ABSTRACT
The transcription factor p73 is a member of the p53 family, of which the transactivation domain containing isoform (TAp73) plays key roles in brain development and neuronal stem cells. TAp73 also facilitates homoeostasis and prevents oxidative damage in vivo by inducing the expression of its target genes. Recently, we found that in addition to its role in regulation of transcription, TAp73 also affects mRNA translation. In cultured cells, acute TAp73 depletion activates eEF2K, which phosphorylates eEF2 reducing mRNA translation elongation. As a consequence, there is a reduction in global proteins synthesis rates and reprogramming of the translatome, leading to a selective decrease in the translation of rRNA processing factors. Given the dramatic effects of Tap73 depletion in vitro it was important to determine whether similar effects were observed in vivo. Here, we report the surprising finding that in brains of TAp73 KO mice there is a reduced level of eEF2K, which allows protein synthesis rates to be maintained suggesting a compensation model. These data provide new insights to the role of TAp73 in translation regulation and the eEF2K pathway in the brain.
Introduction
TAp73 is the longest isoform encoded by the p73 gene, a member of the p53 family of transcription factors [Citation1–Citation3], which plays important roles in tumor suppression [Citation4–Citation8] and cellular homeostasis by promoting the expression of genes to regulate metabolism [Citation9–Citation17]. The p53 family also includes p53 and p63 that share with p73 the capability of promoting cell cycle arrest and apoptosis following DNA damage [Citation18–Citation24]. P53 is mainly known for its powerful tumor suppressor capability [Citation25–Citation29] and for the frequent mutations observed in cancer (approximately 50%) [Citation30–Citation34] that can interfere with the physiological function of all the three family members [Citation35,Citation36]. P63 has an exclusive predominant role in developmental of epidermis and stratified epithelia [Citation37–Citation42], fundamentally contributing to [Citation43] this process with a wide range of downstream targets and interactors [Citation44–Citation50]. Part of p63 function is also mediated by its contribution on the cellular metabolism [Citation45,Citation51–Citation55]. TAp73 plays a peculiar function in brain development [Citation56–Citation58] and TAp73 knockout mice exhibit significant neurodevelopmental defects, including hippocampal dysgenesis with truncation of the lower blade of the dentate gyrus [Citation4,Citation59–Citation61]. Concurrent loss of TAp73 and the shorter isoform ΔNp73 results in even more severe neurological phenotype; p73 full KO mice, in addition to defective hippocampus, display reduced cortical thickness and hydrocephalus [Citation62]. Recent studies have highlighted a critical contribution of p73 on the process of ciliogenesis. TAp73 appears necessary for basal body docking, axonemal extension, and motility during the differentiation of multiciliated cell progenitors, by transcriptionally controlling expression of key regulators of this process, FoxJ1, Rfx2, Rfx3, and miR34bc [Citation63–Citation65]. This recent novel insight into the in vivo biological function of p73 might unify the complex phenotype displayed by p73 mutant mice [Citation66].
The most well studied functions TAp73 have been related to its transcriptional activates and induction of target genes through binding promoter elements that are highly similar to those of p53 [Citation67]. Despite extensive studies of TAp73 functions in maintaining homeostasis and in particular, in protecting against oxidative stress, we still do not understand the full spectrum of TAp73 cellular functions.
Recently, we identified a new and surprising TAp73 function, regulation of mRNA translation [Citation68]. Regulation of protein synthesis is a mechanism for cells to readapt to stress conditions and cope with reduced energy/nutrient supply, attempting to optimize the cellular resources [Citation69–Citation71]. Our study indicated that TAp73 depletion is accompanied by increased activity of eukaryotic elongation factor 2 kinase (eEF2K), a negative regulator of mRNA translation elongation, and reduced translation elongation. This results in a reprogramming of the translatome and using gene knock-down in cell culture and polysome profiling, we found that TAp73 promotes the translation of ribosomal RNA processing factors under resting conditions and of mitochondrial proteins under oxidative stress. Indeed, TAp73 depleted cells exhibit reduced protein synthesis under resting conditions and reduced ATP levels, decreased mitochondrial activity and increased cell death following oxidative stress. These findings show that TAp73 not only regulates mRNA transcription but that it is an important regulator of mRNA translation as well. Here we addressed whether TAp73 regulates protein synthesis and/or translation elongation in vivo.
Results and discussion
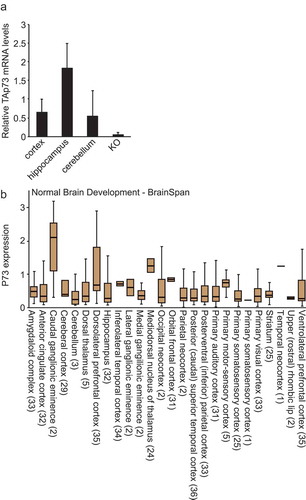
To test whether TAp73 regulates protein synthesis in vivo we used TAp73 KO and WT mice and focused on the brain, since there is a clear brain phenotype in TAp73 KO mice [Citation4] namely, hippocampal dysgenesis [Citation62]. Using qRT-PCR we confirmed that TAp73 is expressed in several regions of the brain () and analyzing published data confirmed that p73 is also expressed in human brain ().
Figure 1. TAp73 is expressed in brain. (a) mRNA levels of TAp73 in the indicated regions of the brain isolated from wild-type mice. KO is used as negative control (RNA derived from the brain of p73 KO mice). (b) The expression of p73 in different regions of human brain was determined using R2.
To measure protein synthesis in mouse brain we used brain slices obtained from TAp73 KO and WT mice and puromycin labeling. The amount of puromycin incorporated proteins in the brain lysates was determined by Western blot and anti-puromycin antibodies [Citation72,Citation73]. Unexpectedly, given our previous in vitro data (where acute depletion of TAp73 resulted in a large decrease in global protein synthesis [Citation68]), we found no significant differences in the rate of protein synthesis between TAp73 KO and WT brains (.
Figure 2. TAp73 KO mice maintain translation in the brain. (a) Brain slices obtained from the indicated genotypes were pulsed with puromycin (45 min; 5 μg/ml). Puromycin incorporation in the brain lysates was determined using Western blot and anti-puromycin antibodies as described [Citation25]. Scanned lanes were quantified using ImageJ. n = 3; *p < 0.05. (b) The levels of the indicated hippocampal proteins and their modifications in lysates obtained from the indicated genotypes were determined using Western blot. (c) Protein levels were quantified using ImageJ. n = 3; *p < 0.05. (d) The correlation between the expression of p73 and eEF2K in brain was determined using R2.
![Figure 2. TAp73 KO mice maintain translation in the brain. (a) Brain slices obtained from the indicated genotypes were pulsed with puromycin (45 min; 5 μg/ml). Puromycin incorporation in the brain lysates was determined using Western blot and anti-puromycin antibodies as described [Citation25]. Scanned lanes were quantified using ImageJ. n = 3; *p < 0.05. (b) The levels of the indicated hippocampal proteins and their modifications in lysates obtained from the indicated genotypes were determined using Western blot. (c) Protein levels were quantified using ImageJ. n = 3; *p < 0.05. (d) The correlation between the expression of p73 and eEF2K in brain was determined using R2.](/cms/asset/6cdb4375-36fe-4166-bc8b-32f327bc618b/kccy_a_1553341_f0002_c.jpg)
We then hypothesized that there could be a compensation mechanism allowing TAp73 KO brains, which have developed in conditions of chronic TAp73 depletion, to maintain translation. Such a compensation mechanism has been described in cell lines that were engineered to exhibit defects in rRNA processing and, despite having a ribosomal defect, maintain protein synthesis rates through a mechanism involving reduction of eEF2K levels [Citation74]. To address this possibility, we analyzed the levels of eEF2K and, as a read-out for eEF2K activity, those of phosphorylated eukaryotic elongation factor 2 (eEF2), in lysates obtained from brains of TAp73 KO and WT mice. In line with our hypothesis, we found a striking reduction in eEF2K protein levels that correlate with reduced phosphorylation of eEF2 (). We did not observe significant differences in the phosphorylation levels of eukaryotic initiation factor 2 alpha (eIF2α) (), a marker of the ER stress pathway, suggesting that the proposed compensation occurs specifically through the translation elongation pathway. Mining published data revealed that there is a high correlation between the expression of p73 and eEF2K in human brain () supporting our premise that TAp73 is involved in regulation of translation elongation pathway in brain as well as in culture.
In previous studies, it has been proposed that to compensate for defective rRNA processing and reduced mRNA translation, cells reduce the expression of eEF2K [Citation74,Citation75]. In agreement with these data, acute TAp73 depletion in cultured cells inhibits the synthesis of nucleolar proteins, rRNA processing and protein synthesis [Citation68]. However, not all cases of defects in rRNA processing lead to reduced protein synthesis. Acute depletion of glutamate-amonia ligase (GLUL) in cell culture resulted in aberrant rRNA processing but sustained protein synthesis [Citation62]. How cells maintained protein synthesis in this case is not known. The transcriptional factor TAp73 was implicated in the metabolism of the glutamine. In particular, our previous studies report that TAp73 transcriptionally controls expression of the glutaminase-2 (GLS-2) a key metabolic enzyme in the hydrolysis of the glutamine in glutamate. The TAp73/GLS-2 axis appears to have multiple implications for neuronal differentiation as well as for the capability of the cancer cells to survive under nutrient deprivation [Citation76,Citation77]. Whether the connection between the function of TAp73 in the glutamine metabolism is connected with its capability of influencing rRNA processing and mRNA translation has not been not clarified. The capability of the cells to cope with oxidative stress is crucially influenced by mechanisms supporting mitochondrial health [Citation78–Citation82]. Dysfunctions in the cellular metabolism result in reduced antioxidant capacity and increased susceptibility to oxygen radicals [Citation83–Citation86]. TAp73 critically contributes to mitochondrial biology and cellular metabolism and under TAp73 acute depletion cells suffers of increased susceptibility to oxidative radicals in connection with a reduced protein synthesis capacity. Overall the current data are suggestive of a connection between TAp73 function on cellular metabolism, including regulation of mitochondrial activity, protein synthesis and oxidative defense. Consistently a readaptation in TAp73 KO mice of the cellular metabolic functions might underline the readaptation of the protein synthesis defect.
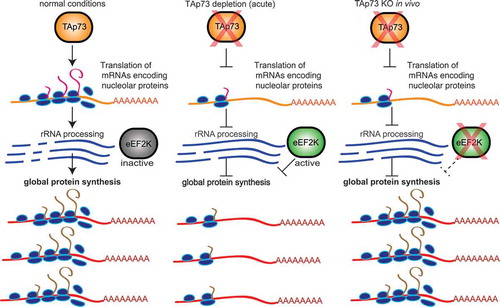
In conclusion, our data strongly suggests that chronic TAp73 depletion in vivo results in reduced eEF2K protein levels that may represent a compensation mechanism for reduced translation capacity (), further implicating TAp73 in regulation of protein synthesis through interaction with the eEF2K pathway.
Figure 3. Proposed model for TAp73 interaction with the translation pathway. Under normal conditions, TAp73 promotes the translation of nucleolar proteins which process rRNA and thus promoting global protein synthesis. Acute TAp73 depletion results in reduced translation of nucleolar proteins, increased activity of eEF2K and reduced global protein synthesis. TAp73 KO in vivo triggers a compensation mechanism in the brain where eEF2K expression is reduced and global protein synthesis is maintained.
Materials and methods
Western blot
Brains were homogenized with RIPA buffer containing phosphatase inhibitors (PhosSTOP Phosphatase Inhibitor Cocktail; Roche), and protease inhibitors (cOmplete Protease Inhibitor Cocktail; Roche). Equal amounts of proteins were run in SDS-PAGE gels, transferred to nitro cellulose membranes (Life Technologies) and probed using the following antibodies: Cell Signaling: eEF2K, Phospho-eEF2, eEF2, Phospho-eIF2α; Santa Cruz: β Tubulin Antibody (H-235); p73: our home made antibody as described [Citation87]; Puromycin (Merk, clone 12D10).
Mice
TAp73 WT and KO mice were maintained as described [Citation60]. All animal work conformed to UK regulations and institutional guidelines and was performed under the authority of a project license granted by the UK Home Office. Western blot analysis of mice hippocampus was performed as described [Citation60]. In situ translation measurements were performed as described [Citation73].
RNA isolation and qPCR
Total RNA from cortex, hippocampus and cerebellum was isolated using Trizol according to the manufacturer’s instructions. RNA samples were treated with RNase-free DNase I (Qiagen). Total RNA was reverse transcribed using RevertAid H Minus First Strand cDNA synthesys kit (Fermentas). qPCR was performed using qPCR Mastermix with SYBR green (Applied Biosystem). The expression of TAp73 was defined from the threshold cycle (Ct), and relative expression levels were calculated by using the 2−ΔΔCt method after normalization with the housekeeping GAPDH and relative to cortex.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Dotsch V, Bernassola F, Coutandin D, et al. p63 and p73, the ancestors of p53. Cold Spring Harb Perspect Biol. 2010;2(9):a004887.
- Van Nostrand JL, Bowen ME, Vogel H, et al. The p53 family members have distinct roles during mammalian embryonic development. Cell Death Differ. 2017;24(4):575–579.
- Vikhreva P, Melino G, Amelio I. p73 alternative splicing: exploring a biological role for the C-terminal isoforms. J Mol Biol. 2018;430(13):1829–1838.
- Tomasini R, Tsuchihara K, Wilhelm M, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22(19):2677–2691.
- Amelio I, Inoue S, Markert EK, et al. TAp73 opposes tumor angiogenesis by promoting hypoxia-inducible factor 1alpha degradation. Proc Natl Acad Sci U S A. 2015;112(1):226–231.
- Vikhreva P, Petrova V, Gokbulut T, et al. TAp73 upregulates IL-1beta in cancer cells: potential biomarker in lung and breast cancer? Biochem Biophys Res Commun. 2017;482(3):498–505.
- Amelio I, Melino G. The p53 family and the hypoxia-inducible factors (HIFs): determinants of cancer progression. Trends Biochem Sci. 2015;40(8):425–434.
- Thakur AK, Nigri J, Lac S, et al. TAp73 loss favors Smad-independent TGF-beta signaling that drives EMT in pancreatic ductal adenocarcinoma. Cell Death Differ. 2016;23(8):1358–1370.
- Rufini A, Niklison-Chirou MV, Inoue S, et al. TAp73 depletion accelerates aging through metabolic dysregulation. Genes Dev. 2012;26(18):2009–2014.
- Amelio I, Antonov AA, Catani MV, et al. TAp73 promotes anabolism. Oncotarget. 2014;5(24):12820–12934.
- Amelio I, Cutruzzola F, Antonov A, et al. Serine and glycine metabolism in cancer. Trends Biochem Sci. 2014;39(4):191–198.
- Du W, Jiang P, Mancuso A, et al. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol. 2013;15(8):991–1000.
- Sharif T, Ahn D-G, Liu R-Z, et al. The NAD(+) salvage pathway modulates cancer cell viability via p73. Cell Death Differ. 2016;23(4):669–680.
- Niklison-Chirou MV, Erngren I, Engskog M, et al. TAp73 is a marker of glutamine addiction in medulloblastoma. Genes Dev. 2017;31(17):1738–1753.
- Agostini M, Romeo F, Inoue S, et al. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016;23(9):1502–1514.
- Nemajerova A, Amelio I, Gebel J, et al. Non-oncogenic roles of TAp73: from multiciliogenesis to metabolism. Cell Death Differ. 2018;25(1):144–153.
- Inoue S, Tomasini R, Rufini A, et al. TAp73 is required for spermatogenesis and the maintenance of male fertility. Proc Natl Acad Sci U S A. 2014;111(5):1843–1848.
- Belyi VA, Ak P, Markert E, et al. The origins and evolution of the p53 family of genes. Cold Spring Harb Perspect Biol. 2010;2(6):a001198.
- Martin-Lopez M, Nikolic A, Kentish SJ, et al. p73 is required for appropriate BMP-induced mesenchymal-to-epithelial transition during somatic cell reprogramming. Cell Death Dis. 2017;8(9):e3034.
- Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370.
- Garcia PB, Attardi LD. Illuminating p53 function in cancer with genetically engineered mouse models. Semin Cell Dev Biol. 2014;27:74–85.
- Murphy ME. Ironing out how p53 regulates ferroptosis. Proc Natl Acad Sci U S A. 2016;113(44):12350–12352.
- Fouchecourt S, Livera G, Messiaen S, et al. Apoptosis of Sertoli cells after conditional ablation of murine double minute 2 (Mdm2) gene is p53-dependent and results in male sterility. Cell Death Differ. 2016;23(3):521–530.
- Belle JI, Petrov JC, Langlais D, et al. Repression of p53-target gene Bbc3/PUMA by MYSM1 is essential for the survival of hematopoietic multipotent progenitors and contributes to stem cell maintenance. Cell Death Differ. 2016;23(5):759–775.
- Pfister NT, Prives C. Transcriptional regulation by wild-type and cancer-related mutant forms of p53. Cold Spring Harb Perspect Med. 2017;7(2).
- Gudkov AV, Komarova EA. p53 and the carcinogenicity of chronic inflammation. Cold Spring Harb Perspect Med. 2016;6(11).
- Jennis M, Kung CP, Basu S, et al. An African-specific polymorphism in the TP53 gene impairs p53 tumor suppressor function in a mouse model. Genes Dev. 2016;30(8):918–930.
- Velletri T, Xie N, Wang Y, et al. P53 functional abnormality in mesenchymal stem cells promotes osteosarcoma development. Cell Death Dis. 2016;7:e2015.
- Pinto M, Pickrell AM, Wang X, et al. Transient mitochondrial DNA double strand breaks in mice cause accelerated aging phenotypes in a ROS-dependent but p53/p21-independent manner. Cell Death Differ. 2017;24(2):288–299.
- Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–317.
- Alexandrova EM, Moll UM. Depleting stabilized GOF mutant p53 proteins by inhibiting molecular folding chaperones: a new promise in cancer therapy. Cell Death Differ. 2017;24(1):3–5.
- Gurpinar E, Vousden KH. Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol. 2015;25(8):486–495.
- Muller PA, Caswell PT, Doyle B, et al. Mutant p53 drives invasion by promoting integrin recycling. Cell. 2009;139(7):1327–1341.
- Charni M, Aloni-Grinstein R, Molchadsky A, et al. p53 on the crossroad between regeneration and cancer. Cell Death Differ. 2017;24(1):8–14.
- Kehrloesser S, Osterburg C, Tuppi M, et al. Intrinsic aggregation propensity of the p63 and p73 TI domains correlates with p53R175H interaction and suggests further significance of aggregation events in the p53 family. Cell Death Differ. 2016;23(12):1952–1960.
- Gebel J, Luh LM, Coutandin D, et al. Mechanism of TAp73 inhibition by DeltaNp63 and structural basis of p63/p73 hetero-tetramerization. Cell Death Differ. 2016;23(12):1930–1940.
- Yang A, Schweitzer R, Sun D, et al. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999;398(6729):714–718.
- Vanbokhoven H, Melino G, Candi E, et al. p63, a story of mice and men. J Invest Dermatol. 2011;131(6):1196–1207.
- Candi E, Amelio I, Agostini M, et al. MicroRNAs and p63 in epithelial stemness. Cell Death Differ. 2015;22(1):12–21.
- Rivetti Di Val Cervo P, Lena AM, Nicoloso M, et al. p63-microRNA feedback in keratinocyte senescence. Proc Natl Acad Sci U S A. 2012;109(4):1133–1138.
- Melino G, Memmi EM, Pelicci PG, et al. Maintaining epithelial stemness with p63. Sci Signal. 2015;8(387):re9.
- Lohrum MA, Vousden KH. Regulation and function of the p53-related proteins: same family, different rules. Trends Cell Biol. 2000;10(5):197–202.
- Berkers CR, Maddocks OD, Cheung EC, et al. Metabolic regulation by p53 family members. Cell Metab. 2013;18(5):617–633.
- Rufini S, Lena AM, Cadot B, et al. The sterile alpha-motif (SAM) domain of p63 binds in vitro monoasialoganglioside (GM1) micelles. Biochem Pharmacol. 2011;82(10):1262–1268.
- Viticchie G, Agostini M, Lena AM, et al. p63 supports aerobic respiration through hexokinase II. Proc Natl Acad Sci U S A. 2015;112(37):11577–11582.
- Vigano MA, Lamartine J, Testoni B, et al. New p63 targets in keratinocytes identified by a genome-wide approach. Embo J. 2006;25(21):5105–5116.
- Lena AM, Cipollone R, Amelio I, et al. Skn-1a/Oct-11 and DeltaNp63alpha exert antagonizing effects on human keratin expression. Biochem Biophys Res Commun. 2010;401(4):568–573.
- Celardo I, Antonov A, Amelio I, et al. p63 transcriptionally regulates the expression of matrix metallopeptidase 13. Oncotarget. 2014;5(5):1279–1289.
- Memmi EM, Sanarico AG, Giacobbe A, et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci U S A. 2015;112(11):3499–3504.
- Celardo I, Grespi F, Antonov A, et al. Caspase-1 is a novel target of p63 in tumor suppression. Cell Death Dis. 2013;4:e645.
- D’Alessandro A, Amelio I, Berkers CR, et al. Metabolic effect of TAp63alpha: enhanced glycolysis and pentose phosphate pathway, resulting in increased antioxidant defense. Oncotarget. 2014;5(17):7722–7733.
- Giacobbe A, Bongiorno-Borbone L, Bernassola F, et al. p63 regulates glutaminase 2 expression. Cell Cycle. 2013;12(9):1395–1405.
- Regina C, Nikolic A, Kentish SJ, et al. DeltaNp63alpha modulates histone methyl transferase SETDB1 to transcriptionally repress target genes in cancers. Cell Death Discov. 2016;2:16015.
- Compagnone M, Gatti V, Presutti D, et al. DeltaNp63-mediated regulation of hyaluronic acid metabolism and signaling supports HNSCC tumorigenesis. Proc Natl Acad Sci U S A. 2017;114(50):13254–13259.
- Amelio I, Melino G, Candi E. p63 adjusts sugar taste of epidermal layers. J Invest Dermatol. 2017;137(6):1204–1206.
- Niklison-Chirou MV, Killick R, Knight RA, et al. How does p73 cause neuronal defects? Mol Neurobiol. 2016;53(7):4509–4520.
- Turnquist C, Horikawa I, Foran E, et al. p53 isoforms regulate astrocyte-mediated neuroprotection and neurodegeneration. Cell Death Differ. 2016;23(9):1515–1528.
- Xie N, Vikhreva P, Annicchiarico-Petruzzelli M, et al. Integrin-beta4 is a novel transcriptional target of TAp73. Cell Cycle. 2018;17(5):589–594.
- Pozniak CD, Radinovic S, Yang A, et al. An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science. 2000;289(5477):304–306.
- Niklison-Chirou MV, Steinert JR, Agostini M, et al. TAp73 knockout mice show morphological and functional nervous system defects associated with loss of p75 neurotrophin receptor. Proc Natl Acad Sci U S A. 2013;110(47):18952–18957.
- Agostini M, Tucci P, Killick R, et al. Neuronal differentiation by TAp73 is mediated by microRNA-34a regulation of synaptic protein targets. Proc Natl Acad Sci U S A. 2011;108(52):21093–21098.
- Yang A, Walker N, Bronson R, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404(6773):99–103.
- Nemajerova A, Kramer D, Siller SS, et al. TAp73 is a central transcriptional regulator of airway multiciliogenesis. Genes Dev. 2016;30(11):1300–1312.
- Marshall CB, Mays DJ, Beeler JS, et al. p73 is required for multiciliogenesis and regulates the Foxj1-associated gene network. Cell Rep. 2016;14(10):2289–2300.
- Gonzalez-Cano L, Fuertes-Alvarez S, Robledinos-Anton N, et al. p73 is required for ependymal cell maturation and neurogenic SVZ cytoarchitecture. Dev Neurobiol. 2016;76(7):730–747.
- Jackson PK, Attardi LD. p73 and FoxJ1: programming multiciliated epithelia. Trends Cell Biol. 2016;26(4):239–240.
- Candi E, Agostini M, Melino G, et al. How the TP53 family proteins TP63 and TP73 contribute to tumorigenesis: regulators and effectors. Hum Mutat. 2014;35(6):702–714.
- Marini A, Rotblat B, Sbarrato T, et al. TAp73 contributes to the oxidative stress response by regulating protein synthesis. Proc Natl Acad Sci U S A. 2018;115(24):6219–6224.
- Blagden SP, Willis AE. The biological and therapeutic relevance of mRNA translation in cancer. Nat Rev Clin Oncol. 2011;8(5):280–291.
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42.
- Delaidelli A, Negri GL, Jan A, et al. MYCN amplified neuroblastoma requires the mRNA translation regulator eEF2 kinase to adapt to nutrient deprivation. Cell Death Differ. 2017;24(9):1564–1576.
- Schmidt EK, Clavarino G, Ceppi M, et al. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods. 2009;6(4):275–277.
- Trinh MA, Ma T, Kaphzan H, et al. The eIF2alpha kinase PERK limits the expression of hippocampal metabotropic glutamate receptor-dependent long-term depression. Learn Mem. 2014;21(5):298–304.
- Liu R, Iadevaia V, Averous J, et al. Impairing the production of ribosomal RNA activates mammalian target of rapamycin complex 1 signalling and downstream translation factors. Nucleic Acids Res. 2014;42(8):5083–5096.
- Pelletier J, Thomas G, Volarevic S. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18(1):51–63.
- Velletri T, Romeo F, Tucci P, et al. GLS2 is transcriptionally regulated by p73 and contributes to neuronal differentiation. Cell Cycle. 2013;12(22):3564–3573.
- Amelio I, Markert EK, Rufini A, et al. p73 regulates serine biosynthesis in cancer. Oncogene. 2014;33(42):5039–5046.
- Raimundo N. Mitochondrial pathology: stress signals from the energy factory. Trends Mol Med. 2014;20(5):282–292.
- Wang SB, Murray CI, Chung HS, et al. Redox regulation of mitochondrial ATP synthase. Trends Cardiovasc Med. 2013;23(1):14–18.
- Garedew A, Henderson SO, Moncada S. Activated macrophages utilize glycolytic ATP to maintain mitochondrial membrane potential and prevent apoptotic cell death. Cell Death Differ. 2017;24(6):1132.
- Van Aken O, Pogson BJ. Convergence of mitochondrial and chloroplastic ANAC017/PAP-dependent retrograde signalling pathways and suppression of programmed cell death. Cell Death Differ. 2017;24(6):955–960.
- Prola A, Da Silva JP, Guilbert A, et al. SIRT1 protects the heart from ER stress-induced cell death through eIF2alpha deacetylation. Cell Death Differ. 2017;24(2):343–356.
- Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947.
- Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat Chem Biol. 2008;4(5):278–286.
- Dong Z, Huang M, Liu Z, et al. Focused screening of mitochondrial metabolism reveals a crucial role for a tumor suppressor Hbp1 in ovarian reserve. Cell Death Differ. 2016;23(10):1602–1614.
- Caputa G, Zhao S, Criado AEG, et al. RNASET2 is required for ROS propagation during oxidative stress-mediated cell death. Cell Death Differ. 2016;23(2):347–357.
- Sayan AE, Paradisi A, Vojtesek B, et al. New antibodies recognizing p73: comparison with commercial antibodies. Biochem Biophys Res Commun. 2005;330(1):186–193.
