved in almost every biological process of multicellular life.
Identified as the first mammalian miRNAs to have functional and sequence conservation to a C. elegans counterpart, the mammalian let-7 family comprises twelve members expressed from eight distinct loci (let-7a-1,-2,-3; let-7b; let-7c; let-7d; let-7e; let-7f-1,-2; let-7g; let-7i; miR-98).[Citation8,Citation9] Numerous reports have shown that the expression levels of let-7 are frequently low and the chromosomal clusters of let-7 are often deleted in many cancers. Let-7 is also a very attractive potential therapeutic target for anticancer drug development.[Citation10,Citation11] Intranasal administration of let-7 has already been found effective in reducing tumor growth in a transgenic mouse model of lung cancer.[Citation12] Similar restoration of let-7 was also shown to inhibit cell proliferation in breast, colon and hepatic cancers, lymphoma, and uterine leiomyoma.[Citation13–Citation15] So let-7 has been characterized as tumor suppressor in several human cancers. It targets a number of proto-oncogenes, including Ras, HMGA2, c-Myc, IL6, and Lin28 and Lin28B.[Citation8,Citation16,Citation17] Lin28/Lin28B function to block the post-transcriptional processing of let-7 to its mature, active cytoplasmic form by binding the primary and precursor forms of let-7. Hence, Lin28/Lin28B and let-7 are mutual antagonists; when expression and activity of one is high, the other is suppressed.[Citation13–Citation15,Citation18]
Recent studies have pointed to a role of microRNAs (miRs) in post-transcriptional regulation of IGF1R.[Citation19–Citation25] While a group of miRs including miR-470, miR-669b and miR-681 are involved in repression of IGF1R in long-lived mutant mice [Citation19], miR-375 and miR-7 target IGF1R in oesophageal and tongue squamous cell carcinoma cells, respectively.[Citation20,Citation21] miR-145 targets the docking protein of IGF1R, insulin receptor substrate-1, thus inhibiting IGF1R signaling.[Citation22] In addition, miR-497 and miR-139 have been reported to target IGF1R and has a tumor suppressive role in human colorectal cancer.[Citation24,Citation25]
In the present study, through immunohistochemistry (IHC) assay, we found IGF1R were significantly over-expressed in CRC tissues compared with adjacent normal tissues. Using the online miRNA target prediction tool, we found let-7 has the highest probability score among the predicted miRNA targeting IGF1R (http://www.targetscan.org). There were three potential let-7-target sites with higher probability of preferential conservation among the 3ʹUTR of IGF1R. Moreover, IGF1 was also a potential target of let-7 according to the analysis of TargetScan. Through real-time RT-RCR, let-7a, let-7b, and let-7e were shown to be significantly down-regulated in CRC tissues compared with adjacent normal tissues. Furthermore, our data also showed that IGF1 stimulation can down-regulate the expression of let-7b and let-7e in colorectal cancer cell HCT-116. Over-expression of let-7e can significantly down-regulate the expression of IGF1R, and attenuate the proliferation, migration and invasion of HCT-116 cells.
Materials and methods
Tissue samples
All human tissue samples were obtained from patients undergoing surgical resection in Shaoxing People’s Hospital. Informed consent was obtained from each patient before tissue collection. The tumor and its adjacent normal tissues were also obtained. Each specimen was divided into 2 parts. One of them was fixed in 10% phosphate-buffered formalin, embedded in paraffin and subjected to Immunohistochemical (IHC) analysis, and the other was used for extraction of miRNA.
IHC analysis
Sections were stained with Envision System method (DakoCytomation, Carpinteria, CA), according to manufacturer’s instructions. Mouse monoclonal antibody against human IGFR1 was from Abcam. Saturation and intensity of immunostained cells was evaluated over 8 visual fields under a light microscope (Olympus Optical, Tokyo, Japan). In statistical analysis, with reference to previous study [Citation26], total staining of IGF1R were scored as the product of the staining intensity (on a scale of 0–3: negative = 0, weak = 1, moderate = 2, strong = 3) × the percentage of cells stained (0 = zero, 1 = 1–25%, 2 = 26–50%, 3 = 51–100%), which resulted in a scale of 0–9. two independent investigators scored each sample without any prior knowledge of each patient’s clinical information and outcome.
Cell culture, IGF1 treatment and transfection
The human colorectal cancer cell line HCT-116, LoVo, SW480 and SW620 were obtained from the American Type Culture Collection (ATCC) and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mmol/L L-glutamine, 100 U/mL penicillin, and 100 U/mL streptomycin in a 5% CO2 incubator at 37 °C. The normal human fetal colon cells (CRL-1831) were from ATCC and cultured in a 1:1 mixture of Ham’s F12 and DMEM containing HEPES (25 mM), cholera toxin (10 ng/ml), insulin (5 μg/ml), transferring (5 μg/ml) and cortisone (100 ng/ml), and supplemented with 10% FBS. The cultures were maintained at 37°C in 5% CO2.
For IGF1 treatment, when cells were grown to 30% confluency, and then medium was replaced with fresh serum-free medium containing 50 ng/ml IGF1 (PeproTech), and PBS were set as vehicle group. Cells were collected at 6 h, 12 h, and 24h after treatment for miRNA analysis.
Let-7e mimics and control mimics, let-7e antagomir and control antagomir were from Ribobio (Guangzhou, China). IGF1R-specific siRNA (On-TargetPlus SMARTpool) were from Dharmacon. miRNA transfection was performed using DharmaFECT (Thermo Scientific) reagent according to manufacturer’s instructions. siRNA transfection was carried out with X-tremeGENE siRNA reagent from Roche according to manufacturer’s instructions.
Western blot analysis
Antibody against IGF1R (#3027), pan Akt (#4691), p-Akt (Thr308, #4056) and p-Akt (Ser473, #4060) were purchased from Cell Signaling Technology. β-Actin (beyotime, #AF0003) was used as an internal control. In brief, after washing with ice-cold PBS three times, cells were homogenized and sonicated in RIPA buffer (50 mM Tris-base, 150 mM NaCl, 1.0 mM EDTA, 0.1% SDS, 1% Triton X-100, and 1% sodium deoxycholate) containing protease inhibitor cocktail (Sigma). Sample concentration was determined using the Bradford assay (Bio-Rad). Extracted proteins were separated by 12% SDS-PAGE and transferred to PVDF membranes. Membranes were blocked overnight with TBS containing 0.1% Tween 20 in 5% skimmed milk at 4°C, and then incubated with primary antibodies at room temperature for 1 h or overnight at 4°C. After three washes in TBST, the membranes were incubated with a HRP-conjugated secondary antibody for 2 h at room temperature, and the protein bands were then visualized using enhanced chemiluminescence reagents (Millipore).
miRNA extraction and real-time RT–PCR analysis
miRNA were extracted from freshly isolated clinical samples or from cultured cell lines by using miRcute miRNA Extraction Kit (TIANGEN Biotech, Beijing, China) according to the manufacturer’s instructions. Reverse transcription was carried out using miRcute miRNA first-strand cDNA Synthesis Kit (TIANGEN). Realtime PCR was performed using miRcute miRNA qPCR Detection Kit (SYBR Green, TIANGEN) according to the manufacturer’s instructions. U6 was used as a miRNA internal control. Let-7 family member-specific primer sets were as described previously.[Citation27]
Proliferation and apoptosis assay
Cell proliferation was measured using the standard MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assay. In brief, at 48 h post-transfection, the transfection medium in each well was replaced by fresh serum-free medium with MTT (1 mg/ml). After incubation at 37°C for 4 h, the medium were removed and 150 μl of dimethylsulfoxide were added to dissolve formazan crystals. Absorbance was measured at 570 nm using an ELISA microplate reader.
Cell apoptosis was determined by Flow cytometry analysis using Annexin V-FITC/PI apoptosis detection kit (beyotime) according to manufacturer’s instructions. Briefly, at 48 h post-transfection, cells (1 × 105) were harvested and suspended in 195 μl binding buffer with 5 μl Annexin V-FITC, and incubated for 10 min at room temperature. After centrifugation, cells were re-suspended in 190 μl binding buffer with 10 μl propidium iodide. Samples were then analyzed by flow cytometry (Becton Dickinson).
Migration and invasion analysis
The ability of migration and invasion of colorectal cells were examined by using non-coated or Matrigel-coated transwell culture chamber (8 μm pore size, Corning) 48 h after transfection. For migration assays, 1 × 105 cells were seeded in the top non-coated chamber and incubated at 37°C for 8 h. For invasion assays, 5 × 104 cells were seeded in the top Matrigel-coated (BD Biosciences, Bedford, MA) chamber and incubated at 37°C for 24 h. In both assays, cells were suspended in serum-free RPMI 1640 medium in the upper chamber, and the lower chamber was filled with 1640 medium containing 5% FBS. After incubation, the top chambers were wiped with cotton wool to remove the non-migratory or non-invasive cells. Cells on the underside of the membrane were fixed in methanol for 30 min, stained with 0.1% crystal violet, and counted under a microscope (Eclipse TS100, Nikon, Japan).
Statistical analysis
All statistical analyzes were performed by Student’s t test. Values of p < 0.05 were considered statistically significant.
Results
Up-regulation of IGF1R in colorectal cancer
IHC analysis was performed to examine the expression of IGF1R in clinical colorectal cancer samples ( & ). As shown, IGF1R was weekly or moderately stained in the well-differentiated CRC tissues, whereas it was moderately or strongly stained in moderately- and poorly-differentiated CRC tissues. Of the 22 adjacent normal tissues, IGF1R was weekly or negatively stained. These result indicated that elevated expression of IGF1R was paralleled with increased severity of colorectal cancer ((a)).
Table 1. IGF1R immunostaining results of patients with colorectal cancer
Figure 1. Expression of IGF1R in colorectal cancer, IGF1R-targeted miRNA prediction, as well as the expression of let-7 miRNA in CRC. (a) Immunohistochemical analysis of the expression of IGF1R in: a, adjacent normal tissues; b, moderately-differentiated CRC and c, pooly-differentiated CRC. (b) Let-7 miRNA family has the highest probability score among the predicted miRNA targeting IGF1R using TargetScanHuman 6.2. (c) qRT–PCR analysis of let-7 miRNA family in total RNA from CRC tissues and paired adjacent normal tissues. The data shown are average fold changes (the mean ± SD of three independent experiments) of individual miRNA expression CRC tissues relative to normal tissues. (d) qRT–PCR analysis of let-7 miRNA family in total RNA from indicated colon cancer cell lines and the normal colon epithelial line CRL-1831. The data shown are average fold changes (the mean ± SD of three independent experiments) of individual miRNA expression in each colon cancer line relative to CRL-1831 cells. *P < 0.05; **P < 0.01. Scale bar, 25 μm
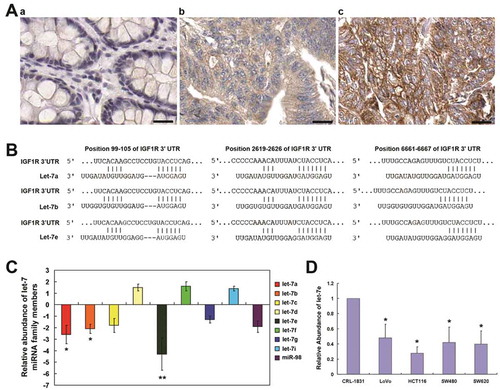
IGF1R-targeted miRNA prediction and the down-regulation of let-7 miRNA in CRC
IGF1R-targeted candidate miRNA were predicted using using TargetScanHuman 6.2. Among the predicated candidate miRNA, let-7 miRNA family has the highest probability score ((b)). As shown, there were three potential let-7-target sites with higher probability of preferential conservation among the 3ʹUTR of IGF1R. So we examined the expression of the nine let-7 members in CRC tissues through real-time RT-RCR. As shown in , let-7a, let-7b, and let-7e were significantly down-regulated in CRC tissues compared with adjacent normal tissues. Among them, let-7e was the most significantly down-regulated member, and so it was selected for further research into the roles of let-7 dysregulation in CRC. showed that let-7e was down-regulated in four colorectal cancer cell lines (HCT-116, LoVo, SW480 and SW620) compared with the normal colon epithelial line CRL-1831.
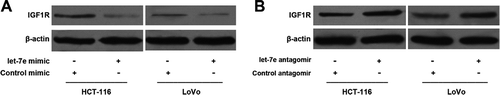
Let-7e down-regulated the expression of IGF1R
As shown in , transfection with let-7e mimic can reduce the protein levels of IGF1R in HCT-116 and LoVo colorectal cancer cells compared with control group. Conversely, knockdown of let-7e by let-7e-specific antagomir can cause an increase in the expression of IGF1R in HCT-116 and LoVo cells.
Figure 2. Effects of Let-7e on the expression of IGF1R. HCT-116 and LoVo human colorectal cancer cells were transfected with let-7e mimic or antagomir for 48 h. The expression levels of IGF1R were examined by Western blotting as described in the Methods section. Results shown are representative of at least three independent experiments
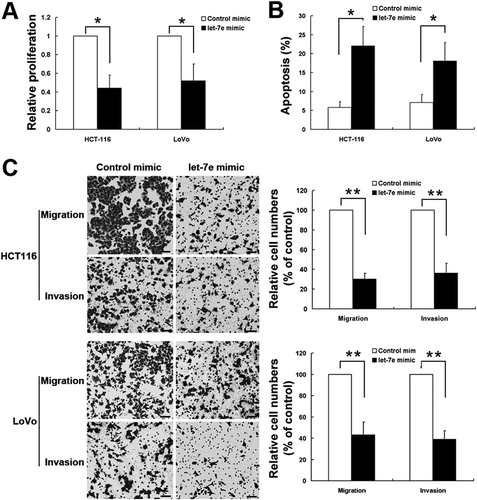
Let-7e promoted apoptosis and inhibited proliferation, migration and invasion in CRC cells
The changes in cell proliferation, apoptosis, migration and invasion in CRC cells were examined by transfection of let-7e mimics. As shown in , compared with control groups, let-7e mimics transfection can significantly inhibited the proliferation of both HCT-116 and LoVo cells using MTT assays. Though flow cytometry analysis using annexin V-FITC/PI double staining, it was shown that let-7e can promoted apoptosis both in HCT-116 and in LoVo cells. Moreover, transwell assays indicated the ability of migration and invasion in HCT-116 and LoVo cells were also significantly reduced by let-7e compared with control groups.
Figure 3. The effects of let-7e on cell proliferation, apoptosis, migration and cell invasion. After transfected with let-7e mimic, the changes in cell proliferation (a), cell-apoptosis rate (B), cell migration and invasion (c) were examined as described in the Methods section. All results shown are means ± SD from at least three independent experiments. *P < 0.05; **P < 0.01. Scale bar, 50 μm
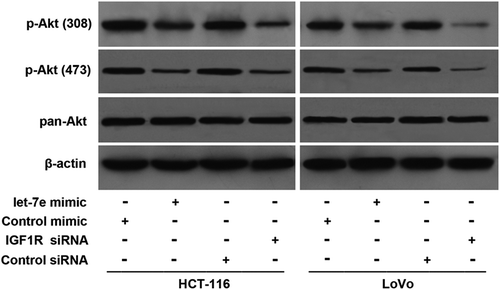
Let-7e inhibited the activation of Akt
Since IGF1R is a major player in the activation of the Akt signalling pathway and Akt activation play essential roles in carcinogenesis, we investigated the effects of let-7e over-expression or IGF1R knockdown on Akt phosphorylation. As shown in , although the total Akt level was not changed, the levels of p-Akt at both residues (Thr308 and Ser473) were reduced after transfection with let-7e mimic for 48 h both in HCT-116 cells and in LoVo cells and similar results were observed in IGF1R siRNA groups. These results suggest that let-7e and IGF1R play important roles in regulating Akt pathway in CRC cells.
Figure 4. Effects of let-7e on activation of Akt. HCT-116 and LoVo human colorectal cancer cells were transfected with let-7e mimic or IGF1R siRNA for 48 h. The levels of total Akt (pan-Akt) and p-Akt (Thr308 and Ser473) were examined by Western blotting as described in the Methods section. Results shown are representative of at least three independent experiments
Negative feedback regulation between microRNA let-7e and IGF1/IGF1R
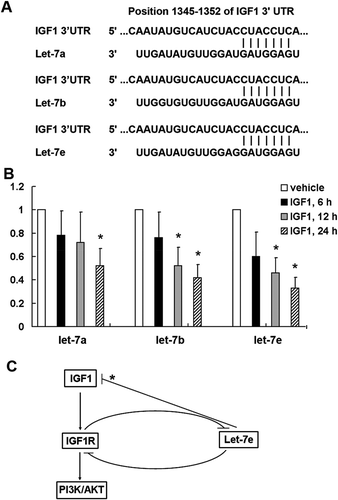
Through bioinformatics analysis we found that let-7 miRNA family was also among the predicted miRNA targeting IGF1 using TargetScanHuman 6.2 (). We also investigated whether IGF1/IGF1R activation can influence the expression of let-7 miRNA in CRC cells. As shown in , let-7a, let-7b and let-7e were significantly down-regulated following treatment with IGF1 in HCT-116 colorectal cancer cells by qRT–PCR analysis. Finally, schematic diagram showed a negative feedback regulation relationship between microRNA let-7e and IGF1/IGF1R ().
Figure 5. Negative feedback regulation between microRNA let-7e and IGF1/IGF1R. (a) Let-7 miRNA family is among the predicted miRNA targeting IGF1 using TargetScanHuman 6.2. (b) qRT–PCR analysis of let-7a, let-7b and let-7e in total RNA from HCT-116 colorectal cancer cell line after stimulated with IGF1 (50 ng/ml) for the indicated time. The data shown are average fold changes (the mean ± SD of three independent experiments) of individual miRNA expression relative to vehicle-treated cells. *P < 0.05. (c) Schematic diagram showing that activation of IGF1/IGF1R pathway can inhibit let-7e expression, and on the other hand over-expression of let-7e can inhibit the expression of IGF1 and IGF1R. * indicates a let-7e target predicted by TargetScanHuman 6.2 tool, but not validated experimentally in the present study
Discussion
The type 1 IGF receptor (IGF1R) is a transmembrane tyrosine kinase that is frequently overexpressed in human cancers. IGF1 binding to IGF1R stimulates downstream two main proliferating pathways: the PI3K-Akt pathway and the Ras-MAPK pathway.[Citation3,Citation4] The result of MAPK pathway activation is increased cellular proliferation, whereas activating the PI3K pathway can inhibit apoptosis and stimulate protein synthesis, and thus IGF1R is crucial for tumor transformation and survival of malignant cell. IGF1/IGF1R signaling also plays important roles in hypoxia signalling, vasculogenesis, protease secretion, tumor cell motility and adhesion, and thus can affect cell propensity for invasion and metastasis.[Citation3–Citation5]
Emerging evidence suggests that IGF1/IGF1R pathway play a central role in the development and progression of CRC.[Citation28–Citation31] Oshima T et al. reported that IGF1R expression was correlated with venous invasion and liver metastasis, and thus was a useful predictor of liver metastasis from colorectal cancer.[Citation28] Reinmuth N et al. suggested that the IGFIR plays an important role in multiple mechanisms that mediate the growth, angiogenesis, and metastasis of human colon cancer, and thus is a valid target for the therapy of human colon cancer.[Citation29] It was also reported that amplified IGF-1/IGF1R signaling is not only associated with an increased relative risk for development of CRC, but also contributes to CRC cell survival, invasion, metastasis and resistance to chemotherapeutic drugs.[Citation6,Citation7] In the present study, through IHC analysis, we found that IGF1R was weekly or moderately stained in the well-differentiated CRC tissues, whereas it was moderately or strongly stained in moderately- and poorly-differentiated CRC tissues, indicating elevated expression of IGF1R was paralleled with increased severity of colorectal cancer. Colorectal carcinomas have 10-50 times higher levels of IGF-I and – II compared with adjacent normal colonic mucosa.[Citation30–Citation32] And it is known that the source of the most IGFI is the liver, the most frequent site of metastasis from colorectal cancer. Therefore, IGF1R inhibition may be an effective therapeutic approach against CRC.
MicroRNAs have been shown to be involved in a wide range of biological processes including cell differentiation, proliferation, apoptosis, and motility. Many researchers try to find the key miRNA targeting important oncogenes such as Ras, c-Met, EGFR, VEGFR, c-Myc and so on. Recent studies have found out some miRNA functionally involved in regulation of IGF1R, such as miR-195, miR-7, miR-139, miR-145, miR-375, miR-470, miR-497, miR-669, and miR-681.[Citation19–Citation25] In the present study, using the online miRNA target prediction tool, we found that miRNA let-7, known tumor suppressor, has the highest probability score among the predicted miRNA targeting IGF1R. There were three potential let-7-target sites with higher probability of preferential conservation among the 3ʹUTR of IGF1R. Moreover, IGF1 was also a potential target of let-7 according to the analysis of TargetScan.
Identified as the first mammalian miRNAs, let-7 family members have been widely reported that their expression levels are frequently low and the chromosomal clusters of let-7 are often deleted in many cancers. Let-7 is also a very attractive potential therapeutic target for anticancer drug development.[Citation8–Citation11] In the present study, through real-time RT-RCR, let-7a, let-7b, and let-7e were shown to be significantly down-regulated in CRC tissues compared with adjacent normal tissues. Moreover, ectopic transfection of let-7e led to a significant reduction in IGF1R at protein level and their downstream Akt inhibition, as well as a reduction in cell proliferation, migration and invasion in colorectal cancer cells, while inhibition of let-7e enhanced the expression of IGF1R. These results suggested that let-7e dysregulation may play an important role in the regulation of IGF1R expression and cell proliferation and metastasis in CRC.
Finally, through bioinformatics analysis we found that let-7 miRNA family was also among the predicted miRNA targeting IGF1. Conversely, let-7a, let-7b and let-7e were significantly down-regulated following treatment with IGF1 in HCT-116 colorectal cancer cells. So in the present study, we found a negative feedback regulation relationship between microRNA let-7e and IGF1/IGF1R.
In conclusion, the present study provides new insights into the role of IGF1/IGF1R and miRNA let-7e in colorectal cancer. Our data indicated that elevated expression of IGF1R was paralleled with increased severity of colorectal cancer. Let-7e was down-regulated in colorectal cancer. More importantly, ectopic transfection of let-7e led to a significant reduction in IGF1R at protein level and their downstream Akt inhibition, as well as a reduction in cell proliferation, migration and invasion in colorectal cancer cells. On the other hand, IGF1 stimulation can significantly down-regulate the expression of let-7e in colorectal cancer cells. The illustrated negative feedback regulation mechanisms can provide clues for new therapeutic strategies for colorectal cancer.
Acknowledgments
This work was supported by Technological Research Project for Public Welfare from Science and Technology Department of Zhejiang Province (No. 2016C33225).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Parkin DM, Pisani P, Ferlay J. Estimates of the worldwide incidence of 25 major cancers in 1990. Int J Cancer. 1999;80(6):827.
- Siegel R, DeSantis C, Virgo K, et al. Cancer treatment and survivorship statistics, 2012. CA Cancer J Clin. 2012 Jul-Aug;62(4):220–241. Review.
- Fürstenberger G, Senn HJ. Insulin-like growth factors and cancer. Lancet Oncol. 2002 May;3(5):298–302. Review.
- Baserga R, Peruzzi F, Reiss K. The IGF-1 receptor in cancer biology. Int J Cancer. 2003 Dec 20;107(6):873–877.
- Alberobello AT, D’Esposito V, Marasco D, et al. Selective disruption of insulin-like growth factor-1 (IGF-1) signaling via phosphoinositide-dependent kinase-1 prevents the protective effect of IGF-1 on human cancer cell death. J Biol Chem. 2010 Feb 26;285(9):6563–6572. . Epub 2009 Dec 31.
- Reinmuth N, Liu W, Fan F, et al. Blockade of insulin-like growth factor I receptor function inhibits growth and angiogenesis of colon cancer. Clin Cancer Res. 2002 Oct;8(10):3259–3269.
- Sekharam M, Zhao H, Sun M, et al. Insulin-like growth factor 1 receptor enhances invasion and induces resistance to apoptosis of colon cancer cells through the Akt/Bcl-x(L) pathway. Cancer Res. 2003 Nov 15;63(22):7708–7716.
- Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008 Oct;18(10):505–516. . Epub 2008 Sep 4.
- Pasquinelli AE, Reinhart BJ, Slack F, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000 Nov 2;408(6808):86–89.
- Jérôme T, Laurie P, Louis B, et al. Enjoy the silence: the story of let-7 MicroRNA and cancer. Curr Genomics. 2007 Jun;8(4):229–233.
- Barh D, Malhotra R, Ravi B, et al. MicroRNA let-7: an emerging next-generation cancer therapeutic. Curr Oncol. 2010 Feb;17(1):70–80.
- Esquela-Kerscher A, Trang P, Wiggins JF, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008 Mar 15;7(6):759–764. Epub 2008 Mar 3.
- Wu L, Nguyen LH, Zhou K, et al. Precise let-7 expression levels balance organ regeneration against tumor suppression. Elife. 2015 Oct 7;4:e09431.
- Hikasa H, Sekido Y, Suzuki A. Merlin/NF2-Lin28B-let-7 is a tumor-suppressive pathway that is cell-density dependent and hippo independent. Cell Rep. 2016 Mar 29;14(12):2950–2961.
- Johnson CD, Esquela-Kerscher A, Stefani G, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007 Aug 15;67(16):7713–7722.
- Johnson SM, Grosshans H, Shingara J, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005 Mar 11;120(5):635–647.
- Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007 Mar 16;315(5818):1576–1579. Epub 2007 Feb 22.
- Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008 Aug;14(8):1539–1549. . Epub 2008 Jun 19.
- Wei W, Zhang WY, Bai JB, et al. The NF-κB-modulated microRNAs miR-195 and miR-497 inhibit myoblast proliferation by targeting Igf1r, Insr and cyclin genes. J Cell Sci. 2016 Jan 1;129(1):39–50.
- Kong KL, Kwong DL, Chan TH, et al. MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut. 2012 Jan;61(1):33–42. . Epub 2011 Aug 3.
- Jiang L, Liu X, Chen Z, et al. MicroRNA-7 targets IGF1R (insulin-like growth factor 1 receptor) in tongue squamous cell carcinoma cells. Biochem J. 2010 Nov 15;432(1):199–205. .
- La Rocca G, Badin M, Shi B, et al. Mechanism of growth inhibition by MicroRNA 145: the role of the IGF-I receptor signaling pathway. J Cell Physiol. 2009 Aug;220(2):485–491. .
- Lerman G, Avivi C, Mardoukh C, et al. MiRNA expression in psoriatic skin: reciprocal regulation of hsa-miR-99a and IGF-1R. PLoS One. 2011;6(6):e20916. Epub 2011 Jun 7.
- Guo ST, Jiang CC, Wang GP, et al. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. Oncogene. 2012 Jun 18. [Epub ahead of print]. DOI:10.1038/onc.2012.214
- Shen K, Liang Q, Xu K, et al. MiR-139 inhibits invasion and metastasis of colorectal cancer by targeting the type I insulin-like growth factor receptor. Biochem Pharmacol. 2012 Aug 1;84(3):320–330. . Epub 2012 May 3.
- Kreisberg JI, Malik SN, Prihoda TJ, et al. Phosphorylation of Akt (Ser473) is an excellent predictor of poor clinical outcome in prostate cancer. Cancer Res. 2004;64:5232–5236.
- Jiang J, Lee EJ, Gusev Y, et al. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005 Sep 28;33(17):5394–5403. Print 2005.
- Oberthür R, Seemann H, Gehrig J, et al. Simultaneous inhibition of IGF1R and EGFR enhances the efficacy of standard treatment for colorectal cancer by the impairment of DNA repair and the induction of cell death. Cancer Lett. 2017 Oct 28;407:93–105.
- Sanchez-Lopez E, Flashner-Abramson E, Shalapour S, et al. Targeting colorectal cancer via its microenvironment by inhibiting IGF-1 receptor-insulin receptor substrate and STAT3 signaling. Oncogene. 2016 May 19;35(20):2634–2644.
- Hakam A, Yeatman TJ, Lu L, et al. Expression of insulin-like growth factor-1 receptor in human colorectal cancer. Hum Pathol. 1999 Oct;30(10):1128–1133.
- Freier S, Weiss O, Eran M, et al. Expression of the insulin-like growth factors and their receptors in adenocarcinoma of the colon. Gut. 1999 May;44(5):704–708.
- Michell NP, Langman MJ, Eggo MC. Insulin-like growth factors and their binding proteins in human colonocytes: preferential degradation of insulin-like growth factor binding protein 2 in colonic cancers. Br J Cancer. 1997;76(1):60–66.
