ABSTRACT
Paip2 (Poly(A)-binding protein – interacting protein 2) is a conserved metazoan-specific protein that has been implicated in regulating the translation and stability of mRNAs. However, we have found that Paip2 is not restricted to the cytoplasm but is also found in the nucleus in Drosophila embryos, salivary glands, testes, and tissue culture cells. Nuclear Paip2 is associated with chromatin, and in chromatin immunoprecipitation experiments it maps to the promoter regions of active genes. However, this chromatin association is indirect, as it is RNA-dependent. Thus, Paip2 is one more item in the growing list of translation factors that are recruited to mRNAs co-transcriptionally.
Keywords:
Introduction
Paip2 is a metazoan-specific factor that was first described in human cells as a translational repressor capable of inhibiting cap-dependent translation [Citation1]. Paip2 interacts with cytoplasmic polyA-binding protein (PABP) through two motifs, PAM1 and PAM2, located in the center and in the C-terminal half of protein, respectively [Citation2]. Paip2 is thought to negatively regulate translation via two distinct molecular mechanisms. First, it displaces PABP from the poly(A) tail of mRNA; second, it disrupts PABP–eIF4G interactions that are important in promoting translation [Citation3,Citation4]. Paip2 also has a negative effect on the translation of mRNAs lacking a polyA tail [Citation5].
In addition to downregulating translation, Paip2 also plays a role in mRNA stability. Though Paip2 does not contain any RNA-binding motifs, it binds to the 3ʹ-UTR of VEGF mRNA and stabilizes this messenger RNA. As a consequence, the level of VEGF protein increases [Citation6]. On the other hand, the effects of Paip2 on the stability of Per1 and Per2 mRNAs in the nervous system are exactly the opposite: Paip2 increases the rate of decay of these transcripts [Citation7]. Paip2 also binds to the 3ʹ-UTR of GLUT5 mRNA and modulates its stability in human cells [Citation8]. Thus, Paip2 controls two important aspects of post-transcriptional regulation of gene expression, namely, mRNA stability and translation efficiency.
Paip2 has been implicated in multiple biological processes, including cell proliferation and differentiation [Citation9,Citation10]; synaptic plasticity, learning, and memory formation in the nervous system [Citation11]; and spermatogenesis [Citation12,Citation13]. In humans, there are two Paip2 paralogues, Paip2A and Paip2B, with the latter sharing 59% identity and 80% similarity with the former. The two PABP-binding domains, PAM1 and PAM2, show the highest degree of similarity between the two proteins. Although both paralogues can function as translational repressors, they differ in their pattern of tissue and cell-line distribution. Paip2 is also found in the fruit fly Drosophila. There is a single paip2 gene, and it encodes a 14-kDa acidic protein that is most similar to the human Paip2B paralogue [Citation10]. Like its vertebrate counterparts, the Drosophila protein interacts with PABP and interferes with its ability to bind mRNA. In an S2 tissue culture cell in vitro translation extract, Paip2 represses translation [Citation10].
In the studies reported here, we show that Drosophila Paip2 has an unexpected subcellular distribution. Instead of being restricted to the cytoplasm, as would be expected for a generic translational regulator, Paip2 is found at a high level in the nuclei. Nuclear localization is observed in embryos, salivary glands, testes, and tissue culture cells. ChIP experiments demonstrate that Paip2 is chromatin-associated, mapping primarily to the promoter regions of active genes. However, promoter association appears to be indirect as it is RNA-dependent. Since Paip2 is found to be associated not only with mature mRNA but also with unspliced precursors, it would appear that Paip2 initially associates with nascent RNAs soon after transcription is initiated and then remains bound to the fully processed mRNA when it is transported from the nucleus to the cytoplasm and then translated.
Results
Paip2 is localized both in the cytoplasm and in the nucleus of S2 cells
To characterize Paip2 in Drosophila, we raised polyclonal antibodies against the full-length recombinant protein. As Paip2 is known to be expressed in S2 cells [Citation14], we used our antibodies to probe Western blots of S2 cell extracts. Two closely spaced bands with an apparent mobility of about 25 kDa were detected ()). This value is markedly greater than the predicted molecular weight of the Paip2 protein (14.6 kDa). However, an abnormally low mobility in SDS PAGE has previously been reported for the Drosophila Paip2 protein [Citation10]. Moreover, we found that the recombinant Paip2 produced in E. coli also had an abnormal electrophoretic mobility (Suppl. ).
Figure 1. Paip2 is detected in the nuclei of S2 cells. (a) Western blot analysis of total protein extract from S2 cells with anti-Paip2 antibodies. The endogenous Paip2 protein is detected as a closely spaced doublet of about 25 kDa. (b) Immunoprecipitation of S2 cell extract (In) with anti-Paip2 and anti-PABP antibodies and preimmune serum (mock). (c) Knockdown of Paip2 in S2 cells led to specific decrease in the intensity of both bands in the doublet detected by Paip2 antibodies. Tubulin was used as loading control. Folds of cell lysate dilution are indicated above the lanes. In control experiment, dsRNA corresponding to the GFP coding sequence was used. (d) Immunostaining of S2 cells (confocal imaging) shows Paip2 localization both in the nucleus and in the cytoplasm. Lamin marks the nuclear envelope. (e) Immunostaining of S2 cells (confocal imaging) shows Paip2 localization versus cytoplasmic localization of translation factor eIF3-S8. (f) Western blot analysis of the lysate of cells expressing FLAG-tagged Paip2 with anti-FLAG and anti-Paip2 antibodies. Note that, unlike with endogenous Paip2, only a single band is observed for the FLAG-tagged Paip2. (g) Immunostaining of S2 cells expressing Paip2-FLAG with FLAG antibodies. Localization of recombinant protein is similar to that of Paip2 detected with Paip2 antibodies in (d).
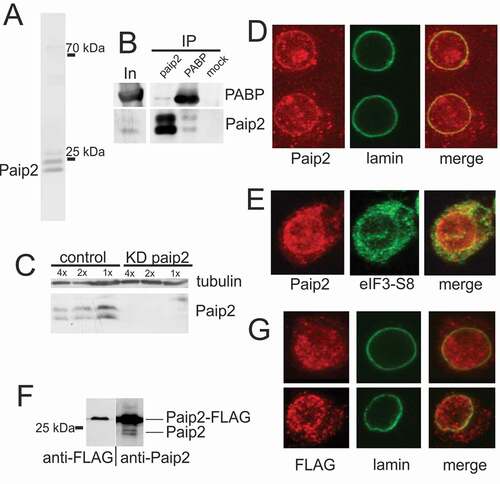
To further confirm the specificity of our antibody, we used two other approaches. First, we performed Western blotting in the presence of excess competing recombinant Paip2 protein and found that the intensity of the Paip2 doublet dropped under such conditions (Suppl. ). Second, we used RNAi to knock down Paip2 expression in tissue culture cells. Treatment of S2 cells with dsRNA against the paip2 coding region resulted in a specific decrease in both the paip2 transcript (100-fold decrease, data not shown) and in the two Paip2 protein isoforms ()). As for the presence of a doublet, this could be due to some type of posttranslational modification, e.g. phosphorylation, as has been suggested for other PAM2-containing proteins [Citation15]. Alternatively, it could be due to proteolytic processing. In IP experiments, our antibodies efficiently precipitated both protein isoforms ()). Antibodies against PABP also co-precipitated both forms of Paip2, while Paip2 antibodies precipitated PABP.
An unexpected result came from analyzing the intracellular localization of Paip2 in S2 cells. Instead of being localized only in the cytoplasm, Paip2 was also detected in the nucleus (), Suppl. ). The results of immunostaining suggest that Paip2 exists in three forms within the cell: (1) diffused in the cytoplasm, (2) concentrated at the nuclear periphery, and (3) localized in multiple bright foci both in the cytoplasm and in the nucleus. Staining of S2 cells with a cytoplasmic marker – translation initiation factor eIF3-S8 – confirmed predominant nuclear localization of Paip2 (), Suppl. ). Localization of Paip2 was found to significantly overlap with TO-PRO-3 and DAPI staining of DNA in the cells (Suppl. , ).
Figure 2. Nuclear Paip2 in Drosophila testis and embryo.(a) Immunostaining of testis with anti-Paip2 (red) and anti-lamin (green) antibodies and counterstaining with DAPI (blue) (confocal imaging), with a merged image shown below: (1) hub and zone of mitotic divisions, (2) zone of primary spermatocytes, (3) zone of meiotic divisions, (4) elongating spermatids (5) seminal vesicle. (b) Immunostaining of the tip of testis (hub and zone of primary spermatocytes) with antibodies against Paip2, FLAG, and lamin (confocal imaging) in wild-type flies (left column) and fly stocks expressing FLAG-Paip2 (central column) and Paip2-FLAG (right column). (c) Immunostaining of an early embryo (stage 3) with antibodies against Paip2 and lamin (confocal imaging). The right column shows images at higher magnification. (d) Immunostaining of the posterior end of an embryo at stage 4–5 (blastoderm) with antibodies against Paip2 and lamin (confocal imaging): SN, somatic nuclei; PGCN, primordial germ cells nuclei.
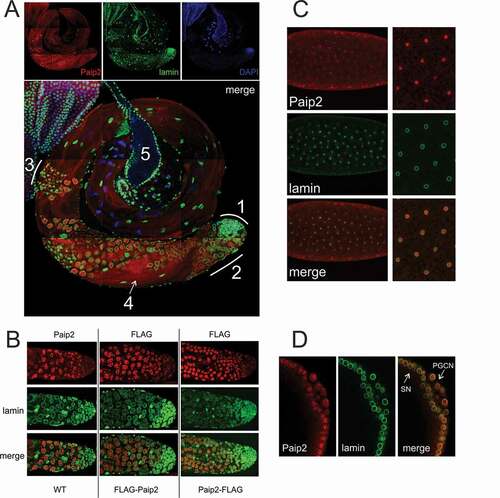
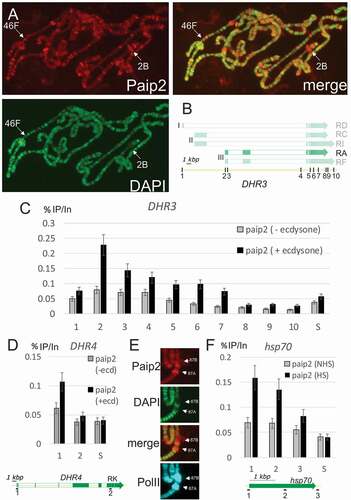
Figure 3. Paip2 is localized to active chromatin.(a) Immunostaining of polytene chromosomes with antibodies against Paip2 and counterstaining with DAPI; a merged image is on the right. Paip2 is detected mainly in DAPI interbands. Loci 46F and 2B are indicated that contain the DHR3 and DHR4 genes, respectively. (b) Scheme of the DHR3 locus and its different transcripts (the ecdysone-inducible RA transcript is highlighted). Roman numerals (I, II, III) indicate three promoters of the gene. Points indicated by Arabic numerals are as follows: (1) RD promoter, (2) RA promoter, (3) 5ʹ-UTR of RA, (4) intron, (5, 6) splice sites, (7, 8) 3ʹ-UTR, (9, 10) polyA signals. (c) ChIP analysis of Paip2 level at different points along the DHR3 gene in untreated cells (grey bars) and cells treated with ecdysone (black bars). The results of ChIP are shown as the percentage of input; S, which corresponds to intergenic spacer which is not transcribed. (d) ChIP analysis of Paip2 level along the DHR4 gene in untreated cells (grey bars) and cells treated with ecdysone (black bars). The structure of the inducible RK transcript is shown. The promoter (point 1) and distal region (point 2) were taken for analysis. (e) Immunostaining of puffs 87A and 87B on polytene chromosomes induced by heat shock. Images show localization of Paip2, DAPI, Paip2 + DAPI (merged), and Pol II. (f) ChIP analysis of Paip2 levels at different sites in the hsp70 transcription. Cells grown at 25°C (grey bars) and heat-shocked cells (black bars) were used for analysis. A scheme of the hsp70 gene is given below, with numerals indicating the locations of primers for ChIP in (1) the promoter, (2) transcribed region, and (3) polyA signal.
Since nuclear localization was not expected, we performed two other experiments to confirm that Paip2 actually localizes to the nucleus. In the first, we stained S2 tissue culture cells with Paip2 antibody in the presence of excess recombinant Paip2. As can be seen in Suppl. , the fluorescent signal was substantially diminished in the presence of the recombinant protein. In the second, we expressed a Paip2 protein with an N-terminal 3×FLAG epitope in S2 cells. As shown in ) and Suppl. , the subcellular distribution of 3×FLAG-Paip2 was similar to that of the endogenous Paip2 protein detected with our rabbit antibody. As might be expected, the 3×FLAG-tagged Paip2 protein migrated more slowly in SDS-PAGE than either of the endogenous protein species, but only a single isoform (instead of two) was detected. There are several possible explanations for this difference (proteolytic processing of the N-terminal tag, post-translational modification), which we are currently evaluating.
Paip2 is detected in the nuclei of several cell types
The unexpected localization pattern of the Paip2 protein in S2 cells raised the question as to whether this subcellular distribution was a peculiarity of these cultured cells, or it would also be observed in the cells of normal tissues. According to FlyBase [Citation14], the levels of paip2 mRNA expression in adult flies are highest in the testes. Immunostaining of the testes revealed a dynamic pattern of Paip2 protein expression. There is little or no Paip2 protein in the germline stem cells at the apical tip of the testes (1 in )), while only relatively low levels are detected in the mitotic cysts. Paip2 expression is substantially upregulated in the primary spermatocyte (2 in )). During this phase of development, many of the gene products needed to support subsequent sperm differentiation are synthesized. The cells in the spermatocyte cysts have large nuclei with mostly decondensed chromatin, which is indicative of high transcriptional activity [Citation16–Citation18]. Paip2 was detected in the nuclei of these cells. At later stages of spermatogenesis, Paip2 was detected in the zone of meiotic divisions and spermatid differentiation (3 in )), where it formed multiple speckles in the cytoplasm. Transcription at this stage is almost shut off in Drosophila [Citation17]. Finally, Paip2 is found in the tails of the elongating spermatids (4 in )).
To confirm the nuclear localization of Paip2, we generated two transgenic fly stocks expressing FLAG-tagged Paip2, with a 3×FLAG epitope introduced at either N- or C-terminus of the protein. The construct carrying the fusion gene under endogenous promoter was integrated into the genome. Staining with anti-FLAG antibody showed that localization pattern of both fusion variants, FLAG-Paip2 and Paip2-FLAG, was similar to that of the endogenous protein: in all three cases, Paip2 was localized to the nuclei of primary spermatocytes ()). Finally, we co-stained the testes with antibodies against Orb2 [Citation19], a cytoplasmic translation factor. As shown in Suppl. , the labeling patterns proved to be almost mutually exclusive: staining for Orb2 was observed in the cytoplasm, while Paip2 was predominantly localized to the nucleus.
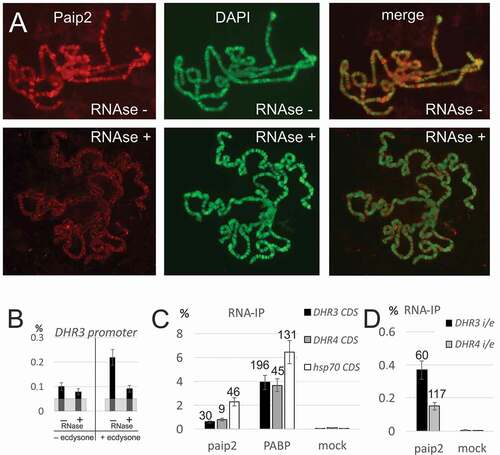
Figure 4. Paip2 is associated with RNA in the nucleus.(a) Immunostaining of polytene chromosomes with antibodies against Paip2 and DAPI counterstaining without and after RNase treatment. (b) ChIP analysis of Paip2 levels in the DHR3 promoter region before and after induction by ecdysone and without and with RNase treatment. Designations are as in . (c, d) RNA immunoprecipitation (RIP) from lysate of S2 cells with antibodies against Paip2 and PABP and with empty beads (mock). The cells were either treated with ecdysone (DHR3 and DHR4 transcripts) or heat shock (hsp70 transcript). Primers for detecting transcripts were located in one exon within the coding region of the corresponding gene (C) or on both sides of the intron/exon boundary of the gene in order to detect nascent transcripts (D). RIP signals are given as percentage of input, enrichment of RNA relative to corresponding mock is indicated above the bars.
Since paip2 is also expressed during embryogenesis [Citation20], we examined the subcellular localization of the Paip2 protein during this phase of development. At the syncytial blastoderm stage, Paip2 had a scattered, diffuse distribution in the cytoplasm but could also be detected in the nuclei ()). In the period just prior to cellularization, at the time when zygotic transcription is being activated [Citation21], we observed high levels of Paip2 in the somatic nuclei [Citation21]. Interestingly, Paip2 was also found in the nuclei of newly formed primordial germ cells (PGCs) ()). Unlike the surrounding soma, PGCs are transcriptionally quiescent. Consequently, it would appear that Paip2 localization in the nuclei at the blastoderm stage is not absolutely dependent upon transcription.
Paip2 binds to chromatin and is recruited onto promoters of active genes
While Paip2 accumulation in the PGC nuclei at the blastoderm stage indicates that high levels of transcription are not needed for nuclear localization, it nevertheless seemed possible that Paip2 might associate with components of the transcriptional or RNA processing machinery. This could potentially explain the bright foci observed in S2 cell nuclei. To further evaluate this possibility, we probed salivary gland polytene chromosomes with Paip2 antibody. ) shows that Paip2 localizes to interbands and puffs on all chromosomal arms [Citation22]. These sites are decondensed and stain only weakly with DAPI. They are also sites of active transcription. For example, the ecdysone inducible gene, DHR4, is located near the tip of X chromosome at 2B. It forms a large puff in polytene chromosomes and is one of the most prominent sites of Paip2 accumulation. It is also decondensed and is barely detectable by DAPI staining ()). In contrast, regions of the chromosome that are condensed and stain strongly with DAPI (e.g. the tip of the X chromosome) contain only little Paip2 protein. Thus, the localization of Paip2 may reflect its association with active genes.
One of polytene chromosome puffs bound by Paip2 is the 46F locus ()). It contains the DHR3 early-late gene of the ecdysone regulatory cascade [Citation23]. This cascade is a convenient model for experiments in S2 cells [Citation24]. Therefore, we used chromatin immunoprecipitation (ChIP) to determine whether Paip2 localizes to specific regions in the DHR3 gene. As indicated in ), the DHR3 gene spans a DNA segment of nearly 40 kb and has three promoters (I, II, and III). The use of these different promoters, together with alternative splicing, is predicted to generate multiple transcripts. Among them, only the RA transcript generated from promoter III is induced by ecdysone in S2 cells [Citation24]. The two other promoters, I and II, are not activated by ecdysone induction in S2. We induced DHR3 expression in S2 cells by hormone treatment (Suppl. Fig. 5A) and then assayed for Paip2 association at ten sites within the DHR3 gene. These sites include the most distal promoter I (1), the promoter III for the RA transcript (2,3), a sequence in the intron immediately upstream of the main protein-coding exons (4), several sites within the main protein coding exons (5–7), the 3ʹ UTR and polyadenylation region (8–10) for the RA transcript and downstream of the RA transcript. Unexpectedly, Paip2 proved to be associated mainly with sequences close to promoter (III) for the RA transcript ()), while its association with chromatin in the region of the DHR3 gene encoding the RA 3ʹ UTR and polyA was negligible.
To provide further evidence that Paip2 localizes to the promoter region, we used Paip2 RNAi knockdown. After this treatment, Paip2 levels in the promoter regions dropped to background (Suppl. Fig. 5C). As another control, we used antibodies against cytoplasmic translation factor eIF3-S8 for ChIP experiments. This protein showed a background signal throughout the gene (Suppl. Fig. 5B). Finally, to rule out a potential influence of differential chromatin accessibility, we examined the distribution of Pol II containing the CTD domain elongation marker, phosphorylated Serine 2 [Citation25]. As expected, its amount proved to be higher in the gene body than on the promoter (Suppl. Fig. 5B).
We also used ChIP to examine the association of Paip2 with the X-linked DHR4 gene. The results were similar: upon its induction (Suppl. Fig. 5A), Paip2 was associated with sequences close to the DHR4 promoter, while at a promoter-distal site the level of Paip2 was at or near background ().
To confirm that Paip2 associates with promoter regions of active genes, we examined the hsp70 heat shock genes. The images in ) show that Paip2 accumulated at the 87A and 87C heat shock puffs after heat induction. In ChIP experiments, Paip2 was found to be predominantly associated with the promoter region of the hsp70 gene ()), while lower levels of signal were observed in the middle of the gene and close to the polyA addition signal. Moreover, the extent of promoter association was found to depend on transcriptional induction by heat shock (Suppl. Fig. 5A).
Paip2 is associated with nuclear mRNA
Since Paip2 in the cytoplasm is associated with mRNA, we treated polytene chromosomes with RNase to determine whether Paip2 recruitment depends on RNA. This treatment led to a dramatic drop in the intensity and sharpness of the Paip2 polytene banding pattern, while the bands evident in DAPI staining remained largely untouched ()). The behavior of Paip2 in this experiment resembled that of Thoc5, a component of the mRNP biogenesis factor THO [Citation26], and was opposite to the behavior of DNA-binding Pol II (Suppl. Fig. 6).
These findings indicate that the global association of Paip2 with polytene chromosomes is mediated by RNA and suggest a model to explain the ChIP results. In this model, Paip2 would be loaded onto nascent transcripts in the promoter regions of genes coincident with the initial steps in transcription. As a consequence, the signal for Paip2 in the promoter region would be quite high. While Paip2 would remain associated with sequences near the 5ʹ end of the nascent transcript as transcription proceeds, the efficiency of crosslinking to chromatin (and thus to the gene sequence) would gradually decrease as the transcript becomes longer and longer.
One prediction of this model is that Paip2 association with promoter regions in ChIP should also be RNA-dependent. ) shows that this is the case. RNase treatment has only a slight effect on the level of Paip2 associated with the promoter of the uninduced DHR3 gene, whereas RNase treatments causes a significant reduction on Paip2 association with the promoter in ecdysone-induced cells.
Another prediction of the model is that Paip2 will be associated with nascent transcripts. To address this question, we performed RNA immunoprecipitation (RNA-IP) with antibodies against Paip2 and PABP (as control). We first confirmed that mature mRNA species can be immunoprecipitated with Paip2 antibodies using primers specific for exon sequences ()). We then used primers that amplify RNA sequences spanning intron–exon junctions to detect Paip2 association with nascent RNAs. As shown in ), Paip2 antibodies immunoprecipitated nascent DHR3 and DHR4 transcripts.
Discussion
The synthesis and functioning of mRNA in the cell are regulated by a number of proteins, which are dynamically loaded onto RNA at specific time points of its life cycle [Citation27]. However, the composition of these RNA–protein complexes, the steps in their assembly, and the functions of these RNA-associated proteins are still poorly understood [Citation28]. Here we show that the Paip2 protein, previously implicated in regulating translation and mRNA stability in the cytoplasm, is also localized in the nucleus. Moreover, our data suggest that this factor is actually loaded onto its mRNA targets at an early transcription step.
We detected Paip2 in the nuclei of several cell types and at different stages of development: in Schneider S2 cells, primary spermatocytes, syncytial and blastoderm-stage somatic cells and PGC, and salivary gland cells. In the salivary glands, Paip2 is found to be associated with polytene chromosomes, and it exhibits a quite striking distribution pattern. Paip2 is localized to interbands and puffs but is largely excluded from bands. Interbands and puffs represent decondensed regions containing transcriptionally active genes, while bands are composed of densely packed chromatin and are largely transcriptionally quiescent. The correlation between Paip2 localization in polytene chromosomes and actively transcribed genes is supported by the results of both ChIP experiments and RNA-IP. ChIP experiments show that Paip2 is selectively enriched near promoter sequences of actively transcribed genes. Similarly, RNA-IP experiments show that Paip2 is associated not only with mature mRNAs but also with nascent transcripts.
During spermatogenesis, Paip2 is localized in the cell nucleus at the stage when transcriptional activity is the highest but shows cytoplasmic speckle-like localization at later stages, when transcription is largely turned off. This is in line with the general pathway of mRNA biogenesis during spermatogenesis in Drosophila, in which transcripts are synthesized in primary spermatocytes and then stored in the cytoplasm until required [Citation17]. These changes in Paip2 localization during spermatogenesis can be explained by its loading onto mRNA at the time of synthesis.
While the nuclear localization of Drosophila Paip2 or of its mammalian relatives has not been described previously, there is evidence that they shuttle between the cytoplasm and the nucleus and are possibly involved in mRNA export [Citation6]. Relevant papers published to date are focused on the mammalian Paip2A paralogue, while Paip2B and closely related Drosophila Paip2 can have a distinct mode of functioning. However, a recently developed antibody against Paip2A (RRID: AB_2645179, see Materials) reveals both nuclear and cytoplasmic localization of this protein in HeLa cells. Since our results suggest that the ratio of nuclear/cytoplasmic Paip2 may differ depending on tissue and cell types, it would be of interest to investigate the subcellular distribution of the two mammalian Paip2 proteins in more detail.
Although Paip2 does not carry a conventional NLS, it is possible that it is imported into the nucleus in complex with other factors. It is noteworthy that PABP in mammals also lacks an NLS and shows dual nuclear and cytoplasmic localization [Citation29,Citation30]. The functions of nuclear PABP are still not fully known [Citation31]. Its function as a coregulator of the androgen receptor has been described [Citation32], while our unpublished studies (Graham and Schedl) implicate it, along with another translation factor (eIF4E), in the alternative splicing of Sex-lethal pre-mRNAs in female flies [Citation33]. To date, several other translational regulators have been found in the nucleus. One of the eEF1A isoforms is important for the recruitment of the transcription factor HSF1 to the hsp70 promoter and its activation in human cells [Citation34]. The translation initiation factor eIF4E has a role in mRNA export from the nucleus [Citation35]. Translation factor eIF4G has been found in the nuclei of human cells and shown to interact with the nuclear cap-binding complex [Citation36]. Nuclear eIF4H has been described in Chironomus [Citation37]. Nuclear translation-like factor eIF4AIII is required for splicing and nonsense-mediated decay in human cells [Citation38].
Our data indicate that Paip2 association with sites of active transcription in polytene chromosomes is RNA-dependent. Moreover, since the highest levels of Paip2 detected in ChIP experiments are located close to the promoters of active genes, it is likely that it loads onto the nascent transcripts at or shortly after transcription initiation. However, it appears that, instead of moving along with the transcriptional apparatus, it remains associated with RNA sequences near the 5ʹ end of the nascent transcript. This model can explain why the sequences near the 3ʹ end of the transcription unit are recovered only in low yield in Paip2 ChIP experiments.
A number of important questions remain open. One is how Paip2 gets targeted to the 5ʹ end of the nascent transcript, and how this is coupled to the processes of initiation and/or elongation. Another question is whether there is any specificity in Paip2–promoter association. ChIP data confirm the presence of Paip2 in the promoter regions of all the genes that we have tested; however, it is possible that there are genes in which its association with the promoter is minimal. The growing amount of evidence indicates a role for promoter-dependent imprinting in mRNP composition, which subsequently has an effect on mRNA cytoplasmic fate and translation efficiency [Citation39–Citation42]. Metazoan-specific factor Paip2 has been previously shown to have tissue-specific functions in neurons and spermatids. Promoter-driven loading of Paip2 could be a way to mark specific transcripts involved in RNA-depending tissue-specific events, e.g. cell polarization [Citation43], neuroplasticity [Citation44], etc. Another possibility, not excluding the aforementioned one, is the involvement of Paip2 in some event(s) at the promoter. Indeed, there is increasing evidence that RNA-related activities are important for processes occurring on chromatin templates [Citation45,Citation46]. For example, RNA-binding activity has been recently demonstrated for the DSIF (Spt4/Spt5) factor of RNA polymerase II pausing [Citation47].
Material and methods
Ethics statement
Animal handling for the antibody production was carried out strictly according to the procedures outlined in the NIH (USA) Guide for the Care and Use of Laboratory Animals. The protocols used were approved by the Committee on Bioethics of the Institute of Gene Biology, Russian Academy of Sciences. All procedures were performed under the supervision of a licensed veterinarian, under conditions that minimize pain and distress. Rabbits were purchased from a licensed specialized nursery, Manihino. Soviet chinchilla rabbits used in the study are not endangered or protected. Only healthy rabbits, certified by a licensed veterinarian, were used. The rabbits were individually housed in standard size, stainless steel rabbit cages and provided ad libitum access to alfalfa hay, commercial rabbit food pellets, and water. The appetite and behavior of each rabbit was monitored daily by a licensed veterinarian. The body weight and temperature of each rabbit were monitored prior to and daily following immunization. No animals became ill or died at any time prior to the experimental endpoint. At the end of the study period all rabbits were euthanized by intravenous injection of barbiturate anesthetics.
Fly stocks
Cultivation of flies and genetic crosses were described previously [Citation48]. The pCasper3 vector was used for transgene construction. A 6110-bp fragment containing the Paip2 gene (12,980,357 to 12,986,467 bp fragment of 3R chromosome) was used, with a 3×FLAG epitope sequence introduced either downstream of the initiation codon or immediately upstream of the stop codon. DNA was injected into y1,w1 embryos, and transgenic flies were obtained according to published protocols [Citation49].
Experiments with S2 cell culture
Drosophila Schneider cell line 2 (S2) was maintained in SFX medium (HyClone) at 25°C. DNA fragments encoding Paip2 with the 3×FLAG epitope were cloned into the pAc5.1/V5-HisB vector (Invitrogen) and used for transfection with the Lipofectamine 3000 reagent. RNAi experiments followed published procedures [Citation50]. Treatment with dsRNA (20 μg per million cells) continued for 5 days. In control experiment, dsRNA corresponding to a GFP coding sequence was used. For ecdysone cascade activation, cells were treated with 1 μM ecdysone overnight. Heat shock was induced by incubation at 37°C for 30 min.
Antibodies
Antibodies against whole-length Paip2 and eIF3-S8 (399–553 aa of isoform PA) were produced in our laboratory (Suppl. ). Several animals were used for immunization, specificity of all of antibodies was the same. Other antibodies were against PABP [Citation51], thoc5 [Citation26], Pol II (Abcam ab817, clone 8WG16), Ser2phospho Pol II (Abcam ab5095), and FLAG (Sigma F1804, clone M2). Antibodies E7 against beta-tubulin (developed by M. Klymkowsky), ADL67.10 against lamin (developed by P.A. Fisher), and Orb2-4G8 against Orb2 (developed by P. Schedl) were obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at the Department of Biology, University of Iowa, Iowa City, IA 52,242. Information about antibody Thermo Fisher Scientific Cat# PA5-63,347 RRID: AB_2645179 is available at https://www.thermofisher.com/antibody/product/PAIP2-Antibody-Polyclonal/PA5-63347.
Chip, RIP and Quantitative (q) PCR Analysis
The protocol for ChIP with S2 cells was described previously [Citation52]. Measurements in intergenic regions and ChIP with antibody-free Sepharose beads were used as negative controls in each experiment. RNA-IP was performed as described [Citation53] using antibody-free Sepharose as a negative control. The sequences of the primers are given in the Supplement. Each experiment was performed in three replicates.
Immunostaining
Previously described protocols were used for immunostaining of S2 cells [Citation54], testes [Citation55], embryos [Citation56], and polytene chromosomes [Citation57] with the corresponding secondary antibodies (Molecular Probes). Polytene chromosomes were fixed with 4% FA for 2 min. The salivary glands were permeabilized with 0.5% NP40 (3 min) and treated with RNase A in PBS (500 μg/mL) for 10 min [Citation25]. Heat shock of the salivary glands was induced as described [Citation52].
Western blotting and immunoprecipitation
Antibodies used in WB were diluted 1:500. To extract proteins, S2 cells were lysed in 10 mM HEPES (pH 7.9) containing 5 mM MgCl2, 0.5% Nonidet P-40, 0.45 M NaCl, 1 mM DTT, and complete protease inhibitor mixture (Roche). IP was performed as described [Citation58]. DNase I (Thermo Scientific, 1 U/μL) and RNase A (Thermo Scientific, 10 μg/μL) were added to IP buffer. The Sepharose pellet was washed with buffer containing 300 mM NaCl.
Supplemental Material
Download Zip (18.9 MB)Acknowledgments
We are grateful to L.V. Olenina and A.A. Kotov for their help with staining of the testes, M. Kurshakova for providing plasmid for Paip2 expression, Matthias Hentze (EMBL Heidelberg) for anti-PABP antibodies, and N.A. Gorgolyuk for his help in preparing the manuscript. This study was performed using the infrastructure of the Centre for Collective Use “Biology of the Living Cell and Drug Biomedical Nanotransporters” of the Institute of Gene Biology, Russian Academy of Sciences.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Khaleghpour K, Svitkin YV, Craig AW, et al. Translational repression by a novel partner of human poly(A) binding protein. Paip2 Mol Cell. 2001;7(1):205–216. PMID: 11172725.
- Kozlov G, Trempe JF, Khaleghpour K, et al. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci U S A. 2001;98(8):4409–4413. PMID: 11287632.
- Karim MM, Svitkin YV, Kahvejian A, et al. A mechanism of translational repression by competition of Paip2 with eIF4G for poly(A) binding protein (PABP) binding. Proc Natl Acad Sci U S A. 2006;103(25):9494–9499. PMID: 16772376.
- Khaleghpour K, Kahvejian A, De Crescenzo G, et al. Dual interactions of the translational repressor Paip2 with poly(A) binding protein. Mol Cell Biol. 2001;21(15):5200–5213. PMID: 11438674.
- Polacek C, Friebe P, Harris E. Poly(A)-binding protein binds to the non-polyadenylated 3ʹ untranslated region of dengue virus and modulates translation efficiency. PMID: 19218215 J Gen Virol. 2009;90Pt3:687–692.
- Onesto C, Berra E, Grepin R, et al. Poly(A)-binding protein-interacting protein 2, a strong regulator of vascular endothelial growth factor mRNA. J Biol Chem. 2004;279(33):34217–34226. PMID: 15175342.
- Alvarez-Saavedra M, Antoun G, Yanagiya A, et al. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum Mol Genet. 2011;20(4):731–751. PMID: 21118894.
- Gouyon F, Onesto C, Dalet V, et al. Fructose modulates GLUT5 mRNA stability in differentiated Caco-2 cells: role of cAMP-signalling pathway and PABP (polyadenylated-binding protein)-interacting protein (Paip) 2. Biochem J. 2003;375(Pt1):167–174. PMID: 12820898.
- Rosenfeld AB. Suppression of cellular transformation by poly (A) binding protein interacting protein 2 (Paip2). PMID: 21957478 PLoS One. 2011;69:e25116.
- Roy G, Miron M, Khaleghpour K, et al. The Drosophila poly(A) binding protein-interacting protein, dPaip2, is a novel effector of cell growth. Mol Cell Biol. 2004;24(3):1143–1154. PMID: 14729960.
- Khoutorsky A, Yanagiya A, Gkogkas CG, et al. Control of synaptic plasticity and memory via suppression of poly(A)-binding protein. Neuron. 2013;78(2):298–311. PMID: 23622065.
- Delbes G, Yanagiya A, Sonenberg N, et al. PABP interacting protein 2A (PAIP2A) regulates specific key proteins during spermiogenesis in the mouse. Biol Reprod. 2012;86(3):95. PMID: 22190698.
- Yanagiya A, Delbes G, Svitkin YV, et al. The poly(A)-binding protein partner Paip2a controls translation during late spermiogenesis in mice. J Clin Invest. 2010;120(9):3389–3400. PMID: 20739757.
- Attrill H, Falls K, Goodman JL, et al. FlyBase: establishing a gene group resource for Drosophila melanogaster. Nucleic Acids Res. 2016;44(D1):D786–92. PMID: 26467478.
- Huang KL, Chadee AB, Chen CY, et al. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA. 2013;19(3):295–305. PMID: 23340509.
- Kibanov MV, Gvozdev VA, Olenina LV. Germ granules in spermatogenesis of Drosophila: evidences of contribution to the piRNA silencing. PMID: 22808315 Commun Integr Biol. 2012;52:130–133.
- White-Cooper H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. PMID: 19755484 Reproduction. 2010;1391:11–21.
- White-Cooper H, Davidson I. Unique aspects of transcription regulation in male germ cells. Cold Spring Harb Perspect Biol. 2011;3(7). PMID: 21555408. DOI: 10.1101/cshperspect.a002626
- Xu S, Hafer N, Agunwamba B, et al. The CPEB protein Orb2 has multiple functions during spermatogenesis in Drosophila melanogaster. PLoS Genet. 2012;8(11):e1003079. PMID: 23209437.
- Graveley BR, Brooks AN, Carlson JW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471(7339):473–479. PMID: 21179090.
- Pritchard DK, Schubiger G. Activation of transcription in Drosophila embryos is a gradual process mediated by the nucleocytoplasmic ratio. Genes Dev. 1996;10(9): 1131–1142. PMID: 8654928.
- Armstrong JA, Papoulas O, Daubresse G, et al. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 2002;21(19):5245–5254. PMID: 12356740.
- Carney GE, Wade AA, Sapra R, et al. DHR3, an ecdysone-inducible early-late gene encoding a Drosophila nuclear receptor, is required for embryogenesis. Proc Natl Acad Sci U S A. 1997;94(22):12024–12029. PMID: 9342356.
- Mazina MY, Nikolenko JV, Fursova NA, et al. Early-late genes of the ecdysone cascade as models for transcriptional studies. Cell Cycle. 2015;14(22):3593–3601. PMID: 26506480.
- Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26(19):2119–2137.
- Kopytova DV, Orlova AV, Krasnov AN, et al. Multifunctional factor ENY2 is associated with the THO complex and promotes its recruitment onto nascent mRNA. Genes Dev. 2010;24(1):86–96. PMID: 20048002.
- Mitchell SF, Parker R. Principles and properties of eukaryotic mRNPs. PMID: 24856220 Mol Cell. 2014;544:547–558.
- Oeffinger M, Montpetit B. Emerging properties of nuclear RNP biogenesis and export. Curr Opin Cell Biol. 2015;34:46–53. PMID: 25938908.
- Afonina E, Stauber R, Pavlakis GN. The human poly(A)-binding protein 1 shuttles between the nucleus and the cytoplasm. J Biol Chem. 1998;273(21): 13015–13021. PMID: 9582337.
- Gray NK, Hrabalkova L, Scanlon JP, et al. Poly(A)-binding proteins and mRNA localization: who rules the roost? Biochem Soc Trans. 2015;43(6):1277–1284. PMID: 26614673.
- Hosoda N, Lejeune F, Maquat LE. Evidence that poly(A) binding protein C1 binds nuclear pre-mRNA poly(A) tails. PMID: 16581783 Mol Cell Biol. 2006;268:3085–3097.
- Eisermann K, Dar JA, Dong J, et al. Poly (A) binding protein cytoplasmic 1 is a novel co-regulator of the androgen receptor. PLoS One. 2015;10(7):e0128495. PMID: 26176602.
- Graham PL, Yanowitz JL, Penn JK, et al. The translation initiation factor eIF4E regulates the sex-specific expression of the master switch gene Sxl in Drosophila melanogaster. PLoS Genet. 2011;7(7):e1002185. PMID: 21829374.
- Vera M, Pani B, Griffiths LA, et al. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. eLife. 2014;(3):e03164. PMID: 25233275. DOI:10.7554/eLife.03164
- Volpon L, Culjkovic-Kraljacic B, Sohn HS, et al. A biochemical framework for eIF4E-dependent mRNA export and nuclear recycling of the export machinery. RNA. 2017;23(6):927–937. PMID: 28325843.
- McKendrick L, Thompson E, Ferreira J, et al. Interaction of eukaryotic translation initiation factor 4G with the nuclear cap-binding complex provides a link between nuclear and cytoplasmic functions of the m(7) guanosine cap. Mol Cell Biol. 2001;21(11):3632–3641. PMID: 11340157.
- Bjork P, Bauren G, Gelius B, et al. The Chironomus tentans translation initiation factor eIF4H is present in the nucleus but does not bind to mRNA until the mRNA reaches the cytoplasmic perinuclear region. J Cell Sci. 2003;116(Pt22):4521–4532. PMID: 14576346.
- Ferraiuolo MA, Lee CS, Ler LW, et al. A nuclear translation-like factor eIF4AIII is recruited to the mRNA during splicing and functions in nonsense-mediated decay. Proc Natl Acad Sci U S A. 2004;101(12):4118–4123. PMID: 15024115.
- Ainaoui N, Hantelys F, Renaud-Gabardos E, et al. Promoter-dependent translation controlled by p54nrb and hnRNPM during myoblast differentiation. PloS One. 2015;10(9):e0136466. PMID: 26332123.
- Slobodin B, Han R, Calderone V, et al. Transcription impacts the efficiency of mRNA translation via co-transcriptional N6-adenosine methylation. Cell. 2017;169(2):326–337e12. PMID: 28388414.
- Trcek T, Larson DR, Moldon A, et al. Single-molecule mRNA decay measurements reveal promoter- regulated mRNA stability in yeast. Cell. 2011;147(7):1484–1497. PMID: 22196726.
- Zid BM, O’Shea EK. Promoter sequences direct cytoplasmic localization and translation of mRNAs during starvation in yeast. PMID: 25119046 Nature. 2014;5147520:117–121.
- Barr J, Yakovlev KV, Shidlovskii Y, et al. Establishing and maintaining cell polarity with mRNA localization in Drosophila. BioEssays. 2016;38(3):244–253. PMID: 26773560.
- Zappulo A, van den Bruck D, Ciolli Mattioli C, et al. RNA localization is a key determinant of neurite-enriched proteome. Nat Commun. 2017;8(1):583. PMID: 28928394.
- Hendrickson D, Kelley DR, Tenen D, et al. Widespread RNA binding by chromatin-associated proteins. Genome Biol. 2016;17:28. PMID: 26883116.
- He C, Sidoli S, Warneford-Thomson R, et al. High-resolution mapping of RNA-binding regions in the nuclear proteome of embryonic stem cells. Mol Cell. 2016;64(2):416–430. PMID: 27768875.
- Blythe AJ, Yazar-Klosinski B, Webster MW, et al. The yeast transcription elongation factor Spt4/5 is a sequence-specific RNA binding protein. Protein Sci. 2016;25(9):1710–1721. PMID: 27376968.
- Georgiev PG, Gerasimova TI. Novel genes influencing the expression of the yellow locus and mdg4 (gypsy) in Drosophila melanogaster. Mol Gen Genet. 1989;220(1): 121–126. PMID: 2558282.
- Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218(4570): 348–353. PMID: 6289436.
- Clemens JC, Worby CA, Simonson-Leff N, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97(12):6499–6503. PMID: 10823906.
- Duncan KE, Strein C, Hentze MW. The SXL-UNR corepressor complex uses a PABP-mediated mechanism to inhibit ribosome recruitment to msl-2 mRNA. PMID: 19941818 Mol Cell. 2009;364:571–582.
- Lebedeva LA, Nabirochkina EN, Kurshakova MM, et al. Occupancy of the Drosophila hsp70 promoter by a subset of basal transcription factors diminishes upon transcriptional activation. Proc Natl Acad Sci U S A. 2005;102(50):18087–18092. PMID: 16330756.
- Kachaev ZM, Gilmutdinov RA, Kopytova DV, et al. RNA immunoprecipitation technique for Drosophila melanogaster S2 cells. Mol Biol (Moscow). 2017;51(1):72–79. PMID: 28251970.
- Vorobyeva NE, Soshnikova NV, Nikolenko JV, et al. Transcription coactivator SAYP combines chromatin remodeler Brahma and transcription initiation factor TFIID into a single supercomplex. Proc Natl Acad Sci U S A. 2009;106(27):11049–11054. PMID: 19541607.
- Kibanov MV, Kotov AA, Olenina LV. Multicolor fluorescence imaging of whole-mount Drosophila testes for studying spermatogenesis. PMID: 23357237 Anal Biochem. 2013;4361:55–64.
- Shidlovskii YV, Krasnov AN, Nikolenko JV, et al. A novel multidomain transcription coactivator SAYP can also repress transcription in heterochromatin. EMBO J. 2005;24(1):97–107. PMID: 15616585.
- Johansen KM, Cai W, Deng H, et al. Polytene chromosome squash methods for studying transcription and epigenetic chromatin modification in Drosophila using antibodies. Methods. 2009;48(4):387–397. PMID: 19272452.
- Georgieva S, Kirschner DB, Jagla T, et al. Two novel Drosophila TAF(II)s have homology with human TAF(II)30 and are differentially regulated during development. Mol Cell Biol. 2000;20(5):1639–1648. PMID: 10669741.
