Abstract
Circular RNAs (circRNAs) are long, non-coding RNAs that result from the non-canonical splicing of linear pre-mRNAs. However, the characteristics and the critical role of circRNA in co-/post-transcriptional regulation were not well recognized until the “microRNA sponge” function of circRNA is discovered. Recent studies have mainly been devoted to the function of the circular RNA sponge for miR-7 (ciRS-7) and sex-determining region Y (SRY) by targeting microRNA-7 (miR-7) and microRNA-138 (miR-138), respectively. In this review, we illustrate the specific role of circRNAs in a wide variety of cancers and in regulating the biological behavior of cancers via miR-7 or miR-138 regulation. Furthermore, circRNA, together with its gene silencing ability, also shows its potential in RNA interference (RNAi) therapy by binding to target RNAs, which provides a novel perspective in cancer treatment. Thus, this review concerns the biogenesis, biological function, oncogenesis, progression and possible therapies for cancer involving circRNAs.
Introduction
Circular RNAs (circRNAs) are thought to arise from non-canonical splicing of linear pre-mRNAs. CircRNAs are long non-coding RNAs (lncRNAs) with circular forms and are widely observed in virus-like infectious particles, such as viroids, circular satellite viruses, or hepatitis delta virus.Citation1–3 Seemingly the result of mistaken RNA splicing, the crucial roles and functions of circRNAs have not be well recognized until very recently and such discovery have permanently altered our perspectives toward cancer, especially in carcinogenesis and cancer progression.Citation4–6 To date, investigators have discovered 2 major kinds of circRNAs, the intronic and exonic circRNAs. Either containing introns or exons, both types of circRNAs can perform a variety of functions.
Several experiments have been performed to determine the specific function(s) of circRNAs. The discovery of the RNA sponge function involved in carcinogenesis and the malignant behavior of cancer has given us a novel recognition concerning the molecular mechanism in cancer. In this review, we concentrate mainly on demonstrating the unique role of circRNAs in the process of carcinogenesis, as well as the relationship between circRNAs and the progression, deterioration, migration and recurrence of cancer. In addition, the improvement in our understanding of circRNAs may provide potential targets for the treatment of cancer, which might lead to novel transcription therapeutic approaches to suppress or regulate the malignant behavior of cancer cells.
Classification of circRNAs
CircRNAs are a small group of competing endogenous RNAs (ceRNAs) in the large family of non-coding RNAs (ncRNAs).Citation7–9 Different from other RNAs, the ncRNAs demonstrate their indispensable biological function mostly in gene regulation during differential process, tumorigenesis and tumor progression.Citation10 According to the length of non-coding RNAs, several subgroups have been classified, including short ncRNAs, mid-size ncRNAs and long ncRNAs. Long ncRNAs (>200 nucleotides in size) comprise long intervening non-coding RNA (lincRNA), transcribed ultraconserved regions (T-UCRs) and ceRNAs.Citation11
So far our understanding of the biological characteristics and function of circRNAs is limited; therefore, classification of circRNAs can only be achieved by focusing on their components. Two types of circRNAs are discussed, intronic circRNAs and exonic circRNAs. Intronic circRNAs are produced by connecting 2 or more introns, which is rare in eukaryotic cells. Correspondently, exonic circRNAs are large molecules comprising exons and are considered as by-products of exon skipping, either in pre-mRNA splicing or in mature mRNA re-splicing.Citation12
Biogenesis and function
The biogenesis of circRNA
With their significant role in cancer, circRNAs are widely explored in different aspects. And exploration in their biogenesis has profoundly changed our perspectives in gene transcription. Generally, as a result of canonical linear mRNA splicing, circRNAs are regarded as co-transcriptional products, and this process shows a strong dependence on short intronic repeat sequences.Citation13 In eukaryotic cells, precursor mRNAs (pre-mRNAs), comprising exons as well as introns, are first synthesized in the nucleus in the pattern of a radial arrangement, mostly by RNA polymerase II from DNA.Citation14 Pre-mRNAs then move to the cytoplasm, where they are precisely dissected into pieces containing one complete isolated intron or exon, which is catalyzed by spliceosomal machinery or by groups I and II ribozymes (only responsible for the production of intronic circRNAs). Subsequently, under the catalysis of a spliceosomal ribozyme, all the exonic circRNAs and part of intronic circRNAs are synthesized. The exons in pre-mRNAs that possess flanking intronic complementary sequences may present a preference for the 3′ end of an upstream exon, which results in the circularization of exons.Citation15 This unique sequence-dependent feature is essential for the co-transcriptional biogenesis of circRNAs, because the production rate and the balance between splicing and circularization are also determined by the intronic sequences.Citation16,17 The rate can be elevated by binding to muscle blind-like (MBL/MBNL1) protein, an RNA binding protein (RBP) that stimulates circRNA biogenesis, or by binding to several specific repeated RNA sequences such as poly (CUG) and poly (CCUG).Taking the Alu repeats as an example, they are found throughout the genome and thought to actively participate in circRNAs genesis. Such repeats retro-transpose by hijacking a reverse transcriptase and an endonuclease from autonomous retro-transposons for re-insertion into the genome.Citation18 A recent study indicated that the majority of circRNAs are formed under the regulation of human epithelial mesenchymal transition (EMT), and over one-third of abundant circRNAs are dynamically regulated by the alternative splicing factor, Quaking (QKI), which is regulated during the EMT process.Citation19 Given the fact that EMT participates in the tumorigenesis and provide important indications for cancer progression, we may get inspirations on the therapeutic role of circRNA by targeting microRNA involved in EMT.Citation20,21
Meanwhile, intronic circRNAs are produced when lariats that escape the debranching enzymes are processed by exonucleases. The formation of circular intronic RNAs (ciRNAs) can be recapitulated using expression vectors. This procedure depends on a consensus motif containing a 7 nt GU-rich element near the 5′ splice site and an 11 nt C-rich element close to the branchpoint site.Citation22 And the integral production rate of circRNA in vivo can be enhanced using the original permuted intron-exon (PIE) method by inducing an induction-free vector system utilizing the highly efficient constitutive lipoprotein (lpp) promoter.Citation23
The function of circRNAs
Different from the majority of RNAs that aid the process of translation, circRNAs often have role in controlling gene expression. And the abnormality of gene expression and biological behavior observed in cancer cells may be considered the result of deregulated circRNA function.Citation11,38 Interestingly, the versatility of circRNA expression mainly lies in post-transcriptional regulation, which is frequently achieved by RNA-RNA interactions.Citation24 CircRNAs, together with some other ceRNAs, are believed to serve as microRNA reservoirs, but with much more binding sites, which enable the circRNAs to specialize in harboring microRNAs.Citation25
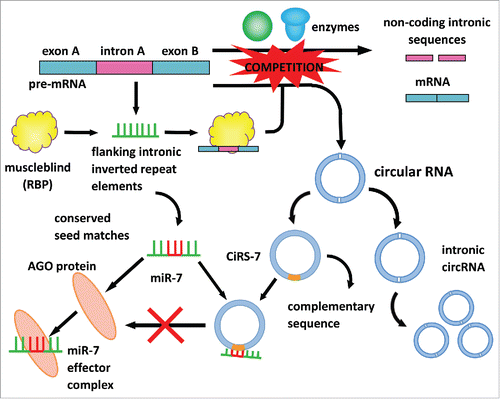
Exonic circRNAs are believed to possess co-transcriptional regulatory potency and have mainly been investigated in neuronal tissues so far.Citation26 Memczak demonstrated that a human circRNA, ciRS-7 transcript (antisense to the cerebellar degeneration-related protein 1), also known as ciRS-7, is densely bound by argonaute proteins (AGO).Citation7 Also, ciRS-7 acts as a miR-7 regulator by binding densely to the microRNA effector complexes via its conserved binding sites. The combination of ciRS-7 and miR-7 enables the circRNA ciRS-7 to inactivate the functional site of the microRNA, which is also known as “microRNA sponge”.Citation27,28 The overlapping co-expression of ciRS-7 and miR-7 in mouse brain revealed the endogenous interaction between ciRS-7 and miR-7 in neocortical and hippocampal neurons. Besides, testis-specific circRNAs were observed as a miR-138 sponge, with its sex-determining region Y (SRY) revealing the generality of microRNA sponge effects achieved by circRNA formation.Citation8 And since both miR-7 and SRY have a crucial impact in the occurrence and progression of cancer, we assume that circRNA must be densely involved in such process as well.
In addition to exonic circRNAs, intronic circRNAs show unique roles as well. Investigators found that intronic circRNAs can be duplicated in plasma and directly passed on to off-spring via mitosis, which hinted at the potential roles of intronic circRNAs in epigenetics and RNA-mediated inheritance.Citation29 Given the fact that the introns in the linear form are engaged in the mechanism to minimize tissue-inappropriate gene expression, the biological function of intronic circRNAs is close to being discovered in the near future. Citation30
Beyond regulating microRNAs, circRNAs may bind to and sequester RNA-binding proteins (RBPs) via their conserved seed matches, resulting in the formation of large RNA-protein complexes. By detecting the covalent UV crosslinking between RNAs and RBPs, Memczak et al. and Hansen et al. proved the existence of the ribonucleoprotein (RNP) complex in plasma and further proved that the RNP complex was synthesized by combining circRNAs with miR-7 and the AGO protein. Generally, RBPs can determine the destiny of RNAs, from synthesis to decay.Citation31 Meanwhile, circRNAs may also function to store, sort or localize RBPs.Citation32 However, further exploration is still needed to unveil the roles of circRNA-RBP interaction and the functions of circRNA-RBP complexes ().
Figure 1. The biogenesis and the microRNA-sponge function of circular RNAs In eukaryotic cells, pre-mRNA comprises intermittently arranged exons and introns. Typically, under the catalysis of spliceosomal machinery or groups I and II ribozymes, exons are spliced out from introns and rejoined into mature mRNAs, while introns are metabolized in cytoplasm. On certain occasions, pre-mRNA with flanking intronic inverted repeat elements can bind with muscleblind, a type of RNA binding protein (RBP), and form a pre-mRNA-RBP complex. The pre-mRNA-RBP complex competes with pre-mRNAs as the catalytic site of spliceosomal machinery or groups I and II ribozymes and consequently is catalyzed into circular RNAs (circRNAs), including intronic circRNA and exonic circRNA. Intronic circRNA exists in the cytoplasm and can are passed on to offspring, affecting RNA-mediated inheritance and epigenetics. Exonic circRNAs function as microRNA sponges in the cytoplasm, regulating the function of microRNAs. For example, miR-7 exerts its biological function by binding with the AGO protein to form an miR-7 effector complex. A circular RNA sponge for miR-7 (ciRS-7) can harbor the conserved seed matches of miR-7 in its complementary sequence, which inhibits the formation of the miR-7 effector complex. Therefore, the function of miR-7 is regulated by ciRS-7.
Theoretically, circRNA may encode and be translated to protein by eukaryotic ribosomes. However, researchers so far have demonstrated that only the synthesized, internal ribosome entry site (IRES) element-containing circRNA could direct the synthesis of a defined protein in vitro, albeit less efficiently compared with the linear RNAs.Citation33 Although synthetic circRNAs containing IRES elements can be translated by ribosomes in eukaryotic cells in vitro, there's no concrete evidence that natural circRNAs in eukaryotic cells contain IRES elements and therefore are not likely to direct protein synthesis.
Roles of circRNAs in the oncogenesis and the malignant behavior of cancer cells
Some endogenous circRNAs have been revealed in mammalian cells and they can be highly abundant and evolutionarily conserved in oncogenesis. To date, the evidence suggests that circRNAs may regulate transcription and pathways by manipulating microRNAs.Citation34,35 The resulting microRNA profiles can act as a distinctive cursor in dividing the induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs); the state of pluripotent stem cell subdivision is of great potential in distinguishing cancer cells from differentiated cells in a p53 network status-related manner.Citation36 Besides, a negative correlation between global circular RNA abundance and proliferation has been confirmed. A circular RNA encoded as Hsa_circ_002059 has already been determined to be significantly downregulated in gastric cancer.Citation37,38 In addition, several circRNAs are considered responsible for the malignant biological behavior of cancer cells. Thus, in this review, we mainly discuss the roles of circRNAs in cancer tissues by engaging in regulation of microRNA. Hopefully, we will further understand the underlying mechanism of malignant biological behavior of cancer cells and improve the diagnosis and prevention of oncogenesis.
CircRNA participates in the occurrence and malignant behavior of cancer cells by regulating the function of miR-7
MicroRNAs are critical components in eukaryotic cell plasma for fine-turning gene expression, playing oncogenic roles or performing tumor-suppressing functions. The changes in the levels of a series of microRNAs in cancer cells are considered to be distinctive. According to the up/downregulation of microRNAs in plasma, microRNAs are divided into 2 types: tumor suppressive microRNAs (tumor-suppressor-miRs), which are usually downregulated in alimentary cancer cells; and oncogenic microRNAs (oncomiRs), which are usually upregulated. For example, the expressions of miR-222–221 and miR-106b-25 clusters are elevated in gastric cancer, resulting in the suppression of p21 family of cyclin-dependent kinase (CDK) inhibitors (p57KIP2, p21CIP1 and p27KIP1) causing the impaired cell cycle. Meanwhile, miR-125a is reduced in gastric cancer cells, leading to the suppression of CDK6, together with reduced cell growth and lack of apoptosis.Citation39–41 Meanwhile, certain microRNAs contain several conserved binding sites for circRNAs, which hint at a possible role of circRNAs in controlling the biological function of microRNAs. Within ncRNA-mediated networks, the crosstalk and competence between circRNAs and microRNAs is the key to our understanding of carcinogenesis.
The novel circular transcript ciRS-7 is confirmed to bind selectively to miR-7 and, with the help of the AGO protein, inhibit the biological function of miR-7.Citation8 At the same time, microRNA-7 regulates epidermal growth factor receptor (EGFR) signaling in human cancer cells. According to bioinformatic prediction, miR-7 functions to reduce EGFR mRNA and protein expression mainly by acting at 2 of the 3 target sites in the human EGFR mRNA 3′-untranslated region.Citation42 Furthermore, miR-7 inhibits EGFR signaling by attenuating the transcription of Raf1 and other genes, including PSME3, PLEC1, CKAP4, CNOT8, CNN3, CAPZA1, PFN2 and ARF4. MiR-7 also suppresses the effector of EGFR signaling by inactivating protein kinase B (PKB/Akt) and extracellular signal-regulated kinase 1/2 (ERK1/2).Citation43–45 Therefore, the regulatory function of circRNAs via their influence on miR-7 is clear: overexpression of ciRS-7 could act as a microRNA sponge to restrain the expression miR-7 and therefore elevate EGFR expression. EGFR overexpression is associated with disease advancement, resistance to chemotherapy and radiation therapy, and poor prognosis, and is closely observed in a series of solid tumors, including cancer stem-like cells (CSC), gastric cancer, hepatocellular carcinoma and colorectal carcinoma.Citation45–47
In gastric cancer, miR-7 effectively binds to an insulin-like growth factor-1 receptor (IGF1R) oncogene, leading to the inhibition of IGF1R expression. The reduced expression of the EGF1R oncogene inhibits the function of E-cadherin suppressor Snail, increases E-cadherin expression and partially reverses the EMT. As a result, cancer cell metastasis and proliferation is blocked, delaying the process of cell growth and tumor invasion. Therefore, downregulation of miR-7 by ciRS-7 results in disinhibition of the IGF1R oncogene and induces enhanced cell migration and multiplication.Citation48
In hepatocellular carcinoma (HCC), miR-7 is decreased in HCC cell plasma and several studies have confirmed its outstanding role in cell cycle regulation.Citation42–44 MiR-7 directly silences the cyclin E1 (CCNE1) gene, and CCNE1 forms a complex with and functions as a regulatory subunit of CDK2, which is required for G1 to S transition.Citation49 Therefore, the overexpression of miR-7 arrests the cell cycle in HCC. In addition, miR-7 binds at the 3′UTR of CUL5, inhibiting colony formation and inducing G1/S phase arrest.Citation50 Further study found that miR-7 directly targets phosphoinositide 3-kinase catalytic subunit delta (PIK3CD) and 2 novel, putative genes: mTOR and p70S6K. As a result, PIK3CD, the mammalian target of rapamycin (mTOR) and p70S6K, are all repressed by miR-7 overexpression. It is suggested that miR-7 might inhibit cell growth by arresting the G0/G1-phase of the cell cycle and impairing cell migration in the PI3K/Akt/mTOR-signaling pathway.Citation45
As a microRNA with tumor suppression for colorectal cancer (CRC), miR-7 has been proved to suppress the expression of the oncogenic Yin Yang 1 (YY1) mRNA, which inhibits p53, modulates the downstream effectors p15, caspase cascades, C-Jun, and activates the Wnt signaling pathway through activating β-catenin, anti-apoptotic survivin and fibroblast growth factor 4 (FGF-4).Citation51,52 MicroRNA expression arrays showed that miR-7 is associated with prognosis as well as cetuximab sensitivity in CRC, and these phenomena are achieved in an EGFR related manner.Citation53 Hence, miR-7 can suppress colon cancer cell proliferation, induce apoptosis and increase sensitivity to cetuximab.
Except for alimentary system carcinoma, miR-7 is also associated with diversiform cancers. IMiR-7 inhibits tumor angiogenesis and growth in murine xenograft glioblastoma by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Microarrays showed that miR-7 inhibits multiple oncogenic pathways to suppress HER2-mediated breast cancer and reverse trastuzumab resistance in vitro.Citation54–56 Moreover, miR-7 is reported to inhibit cell growth and invasion in pancreatic cancer cells, and could sensitize non-small cell lung cancer cells to paclitaxel.Citation57,58 A study in cervical cancer found that miR-7 inhibited the metastasis and invasion of cervical cancer by targeting focal adhesion kinase (FAK) at the transcriptional and translational levels.Citation59
Biological functions of circRNAs in cancer by manipulating miR-138
Huge efforts have been made to determine the possible role of microRNAs in their interactions with targets to shape the cellular protein landscape during post-transcriptional and translational control.Citation60,61 Not only miR-7, but also other microRNAs, such as SRY, showed significant effects on the occurrence of alimentary tumor. However, because our understanding of circRNAs is limited, other circRNAs that can function as microRNAs may yet be discovered.
SRY is the circular, testis-specific sense transcript of the male sex-determining gene.Citation62,63 As a microRNA sponge, SRY functions to control the biological effects of miR-138 by binding to its 16 conserved binding sites. In cholangiocarcinoma, SRY suppresses the expression of miR-138, leading to reduced mRNA and protein expression of the Ras homolog gene family, member C (RhoC). As a result, the RhoC/p-ERK/MMP-2/MMP-9 pathway is upregulated, which causes enhanced proliferation, migration and invasion of cholangiocarcinoma cells.Citation64 In addition, miR-138 could target directly the Twist basic helix-loop-helix transcription factor 2 gene (TWIST2) and reduce its function, which inhibits migration and invasion in colorectal cancer.Citation65 Besides, miR-138 is ovarian cancer-specific, where it functions to inhibit ovarian cancer cell growth by blocking the proliferating cell nuclear antigen (PCNA) and B-cell lymphoma-2 (Bcl−2) pathway. MiR-138 also suppresses invasion and metastasis by targeting SRY-related high mobility group box 4 (SOX4) and hypoxia-inducible factor-1a (HIF-1a). Suppressing miR-138 aggravates the metastasis of ovarian cancer via activation of the LIM kinase 1 (Limk1)/cofilin signaling pathway.Citation66–68 By targeting histone family 2A variant, member X (H2AX), miR-138 is essential to regulate the DNA damage response in small cell lung cancer (SCLC) cells, the upregulation of which is responsible for the increased radiosensitivity of lung cancer.Citation69,70
MiR-138 plays roles not only in solid tumors, but also in chronic lymphocytic leukemia (CLL). A previous study demonstrated that miR-138 is significantly downregulated in CLL, and is associated with resistance toward CD95-mediated apoptosis. Valeska et al. then showed that miR-138 could control palmitoylation-dependent CD95-mediated cell death by targeting acyl protein thioesterase (APT) 1 and 2.Citation71
As a newly discovered, co-transcriptional as well as post-transcriptional, regulator, most research effort has been devoted to ciRS-7, SRY and their downstream targets, miR-7 and miR-138, respectively. Nevertheless, attempts to identify other circRNAs that act as microRNA sponges are ongoing. Moreover, previous studies have demonstrated that many microRNAs are associated with alimentary system carcinogenesis. Thousands of additional circRNAs with unknown functions have been identified in various species. Considering the titanic number of microRNA and circRNA species, we are confident in finding other circRNAs that functions to regulate corresponding microRNA and participate in different aspect of cancer.
Possible therapies targeting circRNA
Recent studies have shown the associations of numerous novel molecules and signal pathways with oncogenesis or in the control of the malignant behavior of cancer cells, both of which are potential targets for new drugs that improve the conditions of cancer patients. CircRNA increases in the process of cell differentiation to capture the exceedingly high levels of microRNAs in stem cells and also benefits the clean up of the opposite strands of mature microRNAs. The process is surprisingly extensive, and a sudden aberration in microRNA level is usually connected with the change of circRNA level and associated with proliferation, differentiation and carcinogenesis process. Meanwhile, microRNAs are a large group of RNAs with the ability to suppress the expression of a target mRNA, the mechanism of which lies in the AGO-mediated endonucleolytic mRNA cleavage, triggered by complementarity between mRNAs and microRNAs.Citation72 Thus, possible therapies may be achieved by manipulating specific microRNAs and further regulate the function of cell.
Several microRNAs can increase the sensitivity to radiosensitivity for human cancers. Taking miR-7 as an example, knockdown of ciRS-7, or overexpression of a microRNA previously shown to cleave ciRS-7 (i.e. miR-671), may decrease the expression of miR-7 target genes. At the same time, ciRS-7 prevented the knockdown of miR-7 targets.Citation9 This suggested that the potential of targeting microRNA-7 for overcoming radio-resistance of cancer cells is activated by EGFR-associated signaling.Citation73 Similarly, tumor-suppressing function is also observed in miR-138.Citation71,74 Therefore, SYR, another circRNA known to target and eliminate miR-138, may provide another promising approach toward cancer.
Considering the different changes of microRNA level observed in carcinogenesis and cancer progression, we figured out 2 main potential approaches to facilitate the treatment toward cancer. One potential treatment for cancer is achieved mainly by targeting the upregulated microRNAs in cancer cells by mimicking the structure of circRNAs to act as an artificial microRNA sponge. Another way to prevent or suppress the happening and the progression of cancer is to enhance the expression and the biological activity of microRNA that is reduced in cancer. And the basic principle for this approach is the blockage of the microRNA-circRNA combination by saturating the conserved binding sites on circRNA. Using plasmid or viral vectors, therapists could accomplish intracellular expression of circRNA-containing complementary microRNA target sequences and saturate the regulation ability of the targeted microRNA. Therefore, doctors may achieve the regulation of microRNAs in cancer cells and hopefully bring a novel perspective to cancer treatment.Citation75
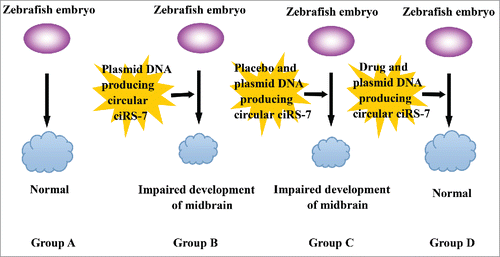
Also, we may get hints on the screening of drugs targeting miR-7/ciRS-7 interaction with the help of zebrafish. Zebrafish is a widely used animal model for the screening of anti-cancer drugs. And the hypothesis relies on the evidence that miR-7 is usually decreased in the event of carcinogenesis and progression and that transgenic expression of mouse ciRS-7 in zebrafish embryos lacking endogenous ciRS-7 substantially reduced the midbrain size and mimicked the phenotype of morpholino knockdown of miR-7 in zebrafish.Citation76,77 By determining the level of miR-7, the therapeutic potential of medication could be assessed. Therefore, a similar system to evaluate the possibility and efficacy of a certain treatment for tumor by targeting miR-7 could be set up. By injecting medications to be evaluated into a zebrafish embryo untreated and treated with plasmid DNA producing circular ciRS-7 previously, the effect of drugs could be performed. Specifically, drugs acting to block “miR-7 sponge” induced by circRNAs are expected to induce improved or normal midbrain development compared with the placebo. ()
Figure 2. The application of zebrafish in drug screening CiRS-7 is associated with impaired midbrain development in zebrafish and can reduce the biological function of miR-7, leading to oncogenesis and a series of malignant behaviors of alimentary cancer. Therefore, drugs that can downregulate ciRS-7 would be promising for the treatment of cancer. The size of the zebrafish midbrain in group A was achieved when cultured with no other influencing factors applied and the size is recorded as the control. In group B, plasmid DNA producing circular ciRS-7 was induced in the culture of zebrafish embryos and the size of the midbrain is smaller than normal, suggesting impaired midbrain development. Upon injecting placebo and plasmid DNA producing circular ciRS-7 in group C, the size of the zebrafish midbrain remained impaired. By applying a drug and plasmid DNA producing circular ciRS-7 in group D, the impaired midbrain development recovered to the normal size, suggesting that the drug is effective in downregulating the function of ciRS-7 and may be useful to treat cancer.
Similarly, such hypothesis could also be applied in the search of inhibitors for cancer metastasis, invasiveness and reoccurrence. Other possible treatment includes searching for drugs that can target and influence the stability of “microRNA-circRNA sponge”, control the expression of circRNAs and affect circRNA degradation etc.
Discussion and perspectives
CircRNAs are a large class of RNAs with as-yet-undetermined functions. Instead of being the products of aberrant splicing events, circRNAs are far more complicated previously considered and play essential and distinctive roles in, for example, alimentary cancers.Citation32,78
Taken together, as non-classical products of transcription, circRNAs are thought to be synthesized co-transcriptionally in the nucleus and are modified in the cytoplasm. CircRNAs can selectively bind to target microRNAs in a sequence specific manner to form the circRNA-RBP complexes that block the functions of microRNAs. Such a biological function is termed a “microRNA sponge” and this negative control of microRNAs can reduce the invasiveness, metastasis and proliferation of alimentary cancers. The expression level of microRNAs is regulated by circRNA; therefore, circRNAs are thought to play an important role in the oncogenesis and malignant behavior of alimentary system cancer. CiRS-7 and SRY bind to the conserved seed matches in miR-7 and miR-138, respectively, to form microRNA sponges that block the functions of miR-7 and miR-138.
Nevertheless, as circRNAs are a newly discovered type of ncRNA, the precise mechanism of reorganizing microRNAs to form circRNAs, the pathway by which circRNAs exert their effects, the role of circRNAs in alimentary carcinogenesis and the possible therapeutic effects of circRNA to treat cancer remain unclear. So far, several theories regarding the biogenesis of circRNAs have been proposed, which explain part of the enigma. Yet there are large numbers of circRNAs whose synthesis mechanism in eukaryotic cell plasma is yet to be fully recognized. Recent studies have shown that several circRNAs exert their effect by forming circRNA-RBP complexes. However, it remains unclear whether other circRNAs or RBPs participate in co-/post-transcription regulation and which pathways are affected by the circRNA-protein complexes. Moreover, only a few circRNAs (i.e., ciRS-7 and SRY) and their association with the occurrence and malignant behavior of alimentary carcinoma are well demonstrated. Considering the huge amount of circRNAs identified, it is possible that certain other types of circRNAs may be confirmed as alimentary carcinoma biomarkers for the early diagnosis and for the prediction of prognosis. In addition, there is great opportunity to identify other circRNAs that function as microRNA sponges in regulating the metastasis, proliferation, invasion and recurrence of alimentary carcinoma.
The functions of circRNAs are providing investigators with a new rote to treat alimentary system cancer. Utilizing the therapeutic effects of microRNA sponges, several potential therapies concerning the regulation of circRNAs and their downstream microRNAs have been proposed. CircRNAs also have the quality to perform gene silencing function, and there is no doubt that circRNAs have potential RNAi functions and that remodeling of the circRNA-microRNA balance might be useful to treat cancer.
In conclusion, circRNAs are a large group of competing endogenous RNAs, with regulatory potency that function as RNA sponges. By binding to the conserved seed matches of microRNAs, circRNAs can affect the transcription and the expression of genes. Dysfunction or changes in circRNAs in cells is considered to be a sign of malignant degeneration. Using their characteristics, circRNAs are proving to be potential targets for the treatment of a variety of cancers.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the National Science Foundation of China (No. 81470820 and No. 81370508)
References
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012; 7:e30733; PMID:22319583; http://dx.doi.org/10.1371/journal.pone.0030733
- Wang KS, Choo QL, Weiner AJ, Ou JH, Najarian RC, Thayer RM, Mullenbach GT, Denniston KJ, Gerin JL, Houghton M. Structure, sequence and expression of the hepatitis delta (delta) viral genome. Nature 1986; 323:508–14; PMID:3762705; http://dx.doi.org/10.1038/323508a0
- Wilusz JE, Sharp, PA. Molecular biology. A circuitous route to noncoding RNA. Science 2013; 340:440–1; PMID:23620042; http://dx.doi.org/10.1126/science.1238522
- Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci U S A 1976; 73:3852–6; PMID:1069269; http://dx.doi.org/10.1073/pnas.73.11.3852
- Burgess DJ. RNA: Dissecting circular RNA biogenesis. Nat Rev Genet 2014; 15:707; PMID:25352302; http://dx.doi.org/10.1038/nrg3852
- Vicens Q, Westhof E. Biogenesis of Circular RNAs. Cell 2014; 159:13–4; PMID:25259915; http://dx.doi.org/10.1016/j.cell.2014.09.005
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013; 495:333–8; PMID:23446348; http://dx.doi.org/10.1038/nature11928
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature 2013; 495:384–8; PMID:23446346; http://dx.doi.org/10.1038/nature11993
- Hansen TB, Wiklund ED, Bramsen JB, Villadsen SB, Statham AL, Clark SJ, Kjems J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J 2011; 30:4414–22; PMID:21964070; http://dx.doi.org/10.1038/emboj.2011.359
- Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014; 15:7–21; PMID:24296535; http://dx.doi.org/10.1038/nrg3606
- Esteller M. Non-coding RNAs in human disease. Nat Rev Genet 2011; 12:861–74; PMID:22094949; http://dx.doi.org/10.1038/nrg3074
- Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci 2014; 15:9331–42; PMID:24865493; http://dx.doi.org/10.3390/ijms15069331
- Liang D, Wilusz JE. Short intronic repeat sequences facilitate circular RNA production. Genes Dev 2014; 28:2233–47; PMID:25281217; http://dx.doi.org/10.1101/gad.251926.114
- Zhou H, Arcila ML, Li Z, Lee EJ, Henzler C, Liu J, Rana TM, Kosik KS. Deep annotation of mouse iso-miR and iso-moR variation. Nucleic Acids Res 2012; 40:5864–75; PMID:22434881; http://dx.doi.org/10.1093/nar/gks247
- Zhang, X.-O., Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary Sequence-Mediated Exon Circularization. Cell 2014; 159:134–147; PMID:25242744; http://dx.doi.org/10.1016/j.cell.2014.09.001
- Fu Y, Ramisetty SR, Hussain N, Baranger AM. MBNL1-RNA recognition: contributions of MBNL1 sequence and RNA conformation. Chembiochem 2012; 13:112–9; PMID:22106026; http://dx.doi.org/10.1002/cbic.201100487
- Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S. circRNA Biogenesis Competes with Pre-mRNA Splicing. Mol Cell 2014; 56:55–66; PMID:25242144; http://dx.doi.org/10.1016/j.molcel.2014.08.019
- Szmulewicz MN, Novick GE, Herrera RJ. Effects of Alu insertions on gene function. Electrophoresis 1998; 19:1260–4; PMID:9694261; http://dx.doi.org/10.1002/elps.1150190806
- Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ. The RNA Binding Protein Quaking Regulates Formation of circRNAs. Cell 2015; 160:1125–34; PMID:25768908; http://dx.doi.org/10.1016/j.cell.2015.02.014
- Garg M. Targeting microRNAs in epithelial-to-mesenchymal transition-induced cancer stem cells: therapeutic approaches in cancer. Expert Opin Ther Targets 2015; 19:285–97; PMID:25563894; http://dx.doi.org/10.1517/14728222.2014.975794
- Ye X, Weinberg R A. Epithelial–Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol 2015; PMID:26437589; http://dx.doi.org/10.1016/j.tcb.2015.07.012
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell 2013; 51:792–806; PMID:24035497; http://dx.doi.org/10.1016/j.molcel.2013.08.017
- Umekage S, Kikuchi Y. In vivo circular RNA production using a constitutive promoter for high-level expression. J Biosci Bioeng 2009; 108:354–6; PMID:19716528; http://dx.doi.org/10.1016/j.jbiosc.2009.04.011
- Guil S, Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem Sci 2015; 40:248–56; PMID:25818326
- Cheng DL, Xiang YY, Ji LJ, Lu XJ. Competing endogenous RNA interplay in cancer: mechanism, methodology, and perspectives. Tumour Biol 2015; 36:479–88; PMID:25604144; http://dx.doi.org/10.1007/s13277-015-3093-z
- Satterlee JS, Basanta-Sanchez M, Blanco S, Li JB, Meyer K, Pollock J, Sadri-Vakili G, Rybak-Wolf A. Novel RNA modifications in the nervous system: form and function. J Neurosci 2014; 34:15170–7; PMID:25392485; http://dx.doi.org/10.1523/JNEUROSCI.3236-14.2014
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, García JA, Paz-Ares J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Geneat 2007; 39:1033–7; PMID:17643101; http://dx.doi.org/10.1038/ng2079
- Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods 2007; 4:721–6; PMID:17694064; http://dx.doi.org/10.1038/nmeth1079
- Talhouarne GJ, Gall JG. Lariat intronic RNAs in the cytoplasm of Xenopus tropicalis oocytes. RNA 2014; 20:1476–87; PMID:25051970; http://dx.doi.org/10.1261/rna.045781.114
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. Wide spread intron retention in mammals functionally tunes transcriptomes. Genome Res 2014; 24:1774–86; PMID:25258385; http://dx.doi.org/10.1101/gr.177790.114
- Castello A, Fischer B, Eichelbaum K, Horos R, Beckmann BM, Strein C, Davey NE, Humphreys DT, Preiss T, Steinmetz LM, et al. Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 2012; 149:1393–406; PMID:22658674; http://dx.doi.org/10.1016/j.cell.2012.04.031
- Hentze MW, Preiss T. Circular RNAs: splicing's enigma variations. EMBO J 2013; 32:923–5; PMID:23463100; http://dx.doi.org/10.1038/emboj.2013.53
- Chen CY, Sarnow P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 1995; 268:415–7; PMID:7536344; http://dx.doi.org/10.1126/science.7536344
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA 2014; 20:1829–42; PMID:25404635; http://dx.doi.org/10.1261/rna.047126.114
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol 2014; 32:453–61; PMID:24811520; http://dx.doi.org/10.1038/nbt.2890
- Neveu P, Kye MJ, Qi S, Buchholz DE, Clegg DO, Sahin M, Park IH, Kim KS, Daley GQ, Kornblum HI, et al. MicroRNA profiling reveals two distinct p53-related human pluripotent stem cell states. Cell Stem Cell 2010; 7:671–81; PMID:21112562; http://dx.doi.org/10.1016/j.stem.2010.11.012
- Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 2015; 444:132–6; PMID:25689795; http://dx.doi.org/10.1016/j.cca.2015.02.018
- Bachmayr-Heyda A, Reiner AT, Auer K, Sukhbaatar N, Aust S, Bachleitner-Hofmann T, Mesteri I, Grunt TW, Zeillinger R, Pils D. Correlation of circular RNA abundance with proliferation–exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. SciRep 2015; 5:8057; PMID:25624062; http://dx.doi.org/10.1038/srep08057
- Rokavec M, Li H, Jiang L, Hermeking H. The p53/microRNA connection in gastrointestinal cancer. Clin Exp Gastroenterol 2014; 7:395–413; PMID:25328413
- Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res 2013; 73:5609–12; PMID:24014594; http://dx.doi.org/10.1158/0008-5472.CAN-13-1568
- Wu HH, Lin WC, Tsai KW. Advances in molecular biomarkers for gastric cancer: miRNAs as emerging novel cancer markers. Expert Rev Mol Med 2014; 16, e1; PMID:24456939; http://dx.doi.org/10.1017/erm.2013.16
- Chen YJ, Chien PH, Chen WS, Chien YF, Hsu YY, Wang LY, Chen JY, Lin CW, Huang TC, Yu YL, et al. Hepatitis B Virus-Encoded X Protein Downregulates EGFR Expression via Inducing MicroRNA-7 in Hepatocellular Carcinoma Cells. Evid Based Complement Alternat Med 2013; 2013:682380; PMID:23840262; http://dx.doi.org/10.1155/2013/682380
- Zhou X, Hu Y, Dai L, Wang Y, Zhou J, Wang W, Di W, Qiu L. MicroRNA-7 inhibits tumor metastasis and reverses epithelial-mesenchymal transition through AKT/ERK1/2 inactivation by targeting EGFR in epithelial ovarian cancer. PLoS ONE 2014; 9, e96718; PMID:24816687; http://dx.doi.org/10.1371/journal.pone.0096718
- Webster RJ, Giles KM, Price KJ, Zhang PM, Mattick JS, Leedman PJ. Regulation of epidermal growth factor receptor signaling in human cancer cells by microRNA-7. J Biol Chem 2009; 284:5731–41; PMID:19073608; http://dx.doi.org/10.1074/jbc.M804280200
- Fang Y, Xue JL, Shen Q, Chen J, Tian L. MicroRNA-7 inhibits tumor growth and metastasis by targeting the phosphoinositide 3-kinase/Akt pathway in hepatocellular carcinoma. Hepatology 2012; 55:1852–62; PMID:22234835; http://dx.doi.org/10.1002/hep.25576
- Arteaga CL. The epidermal growth factor receptor: from mutant oncogene in nonhuman cancers to therapeutic target in human neoplasia. J. Clin. Oncol 2001; 19:32S–40S; PMID:11560969
- Janbabai G, Oladi Z, Farazmandfar T, Taghvaei T, Naghshvar F. The prognostic impact of EGFR, ErbB2 and MET gene amplification in human gastric carcinomas as measured by quantitative Real-Time PCR. J Cancer Res Clin Oncol 2015; 141:1945–52; PMID:25820598; http://dx.doi.org/10.1007/s00432-015-1965-7
- Zhao X, Dou W, He L, Liang S, Tie J, Liu C, Li T, Lu Y, Mo P, Shi Y, et al. MicroRNA-7 functions as an anti-metastatic microRNA in gastric cancer by targeting insulin-like growth factor-1 receptor. Oncogene 2013; 32:1363–72; PMID:22614005; http://dx.doi.org/10.1038/onc.2012.156
- Zhang X, Hu S, Zhang X, Wang L, Zhang X, Yan B, Zhao J, Yang A, Zhang R. MicroRNA-7 arrests cell cycle in G1 phase by directly targeting CCNE1 in human hepatocellular carcinoma cells. Biochem Biophys Res Commun 2014; 443:1078–84; PMID:24370822; http://dx.doi.org/10.1016/j.bbrc.2013.12.095
- Ma C, Qi Y, Shao L, Liu M, Li X, Tang H. Downregulation of miR-7 upregulates Cullin 5 (CUL5) to facilitate G1/S transition in human hepatocellular carcinoma cells. IUBMB Life 2013; 65:1026–34; PMID:24339204; http://dx.doi.org/10.1002/iub.1231
- Chinnappan D, Xiao D, Ratnasari A, Andry C, King TC, Weber HC. Transcription factor YY1 expression in human gastrointestinal cancer cells. Int J Oncol 2009; 34:1417–23; PMID:19360355
- Zhang N, Li X, Wu CW, Dong Y, Cai M, Mok MT, Wang H, Chen J, Ng SS, Chen M, Sung JJ, et al. microRNA-7 is a novel inhibitor of YY1 contributing to colorectal tumorigenesis. Oncogene 2013; 32:5078–88; PMID:23208495; http://dx.doi.org/10.1038/onc.2012.526
- Suto T, Yokobori T, Yajima R, Morita H, Fujii T, Yamaguchi S, Altan B, Tsutsumi S, Asao T, Kuwano H. MicroRNA-7 expression in colorectal cancer is associated with poor prognosis and regulates cetuximab sensitivity via EGFR regulation. Carcinogenesis 2015; 36:338–45; PMID:25503932; http://dx.doi.org/10.1093/carcin/bgu242
- Huynh FC, Jones FE. MicroRNA-7 inhibits multiple oncogenic pathways to suppress HER2Delta16 mediated breast tumorigenesis and reverse trastuzumab resistance. PLoS One 2014; 9, e114419; PMID:25532106; http://dx.doi.org/10.1371/journal.pone.0114419
- Babae N, Bourajjaj M, Liu Y, Van Beijnum JR, Cerisoli F, Scaria PV, Verheul M, Van Berkel MP, Pieters EH, Van Haastert RJ, et al. Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget 2014; 5:6687–700; PMID:25149532; http://dx.doi.org/10.18632/oncotarget.2235
- Liu Z, Jiang Z, Huang J, Huang S, Li Y, Yu S, Yu S, Liu X. miR-7 inhibits glioblastoma growth by simultaneously interfering with the PI3K/ATK and Raf/MEK/ERK pathways. Int J Oncol 2014; 44:1571–80; PMID:24603851; http://dx.doi.org/10.3892/ijo.2014.2322.
- Liu R, Liu X, Zheng Y, Gu J, Xiong S, Jiang P, Jiang X, Huang E, Yang Y, Ge D. MicroRNA-7 sensitizes non-small cell lung cancer cells to paclitaxel. Oncol Lett 2014; 8:2193–2200; PMID:25289099
- Ma J, Fang B, Zeng F, Pang H, Zhang J, Shi Y, Wu X, Cheng L, Ma C, Xia J, et al. Curcumin inhibits cell growth and invasion through up-regulation of miR-7 in pancreatic cancer cells. Toxicol Lett 2014; 231:82–91; PMID:25256401; http://dx.doi.org/10.1016/j.toxlet.2014.09.014
- Hao Z, Yang J, Wang C, Li Y, Zhang Y, Dong X, Zhou L, Liu J, Zhang Y, Qian J. MicroRNA-7 inhibits metastasis and invasion through targeting focal adhesion kinase in cervical cancer. Int J Clin Exp Med 2015; 8:480–7; PMID:25785020
- Bak RO, Mikkelsen JG. miRNA sponges: soaking up miRNAs for regulation of gene expression. Wiley Interdiscip Rev RNA 2014; 5:317–33; PMID:24375960; http://dx.doi.org/10.1002/wrna.1213
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet 2013; 9, e1003777; PMID:24039610; http://dx.doi.org/10.1371/annotation/f782282b-eefa-4c8d-985c-b1484e845855
- Granados-Riveron JT, Aquino-Jarquin G. Does the linear Sry transcript function as a ceRNA for miR-138? The sense of antisense. F1000Res 2014; 3:90; PMID:25580223; http://dx.doi.org/10.12688/f1000research.3872.2
- Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol 2014; 15:409; PMID:25070500; http://dx.doi.org/10.1186/s13059-014-0409-z
- Wang Q, Tang H, Yin S, Dong C. Downregulation of microRNA-138 enhances the proliferation, migration and invasion of cholangiocarcinoma cells through the upregulation of RhoC/p-ERK/MMP-2/MMP-9. Oncol Rep 2013; 29:2046–52; PMID:23446431; http://dx.doi.org/10.3892/or.2013.2304
- Long L, Huang G, Zhu H, Guo Y, Liu Y, Huo J. Down-regulation of miR-138 promotes colorectal cancer metastasis via directly targeting TWIST2. J Transl Med 2013; 11:275; PMID:24171926; http://dx.doi.org/10.1186/1479-5876-11-275
- Ayaz L, Çayan F, Balci Ş, Görür A, Akbayir S, Yıldırım Yaroğlu H, Doğruer Unal N, Tamer L. Circulating microRNA expression profiles in ovarian cancer. J Obstet Gynaecol 2014; 34:620–4; PMID:24911418; http://dx.doi.org/10.3109/01443615.2014.919998
- Chen P, Zeng M, Zhao Y, Fang X. Upregulation of Limk1 caused by microRNA-138 loss aggravates the metastasis of ovarian cancer by activation of Limk1/cofilin signaling. Oncol Rep 2014; 32:2070–6; PMID:25190487; http://dx.doi.org/10.3892/or.2014.3461
- Yeh YM, Chuang CM, Chao KC, Wang LH. MicroRNA-138 suppresses ovarian cancer cell invasion and metastasis by targeting SOX4 and HIF-1alpha. Int J Cancer 2013; 133:867–78; PMID:23389731; http://dx.doi.org/10.1002/ijc.28086
- Yang H, Tang Y, Guo W, Du Y, Wang Y, Li P, Zang W, Yin X, Wang H, Chu H, et al. Up-regulation of microRNA-138 induce radiosensitization in lung cancer cells. Tumour Biol 2014; 35:6557–65; PMID:24691972; http://dx.doi.org/10.1007/s13277-014-1879-z
- Yang H, Luo J, Liu Z, Zhou R, Luo H. MicroRNA-138 Regulates DNA Damage Response in Small Cell Lung Cancer Cells by Directly Targeting H2AX. Cancer Invest 2015; 33:126–36; PMID:25699650; http://dx.doi.org/10.3109/07357907.2015
- Berg V, Rusch M, Vartak N, Jüngst C, Schauss A, Waldmann H, Hedberg C, Pallasch CP, Bastiaens PI, Hallek M, et al. miRs-138 and −424 control palmitoylation-dependent CD95-mediated cell death by targeting acyl protein thioesterases 1 and 2 in chronic lymphocytic leukemia. Blood 2015; 125(19), 2948–2957; PMID:25670628; http://dx.doi.org/10.1182/blood-2014-07-586511
- Kosik KS. Molecular biology: Circles reshape the RNA world. Nature 2013; 495:322–4; PMID:23446351; http://dx.doi.org/10.1038/nature11956
- Lee KM, Choi EJ, Kim IA. microRNA-7 increases radiosensitivity of human cancer cells with activated EGFR-associated signaling. Radiother Oncol 2011; 101:171–6; PMID:21676478; http://dx.doi.org/10.1016/j.radonc.2011.05.050
- Bockhorn J, Prat A, Chang Y F, Liu X, Huang S, Shang M, Nwachukwu C, Gomez-Vega MJ, Harrell JC, Olopade OI, et al. Differentiation and loss of malignant character of spontaneous pulmonary metastases in patient-derived breast cancer models. Cancer Res 2014; 74(24), 7406–7417; PMID:25339353; http://dx.doi.org/10.1158/0008-5472.CAN-14-1188
- Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev 2014; PMID:24859534; http://dx.doi.org/10.1016/j.addr.2014.05.010
- Santoro MM. Antiangiogenic cancer drug using the zebrafish model. Arterioscler Thromb Vasc Biol 2014; 34:1846–53; PMID:24903092; http://dx.doi.org/10.1161/ATVBAHA.114.303221
- Mohseny AB, Hogendoorn PC. Zebrafish as a model for human osteosarcoma. Adv Exp Med Biol 2014; 804:221–36; PMID:24924177; http://dx.doi.org/10.1007/978-3-319-04843-7_12
- Chao CW, Chan DC, Kuo A, Leder P. The mouse formin (Fmn) gene: abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med 1998; 4:614–28; PMID:9848078
