ABSTRACT
In eukaryotic cells, changes in chromatin accessibility are necessary for chromatin to maintain its highly dynamic nature at different times during the cell cycle. Histone chaperones interact with histones and regulate chromatin dynamics. Facilitates chromatin transcription (FACT) is an important histone chaperone that plays crucial roles during various cellular processes. Here, we analyze the structural characteristics of FACT, discuss how FACT regulates nucleosome/chromatin reorganization and summarize possible functions of FACT in transcription, replication, and DNA repair. The possible involvement of FACT in cell fate determination is also discussed.
Abbreviations: FACT: facilitates chromatin transcription, Spt16: suppressor of Ty16, SSRP1: structure-specific recognition protein-1, NTD: N-terminal domain, DD: dimerization domain, MD: middle domain, CTD: C-terminus domain, IDD: internal intrinsically disordered domain, HMG: high mobility group, CID: C-terminal intrinsically disordered domain, Nhp6: non-histone chromosomal protein 6, RNAPII: RNA polymerase II, CK2: casein kinase 2, AID: acidic inner disorder, PIC: pre-initiation complex, IR: ionizing radiation, DDSB: DNA double-strand break, PARlation: poly ADP-ribosylation, BER: base-excision repair, UVSSA: UV-stimulated scaffold protein A, HR: homologous recombination, CAF-1: chromatin assembly factor 1, Asf1: anti-silencing factor 1, Rtt106: regulator of Ty1 transposition protein 106, H3K56ac: H3K56 acetylation, KD: knock down, SETD2: SET domain containing 2, H3K36me3: trimethylation of lysine36 in histone H3, H2Bub: H2B ubiquitination, iPSCs: induced pluripotent stem cells, ESC: embryonic stem cell, H3K4me3: trimethylation of lysine 4 on histone H3 protein subunit, CHD1: chromodomain protein
Graphical Abstract
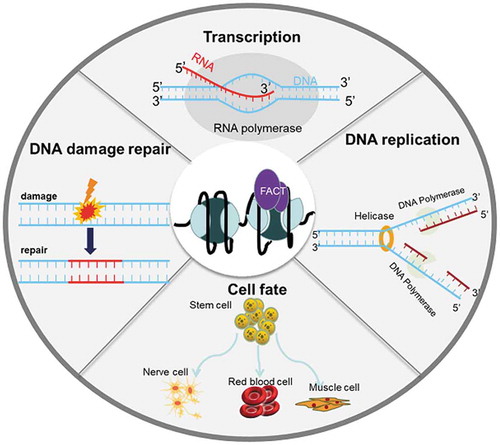
Background
Chromatin is a stable and highly dynamic nuclear protein complex. The fundamental unit of chromatin in eukaryotes is nucleosome, which consists of a histone octamer wrapped by approximately 146 bp of DNA fragments [Citation1]. The dense structure of nucleosomes undeniably protects DNA from damage and at the same time acts as a barrier during cellular processes such as transcription, replication, and DNA repair. Thus, efficient chromatin remodeling is required in all chromatin-related processes [Citation2–4]. Chromatin remodeling refers to a wide range of chromatin changes induced by chromatin-remodeling factors interacting with nucleosomes, which can be achieved by various means, including (i) ATP-dependent remodeling complexes, (ii) histone posttranslational modifications, (iii) histone chaperones, and (iv) the interplay of all three [Citation5–9]. Histone chaperones play crucial roles in the correct remodeling of nucleosomes, first proposed by Ron Laskey [Citation10] to describe an acidic nuclear protein in Xenopus laevis egg extracts, representing a group of proteins that prevent incorrect ionic interactions between histones and DNA. Histone chaperones participate in nucleosome assembly/disassembly without using ATP, and they are not permanent components of nucleosome structures [Citation11]. Among them, facilitates chromatin transcription (FACT) is a highly conserved histone chaperone that was initially discovered to remove histone H2A-H2B dimer during transcription [Citation12,Citation13]. In fact, FACT is capable of binding to all core histones [Citation14] and has been verified to be involved in important cellular processes, including transcription, DNA damage repair, DNA replication, and cell fate determination [Citation15–17].
The structural features of FACT
Human (h) FACT is a heterodimer consisting of two subunits: Suppressor of Ty16 (Spt16) and Structure-Specific Recognition Protein-1 (SSRP1) [Citation18,Citation19]. The two subunits of hFACT are interdependent, and the stability of hFACT depends on the presence of the SSRP1 mRNA [Citation20].
Spt16 is the large subunit of FACT, and has several domains with histone-binding activity: an N-Terminal Domain (NTD), a Dimerization Domain (DD), a Middle Domain (MD), and a C-Terminal Domain (CTD) () [Citation21–23]. Spt16 is very well conserved in yeast and humans (). The NTD of Spt16 is an inactive amino-peptidase-like motif [Citation24]. It has been shown that the NTD has very weak affinity for the H2A-H2B dimer but interacts preferentially with the H3-H4 tetramer [Citation24–26]. The DD of Spt16 interacts with the NTD/DD of SSRP1, and this interaction is crucial for formation of the FACT complex [Citation27]. The MD of Spt16 is essential for recognition of histones H2A-H2B by FACT [Citation28]. Moreover, Spt16 MD also contributes to FACT interaction with H3-H4 tetramers [Citation28,Citation29] and helps dissociate Spt16 from chromatin at the end of transcription [Citation27]. The acidic region of the Spt16 CTD is a highly conserved domain that binds explicitly to the H2A-H2B dimer [Citation30]. The CTD is involved in nucleosome reorganization, and its absence inhibits the activity of FACT [Citation29,Citation31].
Figure 1. Domain organization of hFACT and yFACT. The heterodimeric hFACT complex is composed of the hSpt16 (cyan) and SSRP1 (green) subunits, through an interaction (gray) between the DD domain of hSPT16 (cyan) and the NTD/DD domain of SSRP1. In yeast, ySpt16 (cyan) and Pob3 (blue) subunits form the yFACT complex. (3D structures are from the UniProt website)
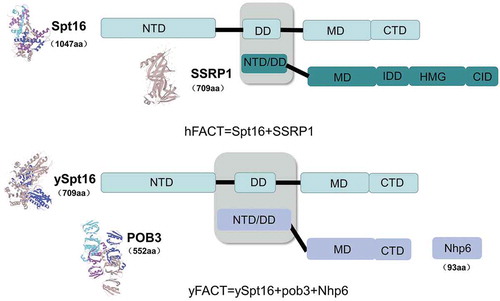
hSSRP1 consists of five different domains: the NTD/DD, MD, internal intrinsically disorder domain (IDD),high mobility group (HMG) domain, and C-terminal Intrinsically disordered (CID) domain () [Citation22,Citation32]. The NTD/DD contains a pleckstrin homology (PH) motif that interacts with Spt16 [Citation28]. The MD of SSRP1 does not bind histones, but binds DNA and functions like HMG [Citation33]. The HMG domain has DNA binding activity [Citation34–36] and stabilizes nucleosome structure by maintaining H3-H4 tetramer binding to DNA [Citation37]. Mouse embryos lacking the HMG domain die before E5.5, suggesting that the HMG domain is indispensable for early embryonic development [Citation36]. Unlike the HMG domain, which binds to bent or kinked DNA, the CID domain of hSSRP1 binds to Z-DNA [Citation38,Citation39]. It has been shown that both the CTD and HMG domains are essential for the nucleosome stabilizing activity of FACT [Citation40].
The homologous protein of SSRP1 in yeast is yPob3 [Citation41,Citation42], which contains the NTD/DD, MD and CTD [Citation22] and forms the yFACT complex with ySpt16 (). yPob3 lacks the HMG domain; however, non-histone chromosomal protein 6 (Nhp6) encodes an HMG1-like motif and provides HMG1-like function for yFACT in yeast [Citation59Citation60Citation64Citation43], enabling yFACT to remodel nucleosomes. It was reported that Nhp6 does not interact directly with yFACT but binds to DNA at both ends of the nucleosomes and bends the structure sharply, which induces a change in nucleosomes accessibility and provides access for yFACT binding [Citation44–46]. In the absence of Nhp6, Spt16 binds to H2B and disrupts the interaction between the histone octamer and DNA, and removes the H2A-H2B dimer by gradually unwrapping/rewrapping DNA from the surface of the octamer [Citation47]. In line with this evidence, the presence of Nhp6 prevents loss of H2A-H2B during FACT-dependent nucleosome remodeling [Citation48].
Functions of FACT in chromatin homeostasis
FACT maintains chromatin homeostasis through its dual functions of assembling and disassembling nucleosomes [Citation14,Citation37]. It has been reported that while Spt16 displaces the H2A-H2B dimer and disassembles nucleosome structure, SSRP1 maintains the H3-H4 tetramer on DNA and promotes deposition of the H2A-H2B dimer [Citation37].
Recent reports have shed new light on the mechanistic insight of FACT in maintaining chromatin homeostasis [Citation14]. FACT seems to preferentially bind histone H2A-H2B, and the affinity between H2A-H2B and FACT is approximately 20-fold higher than that of FACT and H3-H4. The interaction between the CTD of Spt16 and H2A-H2B activates the DNA-binding activity of FACT [Citation30]. In an in vitro nucleosome assembly assay, FACT does not bind to (H3-H4)2 tetrasome in the absence of H2A–H2B dimer. However, when prebound with H2A–H2B, FACT binds to tetrasomes efficiently and forms intermediate complexes (FACT sub-nucleosome) [Citation49]. Thus, it was inferred from the experiment that FACT interacts with H2A-H2B dimer to form H2A-H2B-FACT and uses its DNA-binding activity to dock onto the (H3-H4)2-DNA complex to form the FACT-subnucleosome [Citation49]. The structure of the FACT-subnucleosome looks like a unicycle: (H3-H4)2 is the central part of the wheel; the H2A-H2B dimer is similar to a pedal, and FACT is similar to a frame across the wheel [Citation14,Citation49]. When the FACT-subnucleosome meets the second H2A-H2B, the MD of SSRP1 undergoes structural changes to facilitate docking of the second H2A-H2B onto the histone core and forms nucleosomes [Citation14]. During transcription or DNA replication, DNA temporarily leaves its interaction interface with the (H3-H4)2 tetramer. At the same time, the MD of Spt16 rotates a certain angle (23°) to change the binding modes with the (H3-H4)2 tetramer. The rotation of the Spt16 MD makes its pleckstrin homology domain clash with the H2A docking domain and detach H2A-H2B from the nucleosome. However, the CTD tethers and maintains H2A-H2B nearby to ensure that all components of nucleosomes are intact during the passage of polymerases [Citation14,Citation50].
FACT in transcription
Transcriptional initiation requires a Pre-Initiation Complex (PIC) on promoter to facilitate DNA-binding activity of RNAPII [Citation51]. FACT not only regulates recruitment of PIC components at promoters to facilitate assembly of the PIC but also reorganize nucleosomes to promote accessibility of transcription start sites (TSS) [Citation52,Citation53]. ChIP-seq analysis in mESCs indicated that both of the FACT subunits were strongly associated with actively expressed genes and enriched around the TSS [Citation54]. Moreover, Spt16 was predominantly enriched in the open reading frame (ORF) in yeast [Citation55,Citation56]. Meanwhile, FACT shows the same localization and dynamics with the elongating RNAPII and has been proposed to promote transcriptional elongation [Citation12,Citation18,Citation53,Citation57,Citation61–]. Genome-wide profiling data indicated that FACT is important for RNAPII-mediated transcription in vivo and likely removes nucleosome blocks ahead of RNAPII during transcription and assembles them after RNAPII passage [Citation62]. However, high-resolution mapping of RNAPII traversal of nucleosomes demonstrated that RNAPII could efficiently overcome nucleosome barriers on individual nucleosomes or di-nucleosome substrates in vitro [Citation63,Citation65–]. Two types of working models, the “dimer eviction model” () and the “non-eviction model” (), have been proposed to explain the probable mechanism of FACT-mediated nucleosome reorganization during transcription [Citation55,Citation66,Citation68–]. Of note, both of the models are RNAPII-dependent, although FACT is also found at genomic regions transcribed by RNAPI and RNAPIII [Citation69]. In the “dimer eviction model,” FACT actively removes H2A–H2B from nucleosomes to enhance DNA accessibility. In support of this view, FACT cannot promote transcription when histones are cross-linked to prevent H2A–H2B dimers from being evicted [Citation12,Citation14]. In contrast, recent reports support the “non-eviction model”, where FACT interacts with nucleosomes and increases chromatin accessibility without evicting the H2A–H2B dimer. In accordance with this model, it was found that yFACT promotes transcriptional activation of the GAL1-10 promoter without significant dimer loss [Citation67]. The loss of H2A-H2B dimers, especially H2B, seems to have a great impact on FACT-dependent transcription [Citation31]. Thus, it is likely that FACT binds to the H2A-H2B dimer to ensure the integrity of the nucleosome [Citation70]. Indeed, FACT has been recently proposed to interact with the H2A-H2B dimer through its CTD to ensure nucleosome survival during transcription [Citation31,Citation50]. RNAPII binds to promoter regions and uncoils nucleosome DNA, thereby exposing the DNA-binding surface of the proximal H2A-H2B dimer and enabling its interaction with FACT. In this way, FACT retains the proximal H2A-H2B dimer and maintains the integrity of the nucleosome during transcription. After RNAPII passage, the DNA recoils on the surface of the proximal H2A-H2B dimer, which moves FACT away from the proximal dimer and promotes its interaction with the distal dimer. Taken together, accumulating evidence suggests that FACT causes the instantaneous displacement of H2A-H2B from nucleosome to ensure successful passage of RNAPII during transcriptional elongation [Citation30,Citation49]. FACT also suppresses cryptic transcription buried within the gene body and ensures transcriptional fidelity by maintaining chromatin stability [Citation16,Citation53,Citation71]. The evidence that the absence of FACT is usually accompanied by the loss of nucleosomes after transcription further supports that FACT safeguards the survival of nucleosomes during transcription [Citation14,Citation72,Citation74–]. However, presence of FACT is also accompanied by an occasional eviction of H2A-H2B dimers from nucleosomes, which might due to the transient displacement of H2A-H2B dimer from the nucleosome.
Figure 2. Working models of FACT function. (a) The “dimer eviction model”, in which FACT actively removes H2A–H2B from nucleosomes to enhance DNA accessibility. (b) The “non-eviction model”, in which FACT makes the nucleosome structure more relaxed and dynamic without the eviction of H2A–H2B
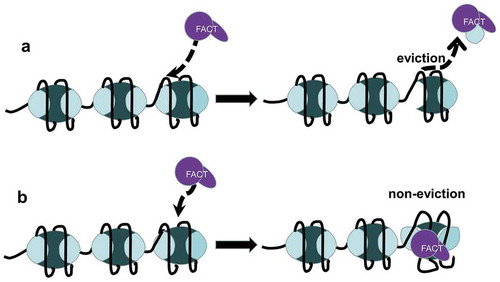
Consistent with the “non-eviction model”, recent reports suggested that FACT cannot bind to intact nucleosomes; Instead, an unstable nucleosome structure during transcription promotes FACT recruitment [Citation29,Citation48,Citation49,Citation68,Citation75–78]. In support of this view, curaxins, an anti-cancer drug that destabilizes nucleosomes, disrupts the co-localization of FACT and RNAPII, and promotes redistribution of FACT into unstable nucleosomes area in non-transcribed regions [Citation55,Citation76,Citation79]. It should be noted that FACT may also regulate transcriptional activation in eukaryotic cells. In Drosophila S2 cells, depletion of FACT releases RNAPII from transcription pause sites and engages in productive elongation, indicating that FACT is able to maintain RNAPII at transcription pause sites and fine-tunes transcriptional activation [Citation80]. Furthermore, FACT knock down (KD) significantly affects gene expression profile in non-small cell lung cancer, and different subsets of genes were affected by SSRP1 and Spt16 KD, indicating that FACT subunits have independent roles in regulating gene expression [Citation81]. Of note, ATP-dependent chromatin-remodeling complexes and histone chaperones were reported to act in concert with FACT during transcription. In yeast, the chromatin-remodeling complex SWI/SNF and the histone chaperone Asf1 work together at heat shock gene promoters during transcription initiation, and yFACT participates in subsequent nucleosome reassembly [Citation73]. For transcriptional elongation, H3K4me3 places the chromatin-remodeling factor CHD1 near the TSS, which in turn recruits FACT to promote transcriptional elongation [Citation82,Citation83].
FACT is also involved in RNAPI and RNAPIII mediated transcription; however, the underlying mechanism is less clear. It was reported that FACT interacts with RNAPI directly and the Spt16 subunit could deposit H2A-H2B dimers onto coding regions of the ribosomal DNA [Citation69,Citation84]. Interestingly, FACT also acts as a chaperone for histone H2A.Z during RNAPIII mediated tDNA transcription and mutation of the subunits (Spt16 and Pob3) increased tDNA transcription in yeast [Citation85].
FACT in DNA damage repair
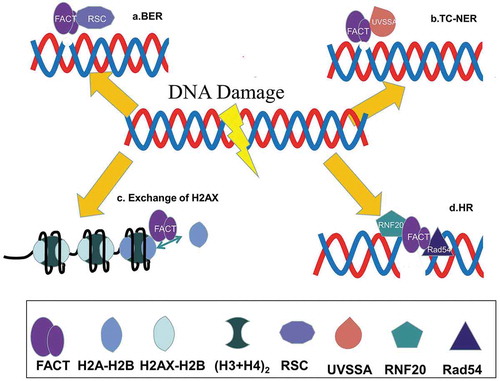
Ionizing radiation (IR), ultraviolet (UV) light and oxidative stress could cause DNA damage [Citation86–88], and numerous studies suggest that FACT is also involved in DNA damage recognition and repair processes. The nucleosome barrier needs to be removed during the early DNA damage repair process. Poly-ADP-Ribosylation (PARlation) promotes removal of histones following DNA Double-Strand Breaks (DDSBs), and FACT is involved in this process [Citation89]. Inhibition of PARlation significantly reduces recruitment of FACT, suggesting that FACT accumulation at DNA damage site is regulated by PARlation. In this process, the C-terminal region of SSRP1 plays a crucial role in recognizing PARylated histones and may serve as a signal for FACT to locate to the DNA damage sites [Citation89]. During UV-induced DNA damage response, the HMG domain of SSRP1 recognizes DNA bending or kinking and recruits SSRP1 to the site of damage [Citation90]. Moreover, accumulating evidence suggests that FACT is the primary regulator of histone H2A.X exchange during DNA damage repair [Citation91]. H2A.X is a histone H2A variant in eukaryotes that contributes to chromatin remodeling during DNA damage repair [Citation92]. H2A.X responds to DDSBs by rapid phosphorylation on the highly conserved serine 139 (γH2A.X) at its C-terminus, and γH2A.X acts as a signal to recruit DNA repair and DNA damage checkpoint proteins, such as INO80, MDC1, and TP53BP1 [Citation93–95]. FACT not only promotes the deposition of newly synthesized H2A.X to nucleosomes [Citation96,Citation97] but also participates in exchange of histone variant H2A.X with canonical H2A in nucleosomes during DNA damage repair. Notably, FACT is responsible for exchanging H2A.X-H2B with H2A-H2B dimer but not with the H2A.Z-H2B dimer [Citation96].
Base excision repair (BER) is an essential and universal DNA repair pathway. However, the presence of nucleosomes strongly interferes with the efficiency of BER [Citation98–100]. FACT participates in BER through its “co-remodeling” activity [Citation101] and acts in concert with Remodeling the Structure of Chromatin (RSC) to facilitate the removal of uracil from nucleoside DNA, indicating that FACT could promote the repair of DNA damage in the initial step of BER [Citation101]. In transcription-coupled nucleotide excision repair (TC-NER), Spt16 has been shown to interact with UV-stimulated scaffold protein A (UVSSA) and promote recovery of subsequent transcription [Citation102].
Homologous Recombination (HR) is another important DNA damage repair mechanism [Citation103]. SSRP1 has been confirmed to interact with HR key repair protein Rad54 both in vitro and in vivo. SSRP1 KD leads to increased HR events resulting in more DNA damage at stalled replication sites. Consistently, overexpression of SSRP1 inhibits spontaneous and Hydroxyurea-induced HR events, indicating that SSRP1 plays an essential role in the HR repair pathway [Citation104]. Studies demonstrated that Spt16 interacts with E3 ubiquitin ligase RNF20 during DNA damage and promotes chromatin remodeling during HR repair through histone H2B ubiquitylation [Citation105–107].
Taken together, the findings suggest that FACT is involved in various DNA damage and repair responses (). FACT interacts with DNA repair proteins and chromatin-remodeling factors at DNA damage sites to create a suitable chromatin environment. However, the exact mechanism of FACT-mediated chromatin remodeling during DNA damage responses remains to be determined.
Figure 3. The role of FACT in DNA damage repair. (a) FACT and RSC work together to promote DNA damage repair during the initial steps of the base excision repair process. (b) FACT acts as an early factor in UV-induced DNA damage and recruits UVSSA during the transcription-coupled nucleotide excision repair (TC-NER) process. (c) FACT participates in the exchange of histone variant H2A.X with conventional H2A during the DDSB repair process. (d) FACT promotes the homologous recombination repair process
FACT in DNA replication
In the process of DNA replication, the nucleosome on the parental DNA in front of the replication fork needs to be disassembled, and the two newly synthesized daughter DNA strands behind the replication fork need to be assembled into chromatin by parental and newly synthesized histones. This process is called DNA replication-coupled nucleosome assembly and is regulated by various histone chaperones [Citation108]. Early genetic research showed that yFACT interacts with various DNA replication factors and participates in DNA replication [Citation22,Citation109–112]. For example, (i) yFACT binds to DNA polymerase α [Citation113,Citation114]; (ii) yFACT interacts with the DNA-replicating protein RPA1 (binds and stabilizes single-stranded DNA intermediates during DNA replication) [Citation22]; and (iii) Pob3 is associated with a variety of replication factors (POL1, CTF4, DNA2, and CHL12) [Citation109] and travels through chromatin with the replisome complex. Recently, it was reported that ubiquitination of Spt16 or histone H2B recruits FACT to the DNA replication origin [Citation115,Citation116]. After recruitment, FACT acts as an important partner for MiniChromosome Maintenance (MCM), which is the catalytic core of replication helicases in eukaryotes, and the instability of the FACT-MCM complex results in defective DNA replication initiation [Citation44,Citation115,Citation117,Citation118]. Since MCM has helicase activity only in the context of naked DNA, but not in the chromatin context, the presence of FACT increases chromatin accessibility and promotes MCM-mediated DNA unwinding on nucleosome templates [Citation117,Citation119]. Furthermore, FACT also increases the speed of replication, suggesting that FACT may promote the progression of replication forks through nucleosome reorganization [Citation70,Citation116,Citation119–121].
As the replication fork progresses, both parental and newly synthesized histones are deposited to the leading and lagging strands [Citation108]. A recent finding suggests that FACT collaborates with Chromatin Assembly Factor 1 (CAF-1) and Regulator of Ty1 Transposition Protein 106 (Rtt106) to participate in new histone deposition. Rtt106 transfers the newly synthesized H3-H4 to FACT, and FACT deposits the H3-H4 onto nucleosomes [Citation120]. The histone modification H3K56ac facilitates this process. Meanwhile, the complex formed by FACT and MCM2 prevents loss of parental histones by retaining parental histones around the replication fork [Citation121]. Collectively, FACT regulates replication initiation and progression through its chromatin-remodeling activity. Meanwhile, FACT is required for the deposition of newly synthesized histones and participates in the recycling process of the parental histones during replication-coupled nucleosome assembly.
FACT and posttranslational modifications
The highly dynamic nature of chromatin is regulated by a complex network consisting of chromatin remodelers, histone chaperones, and posttranslational modifications [Citation122,Citation123]. Among them, histone chaperone FACT has been shown to work in concert with various posttranslational modifications to regulate chromatin dynamics. In particular, H2B Ubiquitylation (H2Bub) and Spt16 have been shown to interplay in each other’s regulation during transcription. While Spt16 regulates the formation of H2Bub, H2Bub is necessary for the stable accumulation of Spt16 at gene-coding regions [Citation124]. Earlier in vitro experiments suggested that RNAPII halts at the first nucleosome upon transcription initiation and recruits FACT, which in turn recruits ubiquitination machinery to induce H2Bub formation [Citation58]. A recent study reported that the binding affinity of FACT to H2A-H2B dimers was reduced approximately 20% in the presence of ubiquitination in vitro, and the loss of H2Bub stabilized interaction between FACT and histones in vivo, suggesting that H2Bub reduces FACT and histone-binding affinity. The acidic path residues on histones are important for FACT binding, and one possibility is that the ubiquitination of H2B could mask certain FACT binding epitopes and block its access to FACT, therefore decreasing the affinity between FACT and the H2A-H2B dimer [Citation125,Citation126]. Moreover, H2Bub enhances the release of histones from FACT and fine-tunes chaperone-histone binding dynamics to facilitate deposition of histones on DNA during nucleosome reassembly following transcription [Citation124,Citation125]. FACT, in turn, is required for the deubiquitinating enzyme Ubp10 to cleave H2Bub from nucleosomes [Citation115,Citation116]. In addition, an increased level of H3K36me3 leads to enhanced recruitment of FACT to the chromatin template, thereby promoting recovery of chromatin structure after RNAPII passage [Citation127]. In contrast, H3K56ac has been shown to impair the ability of FACT to assemble nucleosomes [Citation128]. Of note, FACT also maintains the stability of epigenetic markers by recycling local histones, and its absence results in widespread changes in histone markers during transcription [Citation80,Citation129,Citation130].
Beyond histone modifications, posttranslational modifications of FACT add another mechanistic layer for regulating its activity. Phosphorylation of SSRP1, especially at serine 510, by Casein Kinase 2 (CK2) reduces DNA binding activity of FACT [Citation131]. Phosphorylation also occurs at the IDD of SSRP1, which weakens the binding between DNA and the HMG domain of SSRP1 and inhibits DNA-binding activity of FACT [Citation23]. However, phosphorylation of the C-terminal Acidic Intrinsically Disordered (AID) segment promotes the binding of FACT and nucleosomes [Citation132]. In yeast, Rtt101-mediated ubiquitination of Spt16 is crucial for the interaction between FACT and MCM [Citation115,Citation133]. In contrast, the E3 ubiquitin ligase UBR5-mediated ubiquitination of Spt16 inhibits FACT-dependent transcriptional elongation in human cells [Citation134,Citation135]. Interestingly, PARlation of Spt16, induced by PARP1, inhibits H2A.X exchanging activity of FACT [Citation96].
FACT and cell fate determination
Cell fate determination is mostly regulated by expression of cell type-specific Transcription Factors (TFs) during differentiation [Citation136,Citation137] and requires a permissive chromatin environment to ensure efficient binding of the TFs [Citation138]. In this context, multiple histone chaperones, including FACT, have been shown to regulate cell identity by reshaping chromatin structure [Citation139].
FACT is essential for genomic reprogramming of induced pluripotent stem cells (iPSCs) [Citation140,Citation141] and has been suggested to interact directly with Oct4, a pioneer transcription factor that is essential for iPSC induction [Citation142–144]. Consistently, FACT is abundantly expressed in undifferentiated cells [Citation145]. In addition, FACT is responsible for removing histone H3 after nucleosome demethylation process mediated by Jumonji domain containing 1 C (Jmjd1c) and Oct4 [Citation146]. Collectively, these evidences suggest that by regulating chromatin accessibility, FACT helps TFs to reset gene expression profiles during cell fate determination.
In contrast to abovementioned function, FACT is also able to maintain expression of specific genes during cell differentiation. SSRP1 KD significantly affects activation of the Wnt signaling pathway during differentiation of mesenchymal stem cells to osteoblasts, and depletion of Spt16 affects the anterior pharynx development in C. elegans embryos [Citation147,Citation148]. However, FACT may also act as a negative regulator and restrict expression of other cell-specific genes to protect cell identity [Citation140,Citation141]. In support of this view, FACT prevents mis-activation of transcription start sites in mouse ESCs by regulating structure of the promoter regions [Citation54]. FACT and Tripartite Motif Containing 33 (TRIM33) form a complex on distal regulatory regions in macrophages and restrict transcription of nearby genes [Citation149].
Conclusions and perspective
The histone chaperone FACT is essential for regulating cellular processes involving chromatin, such as transcription, replication and repair, where it interacts directly with nucleosomes and maintains chromatin homeostasis. Unique motifs and domains of the FACT subunits provide structural properties that are conducive to its interaction with histones, especially with the H2A-H2B dimer. While the CTD of the Spt16 subunit is indispensable for the FACT interaction with the H2A-H2B dimer, the MD of Spt16 is essential for FACT interaction with H3-H4 tetramer and is required for recognition of the H2A-H2B dimer in the nucleosome [Citation28,Citation29]. It is likely that FACT dissociates H2A-H2B from nucleosomes but keeps them nearby to maintain relative integrity of nucleosomes during transcriptional elongation. Furthermore, FACT is able to safeguard new histone deposition during DNA replication and fine-tunes histone variants exchange in the DNA damage repair process. Of note, FACT functions together with ATP-dependent chromatin remodelers to reorganize or remodel chromatin architecture during various cellular processes.
Since its discovery in 1999, more than 20 years of studies have deepened our understanding regarding the functions of FACT and the underlying molecular mechanisms. However, many outstanding questions regarding the exact role of FACT in different DNA-templated cellular processes remain to be explored. One is that if the basic role of FACT is to simply maintain a pool of stably folded H2A-H2B dimer, whether and how do other factors regulate its different functions in different cellular contexts? H2B ubiquitination plays an important role in regulating FACT activity, but the interplay between FACT and other posttranslational modifications remain mostly elusive. FACT complex genes in C. elegans include two Ssrp1 orthologs that have redundant functions in embryonic development. However, they are each uniquely required for normal oocyte development and fertility, suggesting that SSRP1 is likely to exert Spt16-independent functions in germ line development [Citation148,Citation150]. Thus, it will be interesting to elucidate the specific role of FACT during the development of mammalian oocytes and preimplantation embryos.
Emerging evidence suggests a novel role of FACT in cell fate determination and differentiation. Expression of FACT in undifferentiated cells is significantly higher than that in somatic cells. However, the higher expression of FACT is not associated with cell proliferation [Citation145,Citation151,Citation152] and decreases with cell differentiation. In line with this, most cells in adult tissues do not have detectable FACT expression at protein level. This line of evidence collectively suggest that high FACT expression is associated with stem cells or less-differentiated cells, whereas low FACT level correlates with differentiated cell status [Citation145]. Notably, FACT is also abundantly expressed in tumor cell, and down regulation of FACT expression greatly suppresses tumor cell growth [Citation152]. Therefore, it is likely that FACT complex also plays important roles in cell fate decisions, and the function and molecular mechanism in the context of totipotency maintenance and carcinogenic transformation need to be explored further in the future.
Availability of data and materials
Not applicable.
Acknowledgments
We thank all members of the Nashun laboratory for stimulating discussions and apologize to all colleagues whose work could not be cited due to the space constraints.
Disclosure statement
The authors declare no conflict of interest.
Additional information
Funding
References
- Luger K, Mader AW, Richmond RK, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997 Sep 18; 389(6648):251–260.
- Talbert PB, Henikoff S. Histone variants on the move: substrates for chromatin dynamics. Nat Rev Mol Cell Bio. 2017 Feb l; 18(2):115–126.
- Clapier CR, Iwasa J, Cairns BR, et al. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat Rev Mol Cell Bio. 2017 Jul l; 18(7):407–422.
- Hammond CM, Stromme CB, Huang H, et al. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Bio. 2017 Mar l; 18(3):141–158.
- Masliah-Planchon J, Bieche I, Guinebretiere JM, et al. SWI/SNF chromatin remodeling and human malignancies. Annu Rev Pathol. 2015;10(10):145–171.
- Petty E, Pillus L. Balancing chromatin remodeling and histone modifications in transcription. Trends Genet. 2013 Nov; 29(11):621–629.
- Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Bio. 2015 Mar l; 16(3):178–189.
- Keck KM, Pemberton LF. Histone chaperones link histone nuclear import and chromatin assembly. Biochim Biophys Acta. 2013 Mar-Apr; 1819(3–4):277–289.
- Tyagi M, Imam N, Verma K, et al. Chromatin remodelers: we are the drivers!! Nucleus. 2016 Jul 3; 7(4):388–404.
- Laskey RA, Honda BM, Mills AD, et al. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978 Oct 5; 275(5679):416–420.
- Avvakumov N, Nourani A, Cote J. Histone chaperones: modulators of chromatin marks. Mol Cell. 2011 Mar 4; 41(5):502–514.
- Orphanides G, Wu WH, Lane WS, et al. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature. 1999 Jul 15; 400(6741):284–288.
- Reinberg D, Sims RJ. 3rd.de FACTo nucleosome dynamics. J Biol Chem. 2006 Aug 18; 281(33):23297–23301.
- Liu Y, Zhou K, Zhang N, et al. FACT caught in the act of manipulating the nucleosome. Nature. 2020 Jan;577(7790):426–431.
- Formosa T, Ruone S, Adams MD, et al. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics. 2002 Dec;162(4):1557–1571.
- Hainer SJ, Pruneski JA, Mitchell RD, et al. Intergenic transcription causes repression by directing nucleosome assembly. Genes Dev. 2011;25(1):29–40. 2011 Jan 1. .
- Gurova K, Chang HW, Valieva ME, et al. Structure and function of the histone chaperone FACT - Resolving FACTual issues. Biochim Biophys Acta Gene Regul Mech. 2018 Jul 25; 1861(9):892–904.
- Belotserkovskaya R, Oh S, Bondarenko VA, et al. FACT facilitates transcription-dependent nucleosome alteration. Science. 301(5636); 2003 Aug 22. 1090–1093.
- Orphanides G, LeRoy G, Chang CH, et al. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 92(1); 1998 Jan 9. 105–116.
- Safina A, Garcia H, Commane M, et al. Complex mutual regulation of facilitates chromatin transcription (FACT) subunits on both mRNA and protein levels in human cells. Cell Cycle. 2013;12(15):2423–2434. 2013 Aug 1. .
- Keller DM, Lu H. p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2.hSPT16.SSRP1 complex. J Biol Chem. 2002;277(51):50206–50213. 2002 Dec 20. .
- VanDemark AP, Blanksma M, Ferris E, et al. The structure of the yFACT Pob3-M domain, its interaction with the DNA replication factor RPA, and a potential role in nucleosome deposition. Mol Cell. 2006 May 5;22(3): 363–74.
- Tsunaka Y, Toga J, Yamaguchi H, et al. Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J Biol Chem. 2009;284(36):24610–24621. 2009 Sep 4. .
- Stuwe T, Hothorn M, Lejeune E, et al. The FACT Spt16 “peptidase” domain is a histone H3-H4 binding module. Proc Natl Acad Sci U S A. 2008 Jul 1 105(26);8884–8889.
- Jiang H, Xu S, Chen Y, et al. The structural basis of human Spt16N-terminal domain interaction with histone (H3-H4)2 tetramer. Biochem Biophys Res Commun. 2019 Jan 15 508(3);864–870.
- Marciano G, Huang DT. Structure of the human histone chaperone FACT Spt16 N-terminal domain. Acta Crystallogr F Struct Biol Commun. 2016 Feb; 72(\(Pt 2)):121–128.
- Myers CN, Berner GB, Holthoff JH, et al. Mutant versions of the S. cerevisiae transcription elongation factor Spt16 define regions of Spt16 that functionally interact with histone H3. PLoS One. 2011;6(6):e20847. .
- Hondele M, Stuwe T, Hassler M, et al. Structural basis of histone H2A-H2B recognition by the essential chaperone FACT. Nature. 2013 Jul 4 499(7456);111–114.
- Tsunaka Y, Fujiwara Y, Oyama T, et al. Integrated molecular mechanism directing nucleosome reorganization by human FACT.. Genes Dev. 2016 Mar 15; 30(6):673–686.
- Winkler DD, Muthurajan UM, Hieb AR, et al. Histone chaperone FACT coordinates nucleosome interaction through multiple synergistic binding events.. J Biol Chem. 2011 Dec 2; 286(48):41883–41892.
- Kemble DJ, McCullough LL, Whitby FG, et al. FACT disrupts nucleosome structure by binding H2A-H2B with conserved peptide Motifs. Mol Cell. 2015 Oct 15; 60(2):294–306.
- Zunder RM, Antczak AJ, Berger JM, et al. Two surfaces on the histone chaperone Rtt106 mediate histone binding, replication, and silencing. Proc Natl Acad Sci U S A. 2012 Jan 17; 109(3):E144–53.
- Zhang W, Zeng F, Liu Y, et al. Crystal structure of human SSRP1 middle domain reveals a role in DNA binding. Sci Rep. 2015 Dec;21(5:):18688.
- Pfab A, Gronlund JT, Holzinger P, et al. The Arabidopsis histone chaperone FACT: role of the HMG-box domain of SSRP1. J Mol Biol. 2018 Aug 17; 430(17):2747–2759.
- Ikeda Y, Kinoshita Y, Susaki D, et al. HMG domain containing SSRP1 is required for DNA demethylation and genomic imprinting in Arabidopsis. Dev Cell. 2011 Sep 13 21(3);589–596.
- Cao S, Bendall H, Hicks GG, et al. The high-mobility-group box protein SSRP1/T160 is essential for cell viability in day 3.5 mouse embryos. Mol Cell Biol. 2003 Aug;23(15):5301–5307. .
- Chen P, Dong L, Hu M, et al. Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Mol Cell. 2018 Jul 19 71(2);284–93 e4.
- Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins.. Cell Mol Life Sci. 2007 Oct 19–20;64:2590–606.
- Safina A, Cheney P, Pal M, et al. FACT is a sensor of DNA torsional stress in eukaryotic cells. Nucleic Acids Res. 2017 Feb 28;45(4):–45.
- Valieva ME, Gerasimova NS, Kudryashova KS, et al. Stabilization of nucleosomes by histone tails and by FACT revealed by spFRET microscopy. Cancers (Basel). 2017 Jan 6 9(12);1.
- Brewster NK, Johnston GC, Singer RA. A bipartite yeast SSRP1 analog comprised of Pob3 and Nhp6 proteins modulates transcription. Mol Cell Biol. 2001 May;21(10):3491–3502.
- Brewster NK, Johnston GC, Singer RA. Characterization of the CP complex, an abundant dimer of Cdc68 and Pob3 proteins that regulates yeast transcriptional activation and chromatin repression.. J Biol Chem. 1998 Aug 21; 273(34):21972–21979.
- Ruone S, Rhoades AR, Formosa T. Multiple Nhp6 molecules are required to recruit Spt16-Pob3 to form yFACT complexes and to reorganize nucleosomes. J Biol Chem. 2003 Nov 14; 278(46):45288–45295.
- Formosa T, Eriksson P, Wittmeyer J, et al. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 2001 Jul 2; 20(13):3506–3517.
- Paull TT, Johnson RC. DNA looping by saccharomyces cerevisiae high mobility group proteins NHP6A/B. consequences for nucleoprotein complex assembly and chromatin condensation. J Biol Chem. 1995 Apr 14; 270(15):8744–8754.
- Stillman DJ. Nhp6: a small but powerful effector of chromatin structure in Saccharomyces cerevisiae. Biochim Biophys Acta. 2010 Jan-Feb;1799(1–2):175–180.
- Zheng S, Crickard JB, Srikanth A, et al. A highly conserved region within H2B is important for FACT to act on nucleosomes. Mol Cell Biol. 2014 Feb;34(3):303–314.
- McCullough LL, Connell Z, Xin H, et al. Functional roles of the DNA-binding HMGB domain in the histone chaperone FACT in nucleosome reorganization. J Biol Chem. 2018 Apr 20 293(16);6121–6133.
- Wang T, Liu Y, Edwards G, et al. The histone chaperone FACT modulates nucleosome structure by tethering its components. Life Sci Alliance. 2018 Aug;1(4):e201800107.
- Hsieh FK, Kulaeva OI, Patel SS, et al. Histone chaperone FACT action during transcription through chromatin by RNA polymerase II. Proc Natl Acad Sci U S A. 2013 May 7 110(19);7654–7659.
- Gupta K, Sari-Ak D, Haffke M, et al. Zooming in on transcription preinitiation. J Mol Biol. 2016 Jun 19; 428(12):2581–2591.
- Petrenko N, Jin Y, Dong L, et al. Requirements for RNA polymerase II preinitiation complex formation in vivo. Elife. 2019 Jan;25(8):
- Biswas D, Yu Y, Prall M, et al. The yeast FACT complex has a role in transcriptional initiation. Mol Cell Biol. 2005 Jul; 25(14):5812–5822.
- Mylonas C, Tessarz P. Transcriptional repression by FACT is linked to regulation of chromatin accessibility at the promoter of ES cells. Life Sci Alliance. 2018 Jun; 1(3):e201800085.
- Pathak R, Singh P, Ananthakrishnan S, et al. Acetylation-dependent recruitment of the FACT complex and its role in regulating pol II occupancy genome-wide in Saccharomyces cerevisiae. Genetics. 2018 Jul; 209(3):743–756.
- True JD, Muldoon JJ, Carver MN, et al. The modifier of transcription 1 (Mot1) atpase and spt16 histone chaperone co-regulate transcription through preinitiation complex assembly and nucleosome organization. J Biol Chem. 2016 Jul 15 291(29);15307–15319.
- Saunders A, Werner J, Andrulis ED, et al. Tracking FACT and the RNA polymerase II elongation complex through chromatin in vivo. Science. 2003 Aug 22;301(5636):1094–1096. .
- Pavri R, Zhu B, Li G, et al. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006 May 19 125(4);703–717.
- Mason PB, The SK. FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol Cell Biol. 2003 Nov; 23(22):8323–8333.
- Kaplan CD, Laprade L, Winston F. Transcription elongation factors repress transcription initiation from cryptic sites.Science. 2003 Aug 22;301(5636):1096–1099. .
- van Bakel H, Tsui K, Gebbia M, et al. A compendium of nucleosome and transcript profiles reveals determinants of chromatin architecture and transcription. PLoS Genet. 2013 May;9(5):e1003479.
- Schwabish MA, Struhl K. Evidence for eviction and rapid deposition of histories upon transcriptional elongation by RNA polymerase II. Mol Cell Biol. 2004 Dec; 24(23):10111–10117.
- Izban MG, Luse DS. Transcription on nucleosomal templates by RNA polymerase II in vitro: inhibition of elongation with enhancement of sequence-specific pausing. Genes Dev. 1991 Apr;5(4):683–696.
- Luse DS, Studitsky VM. The mechanism of nucleosome traversal by RNA polymerase II: roles for template uncoiling and transcript elongation factors. RNA Biol. 2011 Jul-Aug; 8(4):581–585.
- Orphanides G, Reinberg D. RNA polymerase II elongation through chromatin. Nature. 2000 Sep 28;407(6803):471–475. .
- Voth WP, Takahata S, Nishikawa JL, et al. A role for FACT in repopulation of nucleosomes at inducible genes. PLoS One. 2014;9(1):e84092.
- Xin H, Takahata S, Blanksma M, et al. yFACT induces global accessibility of nucleosomal DNA without H2A-H2B displacement. Mol Cell. 2009 Aug 14; 35(3):365–376.
- Valieva ME, Armeev GA, Kudryashova KS, et al. Large-scale ATP-independent nucleosome unfolding by a histone chaperone. Nat Struct Mol Biol. 2016 Dec 23(12);1111–1116.
- Birch JL, Tan BC, Panov KI, et al. FACT facilitates chromatin transcription by RNA polymerases I and III. EMBO J. 2009 Apr 8 28(7);854–865.
- Formosa T. The role of FACT in making and breaking nucleosomes. Biochim Biophys Acta. 2012 Mar; 1819(3–4):247–255.
- Cheung V, Chua G, Batada NN, et al. Chromatin- and transcription-related factors repress transcription from within coding regions throughout the Saccharomyces cerevisiae genome. PLoS Biol. 2008 Nov 11 6(11);e277.
- Hainer SJ, Charsar BA, Cohen SB, et al. Identification of mutant versions of the spt16 histone chaperone that are defective for transcription-coupled nucleosome occupancy in saccharomyces cerevisiae. G3-Genes Genomes Genet. 2012 May 1;2(5):555–567.
- Erkina TY, Erkine A. ASF1 and the SWI/SNF complex interact functionally during nucleosome displacement, while FACT is required for nucleosome reassembly at yeast heat shock gene promoters during sustained stress. Cell Stress Chaperones. 2015 Mar; 20(2):355–369.
- Feng JX, Gan HY, Eaton ML, et al. Noncoding transcription is a driving force for nucleosome instability in spt16 mutant cells. Mol Cell Biol. 2016 Jul 36(13);1856–1867.
- Gasparian AV, Burkhart CA, Purmal AA, et al. Curaxins: anticancer compounds that simultaneously suppress NF-kappaB and activate p53 by targeting FACT. Sci Transl Med. 2011 Aug 10;3(95):95ra. 74.
- Chang HW, Valieva ME, Safina A, et al. Mechanism of FACT removal from transcribed genes by anticancer drugs curaxins. Sci Adv. 2018 Nov;4(11):eaav2131. .
- Nesher E, Safina A, Aljahdali I, et al. Role of chromatin damage and chromatin trapping of FACT in mediating the anticancer cytotoxicity of DNA-binding small-molecule drugs. Cancer Res. 2018 Mar 15 78(6);1431–1443.
- Martin BJE, Chruscicki AT, Howe LJ. Transcription promotes the interaction of the facilitates chromatin transactions (FACT) complex with nucleosomes in Saccharomyces cerevisiae. Genetics. 2018 Nov;210(3):869–881.
- Kantidze OL, Luzhin AV, Nizovtseva EV, et al. The anti-cancer drugs curaxins target spatial genome organization. Nat Commun. 2019 Mar 29 10(1);1441.
- Tettey TT, Gao X, Shao W, et al. A role for FACT in RNA polymerase II promoter-proximal pausing. Cell Rep. 2019 Jun 25 27(13);3770–79 e7.
- Li Y, Zeng SX, Landais I, et al. SSRP1 has Spt16-dependent and -independent roles in gene transcription.. J Biol Chem. 2007 Mar 9; 282(10):6936–6945.
- Sims RJ, Millhouse S, Chen CF, et al. Recognition of trimethylated histone h3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007 Nov 30;28(4):665–676.
- Simic R, Lindstrom DL, Tran HG, et al. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003 Apr 15 22(8);1846–1856.
- Johnson JM, French SL, Osheim YN, et al. Rpd3-and Spt16-mediated nucleosome assembly and transcriptional regulation on yeast ribosomal DNA Genes. Mol Cell Biol. 2013 Jul 33(14);2748–2759.
- Mahapatra S, Dewari PS, Bhardwaj A, et al. Yeast H2A.Z, FACT complex and RSC regulate transcription of tRNA gene through differential dynamics of flanking nucleosomes. Nucleic Acids Res. 2011 May; 39(10):4023–4034.
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010 Oct 22; 40(2):179–204.
- Vitor AC, Huertas P, Legube G, et al. Studying DNA double-strand break repair: an ever-growing toolbox. Front Mol Biosci. 2020(7):24.
- Zhang L, Wang Z, Shi R, et al. RNF126 quenches RNF168 function in the DNA damage response. Genomics Proteomics Bioinformatics. 2018 Dec 16(6);428–438.
- Yang G, Chen Y, Wu J, et al. Poly(ADP-ribosyl)ation mediates early phase histone eviction at DNA lesions. Nucleic Acids Res. 2020 Apr 6 48(6);3001–3013.
- Krohn NM, Stemmer C, Fojan P, et al. Protein kinase CK2 phosphorylates the high mobility group domain protein SSRP1, inducing the recognition of UV-damaged DNA. J Biol Chem. 2003 Apr 11; 278(15):12710–12715.
- Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011 Mar;21(3):396–420.
- Turinetto V, Giachino C. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res. 2015 Mar 11; 43(5):2489–2498.
- Morrison AJ, Highland J, Krogan NJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004 Dec 17 119(6);767–775.
- Stucki M, Clapperton JA, Mohammad D, et al. MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell. 2005 Dec 29; 123(7):1213–1226.
- Kleiner RE, Verma P, Molloy KR, et al. Chemical proteomics reveals a gammaH2AX-53BP1 interaction in the DNA damage response. Nat Chem Biol. 2015 Oct; 11(10):807–814.
- Heo K, Kim H, Choi SH, et al. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol Cell. 2008 Apr 11;30(1):86–97.
- Piquet S, Le Parc F, Bai SK, et al. The histone chaperone FACT coordinates H2A.X-dependent signaling and repair of DNA damage. Mol Cell. 2018 Dec 6; 72(5):888–901e7.
- Meas R, Wyrick JJ, Smerdon MJ. Nucleosomes regulate base excision repair in chromatin. Mutat Res. 2019 Apr-Jun; 780::29–36.
- Wallace SS. Base excision repair: a critical player in many games. DNA Repair (Amst). 2014 Jul; 19:14–26.
- Moor NA, Lavrik OI. protein-protein interactions in DNA base excision repair. Biochemistry (Mosc). 2018 Apr;;83(4):411–422.
- Charles Richard JL, Shukla MS, Menoni H, et al. FACT assists base excision repair by boosting the remodeling activity of RSC. PLoS Genet. 2016 Jul 12(7);e1006221.
- Wienholz F, Zhou D, Turkyilmaz Y, et al. FACT subunit Spt16 controls UVSSA recruitment to lesion-stalled RNA Pol II and stimulates TC-NER. Nucleic Acids Res. 2019 May 7 47(8);4011–4025.
- Wright WD, Shah SS, Heyer WD. Homologous recombination and the repair of DNA double-strand breaks. J Biol Chem. 2018 Jul 6; 293(27):10524–10535.
- Kumari A, Mazina OM, Shinde U, et al. A role for SSRP1 in recombination-mediated DNA damage response. J Cell Biochem. 2009 Oct 1; 108(2):508–518.
- Oliveira DV, Kato A, Nakamura K, et al. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J Cell Sci. 2014 Feb 15 127(\(Pt 4));763–772.
- Nakamura K, Kato A, Kobayashi J, et al. Regulation of homologous recombination by RNF20-dependent H2B ubiquitination. Mol Cell. 2011 Mar 4 41(5);515–528.
- Moyal L, Lerenthal Y, Gana-Weisz M, et al. Requirement of ATM-dependent monoubiquitylation of histone H2B for timely repair of DNA double-strand breaks. Mol Cell. 2011 Mar 4 41(5);529–542.
- Serra-Cardona A, Zhang Z. Replication-coupled nucleosome assembly in the passage of epigenetic information and cell identity. Trends Biochem Sci. 2018 Feb;43(2):136–148.
- Schlesinger MB, Formosa T. POB3 is required for both transcription and replication in the yeast Saccharomyces cerevisiae. Genetics. 2000 Aug;155(4):1593–1606.
- Okuhara K, Ohta K, Seo H, et al. A DNA unwinding factor involved in DNA replication in cell-free extracts of Xenopus eggs. Curr Biol. 1999 Apr 8 9(7);341–350.
- Hertel L, De Andrea M, Bellomo G, et al. HMG protein T160 colocalizes with DNA replication foci and is down-regulated during cell differentiation. Exp Cell Res. 1999 Aug 1; 250(2):313–328.
- Abe T, Sugimura K, Hosono Y, et al. The histone chaperone facilitates chromatin transcription (FACT) protein maintains normal replication fork rates. J Biol Chem. 2011 Sep 2 286(35);30504–30512.
- Wittmeyer J, Formosa T. The Saccharomyces cerevisiae DNA polymerase alpha catalytic subunit interacts with Cdc68/Spt16 and with Pob3, a protein similar to an HMG1-like protein. Mol Cell Biol. 1997 Jul;17(7):4178–4190.
- Wittmeyer J, Joss L, Formosa T. Spt16 and Pob3 of Saccharomyces cerevisiae form an essential, abundant heterodimer that is nuclear, chromatin-associated, and copurifies with DNA polymerase alpha. Biochemistry. 1999 Jul 13; 38(28):8961–8971.
- Han J, Li Q, McCullough L, et al. Ubiquitylation of FACT by the cullin-E3 ligase Rtt101 connects FACT to DNA replication. Genes Dev. 2010 Jul 15; 24(14):1485–1490.
- Nune M, Morgan MT, Connell Z, et al. FACT and Ubp10 collaborate to modulate H2B deubiquitination and nucleosome dynamics. Elife. 2019 Jan;25:8.
- Tan BC, Chien CT, Hirose S, et al. Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 2006 Sep 6;25(17):3975–3985. .
- Zhai Y, Li N, Jiang H, et al. Unique roles of the non-identical MCM Subunits in DNA replication licensing. Mol Cell. 2017 Jul 20; 67(2):168–179.
- Kurat CF, Yeeles JTP, Patel H, et al. Chromatin controls DNA replication origin selection, lagging-strand synthesis, and replication fork rates. Mol Cell. 2017 Jan 5; 65(1):117–130.
- Yang J, Zhang X, Feng J, et al. The Histone chaperone FACT contributes to DNA replication-coupled nucleosome assembly. Cell Rep. 2016 Sep 20 16(12);3414.
- Foltman M, Evrin C, De Piccoli G, et al. Eukaryotic replisome components cooperate to process histones during chromosome replication. Cell Rep. 2013 Mar 28 3(3);892–904.
- Zhang G, Pradhan S. Mammalian epigenetic mechanisms. IUBMB Life. 2014 Apr; 66(4):240–256.
- Lai WKM, Pugh BF. Understanding nucleosome dynamics and their links to gene expression and DNA replication. Nat Rev Mol Cell Biol. 2017 Sep; 18(9):548–562.
- Fleming AB, Kao CF, Hillyer C, et al. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell. 2008 Jul 11; 31(1):57–66.
- Murawska M, Schauer T, Matsuda A, et al. The chaperone FACT and histone H2B ubiquitination maintain S. pombe genome Architecture through genic and subtelomeric functions. Mol Cell. 2020 Feb 6 77(3);501–13e7.
- Hodges AJ, Gloss LM, Wyrick JJ. Residues in the nucleosome acidic patch regulate histone occupancy and are important for FACT binding in Saccharomyces cerevisiae. Genetics. 2017 Jul; 206(3):1339–1348.
- Carvalho S, Raposo AC, Martins FB, et al. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013 Mar 1 41(5);2881–2893.
- McCullough LL, Pham TH, Parnell TJ, et al. Establishment and maintenance of chromatin architecture are promoted independently of transcription by the histone chaperone FACT and H3-K56 acetylation in Saccharomyces cerevisiae. Genetics. 2019 Mar 211(3);877–892.
- Begum NA, Stanlie A, Nakata M, et al. The histone chaperone Spt6 is required for activation-induced cytidine deaminase target determination through H3K4me3 regulation.. J Biol Chem. 2012 Sep 21; 287(39):32415–32429.
- Jeronimo C, Poitras C, Robert F. Histone recycling by FACT and Spt6 during transcription prevents the scrambling of histone modifications. Cell Rep. 2019 Jul 30; 28(5):1206–18e8.
- Li Y, Keller DM, Scott JD, et al. CK2 phosphorylates SSRP1 and inhibits its DNA-binding activity. J Biol Chem. 2005 Mar 25; 280(12):11869–11875.
- Mayanagi K, Saikusa K, Miyazaki N, et al. Structural visualization of key steps in nucleosome reorganization by human FACT. Sci Rep. 2019 Jul 15 9(1);10183.
- Luke B, Versini G, Jaquenoud M, et al. The cullin Rtt101p promotes replication fork progression through damaged DNA and natural pause sites. Curr Biol. 2006 Apr 18 16(8);786–792.
- de Vivo A, Sanchez A, Yegres J, et al. OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res. 2019 Jan 25; 47(2):729–746.
- Sanchez A, De Vivo A, Uprety N, et al. BMI1-UBR5 axis regulates transcriptional repression at damaged chromatin. Proc Natl Acad Sci U S A. 2016 Oct 4;113(40):11243–11248.
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development..Science. 2001 Aug 10;293(5532):1089–1093. .
- Wang JH, Li Y, Deng SL, et al. Recent research advances in mitosis during mammalian gametogenesis. Cells. 2019 Jun 10; 8(6):567.
- Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2014 Feb;15(2):69–81.
- Nashun B, Hill PW, Hajkova P. Reprogramming of cell fate: epigenetic memory and the erasure of memories past. EMBO J. 2015 May 12; 34(10):1296–1308.
- Shen Z, Formosa T, Tantin D. FACT inhibition blocks induction but not maintenance of pluripotency.. Stem Cells Dev. 2018 Dec 15; 27(24):1693–1701.
- Koche RP, Smith ZD, Adli M, et al. Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell. 2011 Jan 7 8(1);96–105.
- Ding J, Xu H, Faiola F, et al. Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 2012 Jan; 22(1):155–167.
- Pardo M, Lang B, Yu L, et al. An expanded Oct4 interaction network: implications for stem cell biology, development, and disease. Cell Stem Cell. 2010 Apr 2;6(4):382–395.
- Strebinger D, Deluz C, Friman ET, et al. Endogenous fluctuations of OCT4 and SOX2 bias pluripotent cell fate decisions. Mol Syst Biol. 2019 Sep; 15(9):e9002.
- Garcia H, Fleyshman D, Kolesnikova K, et al. Expression of FACT in mammalian tissues suggests its role in maintaining of undifferentiated state of cells. Oncotarget. 2011 Oct;2(10):783–796. .
- Shakya A, Callister C, Goren A, et al. Pluripotency transcription factor Oct4 mediates stepwise nucleosome demethylation and depletion. Mol Cell Biol. 2015 Mar 35(6);1014–1025.
- Hossan T, Nagarajan S, Baumgart SJ, et al. Histone chaperone SSRP1 is essential for Wnt signaling pathway activity during osteoblast differentiation. Stem Cells. 2016 May 34(5);1369–1376.
- Suggs BZ, Latham AL, Dawes AT, et al. FACT complex gene duplicates exhibit redundant and non-redundant functions in C. elegans. Dev Biol. 2018 Dec 15; 444(2):71–82.
- Ferri F, Petit V, Barroca V, et al. Interplay between FACT subunit SPT16 and TRIM33 can remodel chromatin at macrophage distal regulatory elements. Epigenetics Chromatin. 2019 Jul 22; 12(1):46.
- Koltowska K, Apitz H, Stamataki D, et al. Ssrp1a controls organogenesis by promoting cell cycle progression and RNA synthesis. Development. 2013 May;140(9):1912–1918. .
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008 Jul 11; 134(1):162–174.
- Garcia H, Miecznikowski JC, Safina A, et al. Facilitates chromatin transcription complex is an “accelerator” of tumor transformation and potential marker and target of aggressive cancers. Cell Rep. 2013 Jul 11 4(1);159–173.
