ABSTRACT
Angiogenin (ANG) is a secreted ribonuclease best known for its ability to promote formation of blood vessels. Extensive research over many years has elucidated its structure and biophysical properties, although our knowledge of molecular mechanisms underlying ANG-associated biologic processes remains limited. Intriguingly, many of processes require the ribonuclease activity of ANG, thus highlighting the importance of identifying and characterizing RNA targets and intermediates of ANG-mediated endonucleolytic cleavage. While ANG demonstrates ribonuclease activity toward many RNA substrates in vitro, specific target of ANG, namely mature tRNA, was only recently identified in vivo. ANG-mediated tRNA cleavage is an unorthodox manner of generating non-coding RNAs with diverse biologic activities. In addition, the ribonuclease activity of ANG has been reported to be crucial for rRNA transcription. Here we critically discuss various aspects of ANG biology related to its RNase activity and discuss areas in need of further investigation.
Historical considerations
Early in vivo tumor transplantation experiments suggested that cancer cells produce a molecule that potently induces new blood vessel formation. Researchers hypothesized that identification of this tumor angiogenesis factor (TAF) would lead to the development of drugs that inhibit tumor growth by preventing recruitment of new blood vessels.Citation1 Years of study identified not a single TAF but several TAFs, including angiogenin (ANG). From over 2000 L of conditioned media from HT-29 human adenocarcinoma cells, Vallee and colleagues purified 1 mg of ANG and showed it potently stimulates angiogenesis.Citation2
ANG is an atypical RNase
Surprisingly, amino acid analysis of ANG (also known as RNase 5) revealed a 35% amino acid similarity with ribonuclease (RNase) ACitation3. ANG has relatively weak ribonuclease activity yet it is required for angiogenesis. In contrast, the more potent RNase A does not promote angiogenesis. Crystallization studies determined that the pyrimidine binding site of ANG is occluded by Gln-117 in comparison to the catalytic site of RNase A; introduction of a Gln117Gly mutation increases the ribonuclease activity of ANG 30-fold.Citation4 Despite its reduced RNase activity, ANG readily cleaves RNA in vitro and in vivo. Replacements of important site residues (e.g. His13 and/or His114) invariably diminish the activity toward RNA substrates (up to 10,000 fold) and abolish angiogenic activities.Citation5-7
ANG is a vertebrate-specific RNase.Citation8 Humans have a single ANG gene whereas mice have 5 ANG orthologes and 3 pseudogenes.Citation9 In contrast, the ANG gene is absent from some mammalian genomes (e.g., guinea pig or dog). ANG is a secreted molecule. In some cells (e.g., vascular endothelial and smooth muscle cells), it is actively secreted and taken up in an autocrine and paracrine manner. Consequently, although initially identified using tumor cells, ANG is also found in normal tissues and fluids including blood, cerebrospinal and amniotic fluids.Citation10,11 Liver is the predominant organ of ANG secretion.Citation12 Interestingly, the human ANG gene encodes a 24 amino acid N-terminal signal peptide, a signature of other RNase superfamily members.Citation10 It is postulated, but not experimentally proven, that this signal peptide is cleaved before secretion from cells. The signal peptide may also be required for processing at the endoplasmic reticulum to ensure correct disulfide bond formation. It should be noted that literature is based on mature ANG lacking the signal peptide.
ANG is internalized through a receptor-mediated process, but the identity of the receptor remains controversial. One study suggests that Syndecan-4 (22 kDa) mediates internalization in astroglia,Citation13 while other studies have identified proteins of ∼170 kDa and ∼49 kDa as mediators of internalization in endothelial cells.Citation14 Regardless of the internalization mechanism, upon cellular entry, ANG is found in both cytoplasmic and nuclear compartments where it exerts distinct functional effects.
Remaining questions
The definitive identification of the ANG receptor (or receptors) is an important goal for future research. It should be noted that due to its extreme positive charge (pI >10.5), ANG can avidly bind the extracellular matrix and cellular membranes. This could allow passive endocytosis in the absence of a high affinity receptor. Nevertheless, the existence of a cell-type specific ANG receptor could distinguish cells that internalize ANG more or less efficiently and also introduce receptor specific functions. Another gap in our knowledge is the presence and functions of the signal peptide. Is signal peptide removal required for secretion or important for other aspects of processing? During maturation, ANG also undergoes cyclization of its N-terminal glutaminyl residue, which contributes to its extreme stability in serum. Thus removal of the signal peptide (by an unknown protease) may regulate protein stability. Hypothetically, the signal peptide may alter stability, protein interactions or RNase activity.
Search for ANG RNA substrates: In vitro situation
ANG possesses the same general catalytic properties as RNase A, cleaving the 3′-side of pyrimidine nucleotides using a transphosphorylation-hydrolysis mechanism that prefers single-stranded RNA molecules.Citation3 However, compared with RNase A, ANG is much less active or even inactive on di- and polynucleotides, with order of the reactivity CpA>CpG>UpA>UpG.Citation7,15 Many conventional in vitro RNase substrates (e.g., Poly(C), Poly(U), yeast or wheat germ RNA) are not efficiently cleaved by ANG.Citation7 Thus, ANG is less promiscuous than RNase A, implying that it has limited cleavage sites within the transcriptome. In HT-29 cancer cells, ANG digests 28S and 18S rRNAs to produce ∼100–500 nt fragments.Citation16 In Escherichia coli and yeast 5S rRNA is preferentially cleaved when a pyrimidine is followed by adenine.Citation17 Although performed exclusively in vitro, these early experiments highlight an important aspect of ANG activity. In contrast to RNase A, which cleaves after any pyrimidine nucleoside, ANG cleaves at selected regions within a given substrate. This conclusion was complimented by studies showing that human ANG mediates non-random cleavage of yeast tRNA. Recent in vitro studies using total RNA from HEK293 cells also suggested that ANG can efficiently cleave tRNAs and other small RNAs (e.g., snRNAs) at specific sites.Citation18 Moreover, mutagenesis analysis of substrate tRNA (tRNALys) suggested that substitution of specific nucleotides in tRNA can affect ANG-mediated tRNA cleavage in vitro.Citation18
The first in vivo hints of ANG specificity came from injection of Xenopus laevis oocytes with recombinant ANG. The authors determined that ANG inhibited protein synthesis due to cleavage of tRNAs, while the stability of other analyzed rRNAs and mRNAs were unaffected.Citation64
Remaining questions
Although “naked RNA” rarely exists in cells, studies of ANG using purified RNA substrates suggest that ANG cleaveage is dependent on both sequence and local secondary structure. Surprisingly, the effects of simple secondary structures (e.g., RNA hairpins, bulges, etc.) on ANG cleavage efficiency have not been reported. Both rRNAs and tRNAs are structured molecules with known topology, thus analysis of ANG-mediated cleavage patterns may suggest how structural motifs impact ANG-RNA recognition.
tRNA as a genuine RNA substrate for ANG in vivo
It took several decades to identify an in vivo ANG-mediated cleavage event. ANG is a stress-responsive RNase that is transcriptionally induced by stressCitation19,20 or androgen treatment.Citation21 Two groups showed that in response to various stresses, mammalian cells reacted by cleaving tRNAs within their anticodon loops, generating 2 small RNAs from either half of the mature tRNA.Citation22,23 These RNAs, alternatively called tRNA-derived stress-induced RNAs (tiRNAs)Citation22 or tRNA halves (tRHs),Citation23 are products of ANG-mediated cleavage. Stress-induced tRNA cleavage is effectively blocked by knockdown of endogenous ANG or stimulated by knockdown of RNH1 (also known as RI), the known inhibitor of ANG.Citation65 Independent of exogenous stresses, recombinant ANG added to cell culture media is rapidly internalized and cleaves mature cytoplasmic tRNAs within anticodon loops. In accordance with it being an RNase A family member, tRNA cleavage results in a 2′–3′ cyclic phosphate at the 3′-end of the 5′- cleavage product (5′tiRNA) and a 5′ hydroxyl on the 5′—end of the 3′ cleavage product (3′tiRNA). Importantly, studies using a tRNA-specific microarrayCitation66 as well as RNA-sequencing (data not shown, our laboratory) demonstrate that any tRNA species can serve as an ANG substrate, although the repertoire of tiRNAs may be stress and cell-type specific.
One study suggests that ANG also targets the CCA end of tRNAs,Citation24 a triplet that is added post-transcriptionally in human cells. In this model, ANG removes the CCA during stress and upon return to homeostatic conditions the CCA-adding enzyme TRNT1 re-adds CCA. Although attractive, this model is based solely on in vitro evidence. Additionally, the 2′–3′ cyclic phosphate left by ANG-mediated cleavage would need to be resolved to a 3′-hydroxyl by an unknown phosphatase for TRNT1-mediated CCA re-addition.
We propose a model where under optimal conditions endogenous ANG is kept inactive via interaction with RNH1 (). When stressed 3 processes occur to increase the cytoplasmic concentration of active ANG (): 1) the ANG:RNH1 complex dissociates, thereby releasing ANG, 2) nuclear ANG is translocated to the cytoplasm and 3) ANG transcription is induced. Increased cytoplasmic uninhibited ANG leads to the cleavage of mature cytoplasmic tRNAs producing tiRNAs (). Additionally, the stressed cell secretes ANG which is taken up by surrounding cells acting as an “interferon/alarmone-like” response (). This model has broad applications toward different biologic phenomena related to RNase functions of ANG (discussed below).
Figure 1. ANG localization regulates its activity. (A) In unstressed cells, ANG (Green) is primarily localized in the nucleus. The cytoplasmic pool of ANG is held in an inactive state through interaction with RNH1 (Purple). A minor pool of ANG is secreted, found in serum and is able to be taken up by surrounding cells. (B) In a stressed cell (red), ANG dissociates from RNH1 and relocalizes from the nucleus to the cytoplasm. ANG expression is also induced, likely as a result of increased transcription. Stressed cells actively secrete ANG to surrounding cells which take up ANG via a receptor mediated endocytosis.
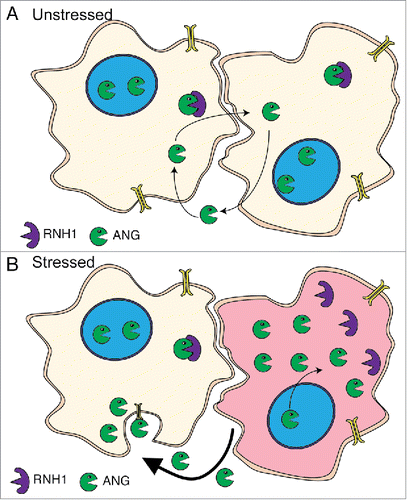
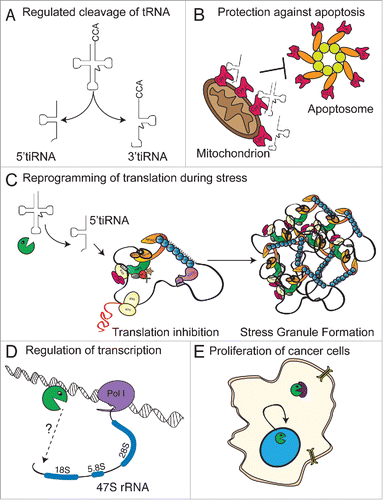
Figure 2. RNase dependent functions of ANG. (A) ANG cleaves the anticodon loop of mature cytoplasmic tRNAs generating 2 smaller RNA species, termed 5′- and 3′- tiRNAs. (B) A subset of 5′ and 3′ tiRNAs bind to Cytochrome C (red) preventing it's interaction with the apoptosome. Failure to assemble the apoptosome promotes survival by preventing apopotosis. (C) A subset of 5′tiRNAs inhibit translation by targeting the initiation step thereby promoting the formation of stress granules. (D) ANG is translocated to the nucleus where it promotes the expression of rRNA. (E) Increased expression of ANG increases proliferation of cancer cells linked to nuclear import.
Remaining questions
Our current knowledge of ANG-mediated tRNA cleavage in vivo is very limited and based mostly on northern blotting and tRNA microarrays. As tRNAs are highly structured and rich in modifications, the use of RNA sequencing to quantify ANG-induced tRNA cleavage has proven problematic. Recent development of tRNA sequencing methodologiesCitation25,26 will hopefully overcome such technical limitations to reveal the complete repertoire of ANG-induced tRNA fragments.
Regulation of ANG-mediated tRNA cleavage
Post-transcriptional tRNA processing, a process necessary for fine-tuning of tRNA function, involves incorporation of various chemical nucleotide modificationsCitation27,28 Recent data suggest that some modifications affect ANG-mediated tRNA cleavage. One of the most conserved and common modifications is methylation of carbon-5 of cytosine by methyltransferases such as DNMT2 or NSun2, generating 5-methylcytosine (m5C). DNMT2 specifically targets cytosines in the anticodon loop at position 38 in tRNAAsp(GUC), tRNAGly(GCC) and tRNAVal(AAC)Citation29,30. Overexpression of DNMT2 inhibits, while its depletion increases, ANG-mediated tRNA cleavage. Similarly, NSun2 is responsible for m5C in the variable loop at position 48–50 of specific tRNAs, and loss of m5C through NSun2 deletion enhances ANG-mediated tRNA cleavage.Citation31 Importantly, this mode of regulation plays important roles in pathological and physiologic conditions. Loss of m5C modifications via NSun2 or DNMT2 deletion enhances sensitivity to oxidative stress in flies, mice and human cells. Mice lacking both Dnmt2 and NSun2 show complete loss of m5C modification in tRNAs and are characterized by reduced overall protein synthesis, developmental defects and lethality, suggesting the importance of m5C modifications in the regulation of protein synthesis.Citation30
In addition, recent work has shown that dynamic regulation of NSun2 activity contributes to stem cell fate.Citation32 Restrained protein synthesis in stem cells contributes to maintaining an undifferentiated state, and upregulation of protein synthesis is necessary for stem cell differentiation. In stem cells, NSun2 activity is downregulated allowing accumulation of tiRNAs and downregulation of cellular translation. Once NSun2 expression is upregulated, m5C modifications protect against ANG-mediated tRNA cleavage, leading to translation of lineage-specific transcripts that drive terminal differentiation.
Remaining questions
Although the role of m5C in ANG-mediated tRNA cleavage is evident, the role of other tRNA modifications remains unclear. Further studies are needed to elucidate the physiologic and pathological significance of how post-transcriptional RNA modifications regulate ANG cleavage. In prokaryotes certain modifications attract tRNA-specific RNases to cleave tRNA substrates:Citation67 whether this occurs for eukaryotic tRNAs and is a mode of ANG cleavage remains to be determined. It would also be interesting to know how RNA-binding proteins facilitate or inhibit ANG-mediated cleavage.
Diverse roles of ANG-induced tRNA fragments: Emerging universe
tRNA cleavage in response to diverse stimuli is a conserved evolutionary process. In mammals, ANG-mediated tRNA cleavage has been implicated in various biologic processes. ANG-induced tRNA fragments have variously been called tRNA-derived stress-induced RNAs (tiRNAs)Citation22, tRNA halves (tRHs)Citation23, tRNA-derived small RNAs (tsRNAs)Citation68 and Sex Hormone-dependent tRNA derived RNAs (SHOT-RNAs).Citation69 These likely represent the same molecules and for the purposes of discussion here, we refer to the molecules generated via ANG-dependent cleavage within the anticodon loop as tiRNAs ().
Cells treated with recombinant ANG demonstrate transient reduction in global protein synthesis.Citation22 tiRNA production is the cause of this inhibition as transfection of endogenously purified or synthetic tiRNAs similarly reduces protein synthesis. Furthermore, transfection of tiRNAs into cells triggers stress granule (SG) formation.Citation33 SGs are cytoplasmic foci consisting of 40S ribosomal subunits, mRNAs, translation initiation factors and RNA-binding proteins.Citation34 During a stress response, SGs are thought to serve as hubs for sorting and storing untranslated mRNAs, RNA-binding proteins and signaling molecules in a way that promotes cell survival.Citation70 Treatment of cells with recombinant wild type but not catalytically dead ANG enhances SG formation. Further characterization of individual tiRNAs showed that 5′tiRNAAla and 5′tiRNACys are potent inhibitors of translation that act by displacing the cap-binding eIF4F complex from the cap structures of mRNAs, thereby blocking translation initiation ().Citation35 They trigger the formation of SGs through a mechanism dependent upon Y-box binding protein 1 (YB-1, YBX1).Citation36 This activity is dependent on a stretch of 5 guanines at the 5′ end of these 2 tiRNAs, termed the 5′ terminal oligoguanine (5′TOG) motif. Mutation or deletion of the 5′TOG motif abolishes the ability to repress translation, trigger SG formation or displace eIF4F from mRNA caps. 5′TOG-containing tiRNAs do not inhibit translation of transcripts bearing IRES (Internal Ribosome Entry Site) elements, which initiate translation in a cap-independent manner. This observation has led to the hypothesis that during a stress response, tiRNAs promote survival by reprogramming cellular translation - repressing cap-dependent translation while promoting translation of mRNAs possessing IRES-like structures, e.g., the anti-apoptotic Bcl2 protein. This hypothesis awaits experimental proof.
Additionally, tiRNAs promote survival through their interaction with Cytochrome c (Cyt c).Citation37 During apoptosis, Cyt c is released from mitochondria and binds the apoptotic protease activating factor 1 protein (Apaf-1) triggering a conformational change that allows for oligomerization to form the apoptosome, a multicomponent complex that facilitates procaspase-9 cleavage, which in turn activates executioner caspase-3Citation38. ANG treatment prevents apoptosome formation, inhibits caspase-3 activation and increases the viability of cells under hyperosmotic stress. Immunoprecipitation experiments showed that Cyt c binds a large portion of tiRNAs (20 different tiRNAs) and it has been hypothesized that this prevents Cyt c interaction with the apoptosome (), thus inhibiting apoptosis. Both 5′tiRNAs and 3′tiRNAs bind cytochrome C to prevent apoptosis, in contrast to the unique role of selected 5′tiRNAs in translation inhibition. In both cases, only a subset of tiRNAs are functional, despite the observation that all tRNAs are cleaved in response to various stresses.
ANG-mediated tRNA cleavage is also triggered by some viral infections, e.g. Respiratory Syncytial Virus (RSV)Citation39,40 or Hepatitis virus.Citation41 During RSV infection, tiRNA expression increases to 36.5% of total small RNA species, while miRNAs account for only 6%, with 5′tiRNAGlu being among the most highly upregulated. Knockdown of a panel of cellular RNases determined that ANG was required for RSV induced tiRNA formation. Surprisingly, 5′tiRNAGlu functions in an RNAi manner analogous to miRNAs or siRNAs relying upon base-pairing.Citation39 This is surprising as 5′tiRNAs are longer than miRNAs and siRNAs (∼30 vs 20–22 nts) which normally catalyze this effect and are considered optimized for length. 5′tiRNAGlu specifically interacts with and downregulates a subset of cellular RNAs during RSV infection, most notably transcripts encoding immunity-related proteins. Blocking 5′tiRNAGlu activity using antisense oligos or via knockdown of ANG reduces the ability of RSV to replicate in cells. Although poorly understood mechanistically, these reports indicate that tiRNAs may work through RNAi-like mechanisms.
Remaining questions
The list of tRNA-derived fragments continues to grow but the mechanism of their biogenesis is largely unknown.Citation42 Another important question is the functions of tiRNAs. Our own studies using a subset of tiRNAs revealed roles in translation inhibition. Can other tiRNAs inhibit or possibly stimulate translation? As many post-transcriptional mechanisms of gene expression coordinate mRNA decay and translation, do tiRNAs also affect transcript stability? Another unanswered question is identification of specific tiRNA targets. Even for the known 5'TOG-bearing tiRNAs we do not know definitively how they reprogram cellular translation or the repertoire of tiRNA-binding proteins. As tiRNAs are very diverse in their predicted secondary structures, they are likely to have diverse functions. An important aspect of tiRNAs is their role in stress response and survival. What is the structure/sequence/Cyt c-binding relationship? An intriguing aspect of tiRNAs is their ability to regulate expression in an RNAi-like manner. It has yet to be determined if this process involves loading 5′tiRNAGlu into a RISC complex and/or a related protein complex. Many questions remain to be answered.
Role of ANG in ribosome biogenesis
ANG plays a role in the biogenesis of rRNAs. Upon translocation to the nucleus, ANG is concentrated in the nucleolusCitation43,44, the site of rRNA transcription and ribosome assembly. Synthesis and assembly of ribosomes is one of the most intricate and energy intensive tasks a cell undertakes. Each functional ribosome contains 79 ribosomal proteins (rps) and 4 rRNAs, and rRNAs account for ∼80% of cellular RNA and mRNAs encoding ribosomal proteins may account for >20% of the total pool of cellular mRNAs. Under stressful conditions, we propose that the egress of ANG from the nucleus halts ribosome assembly to conserve energy for other cellular tasks ().
Humans have over 400 copies of the gene encoding the 47S precursor rRNA (approximately 13 kb), which is processed to generate the mature 18S, 5.8S and 28S rRNAs, while the 5S rRNA is transcribed from a different locus [reviewed inCitation45,46]. A single 47S gene is separated from the next gene by a 30 kb intergenic spacer (IGS). The process of maturation involves several reiterative endonucleolytic cleavage and exonucleolytic trimming generating several intermediate rRNA species (43S, 30S, 21S, 18S-E, etc.). The exosome is implicated in exonucleolytic processing of precursor rRNAs but the identity of endonucleases involved in processing has remained elusive.
Active rRNA transcription depends upon 2 cis elements in the rDNA locus: the core promoter and the upstream control element (UCE) (reviewed inCitation47). The UCE is located at −156 to −107 relative to the transcription start site and is bound by upstream binding factor (UBF). Pol I transcription requires polymerase I ( Pol I) binding to the core promoter and UBF interaction with the UCE. Efficient transcription of the rDNA locus is intrinsically linked with efficient processing of the precursor 47S rRNA to generate the mature 18S, 28S and 5.8S rRNAsCitation48,49
In HUVE cells, ANG increases the expression of 45S rRNA, a precursor generated after initial processing of the 47S rRNA.Citation50 Treatment of isolated nuclei with ANG leads to an increase in run-on transcription of an RNA product approximately 400–500 nts in length. Analysis of a 4.5 kb DNA fragment from the IGS showed that ANG bound this DNA at “CT” rich sequences designated ANG binding elements (ABE).Citation51 ChIP analysis showed that ANG interacts with 2 ABEs, referred as ABE1 and ABE2, located ∼1 kb and ∼9 kb downstream of the 47S rRNA.Citation52 ChIP analysis also suggested that ANG interacts with the UCECitation52,53, although whether ANG affects the binding of factors required for efficient rRNA transcription is unclear. Interestingly, inserting an ABE in a plasmid encoding firefly luciferase increased the expression of the reporter.Citation51 This data and the increased accumulation of 45S rRNA upon ANG treatment are used as evidence to suggest that ANG recruitment to the ABE stimulates rDNA transcription. However, the reporter used in these studies has signals necessary for Pol II dependent transcription (polyA signals and SV40 enhancer). If ANG stimulates rRNA production by acting as a transcription factor for rRNA, it is not clear how Pol I recruitment to a luciferase reporter harboring Pol II processing elements would produce a functional mRNA that is properly processed, exported from the nucleus and translated.
What is clear is that ANG promotes the expression of rRNA and it has been long appreciated that increased ribosome biogenesis is required for cellular proliferation.Citation54 Blocking nuclear import of ANG abolishes its ability to promote proliferation,Citation43,50,55 linking its effect on rRNA expression to its ability to enhance proliferation (). Although nuclease activity is required for ANG to promote proliferation and angiogenesis, it has not been formally shown that nuclease activity is required to enhance rRNA expression, although this seems likely. Therefore, we propose that rather than acting as a bona fide transcription factor that recruits Pol I to rDNA for transcription, ANG promotes rRNA expression by acting as a pre-rRNA processing factor. The field of RNA biology is littered with examples of proteins initially identified as transcription factors that are later shown to be RNA processing factors. For example, FLASH was originally identified as necessary for transcription of replication-dependent histone mRNAsCitation56 but later shown to be essential for histone mRNA 3′-end processing.Citation57 Similarly, tristetraprolin (TTP) was originally classified as a transcription factorCitation58,59, but later shown to regulate the stability of mRNAs that it bound.Citation60
In humans, it is unknown which endonucleases function in rRNA maturation. ANG is able to cleave a synthetic RNA with the A1 cleavage site, which converts the 47S rRNA to the 45 rRNA. These data point to the possibility that ANG functions in the processing of rRNA rather than transcription. It is worth noting that in humans A1 cleavage generates a 415 nt 5′ fragment,Citation61 approximately the same size as the band that was increased after ANG treatment in nuclear run-on assays. We believe that ANG promotes co-transcriptional endonucleolytic cleavage. Further supporting this model, ChIP of ANG localizes it to the extreme 5′ and 3′ ends of the 47S rRNA.Citation21,53
Another possible mechanism by which ANG could regulate rRNA biosynthesis is via regulation of ncRNAs associated with the nucleolus. It is now clear that the IGS is pervasively transcribed and a source of long noncoding RNAs (ncRNAs).Citation71 One such RNA is transcribed approximately 2 kb upstream of the rDNA promoter and processed into smaller (∼150–300 nt) RNAs.Citation62 These RNAs, termed promoter RNAs (pRNAs), interact with the nucleolar remodeling complex (NoRC).Citation63 The pRNA:NoRC complex localizes to the promoter of the pre-rRNA transcript and causes an increase in histone H3K9 and H4K20 methylation, which silence the locus by inhibiting transcription. ANG could increase production of rRNA by cleaving pRNAs, thereby preventing pRNA:NoRC complex formation and resulting in a decrease in inhibitory histone marks. In support of our hypothesis, knockdown of ANG significantly increases H3K9 methylation at the rDNA promoter.Citation53
Remaining questions
The nucleolus is a multifunctional organelle coordinating multiple processes linked to its dynamic architecture. Recent data suggest that ncRNAs are integral parts of the nucleolus that actively participate in both rRNA production and remodeling of the nucleolus. ANG is a nucleolar component and may play a role in rRNA metabolism beyond its proposed functions in rRNA transcription. We specifically speculate that it has a role in rRNA processing and stability of IGS-associated ncRNAs that regulate rRNA production. This hypothesis requires further examination. We noticed that during revision of this manuscript, the paper from Prof. Raines lab has been published that partially validated our hypothesis (NAR, published online December 2nd, 2016).Citation72
Perspectives
ANG is an exciting molecule given its proposed functions in RNA metabolism. Importantly, multiple biologic processes are linked to ANG functions, e.g., angiogenesis and neovascularization, stress adaptation and survival, cell-to-cell signaling and maintenance of stem cell homeostasis. Given established roles of ANG in the etiology of common (cancer, infections) and rare (neurodegenerative diseases such as Amyotrophic Lateral Sclerosis) pathophysiological processes, further studies of ANG-mediated molecular mechanisms are required. Here we propose that many of these mechanisms involve the RNase function of ANG.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank members of the Ivanov and Anderson laboratories for helpful discussion and feedback on this manuscript. This work is supported by the National Institutes of Health [GM111700 and CA168872 to PA, NS094918 to P and F32GM119283 to SML].
References
- Folkman J. Tumor angiogenesis: therapeutic implications. N Engl J Med 1971; 285(21):1182-6; PMID:4938153; http://dx.doi.org/10.1056/NEJM197111182852108
- Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry 1985; 24(20):5480-6; PMID:4074709; http://dx.doi.org/10.1021/bi00341a030
- Strydom DJ, Fett JW, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Amino acid sequence of human tumor derived angiogenin. Biochemistry 1985; 24(20):5486-94; PMID:2866794; http://dx.doi.org/10.1021/bi00341a031
- Russo N, Shapiro R, Acharya KR, Riordan JF, Vallee BL. Role of glutamine-117 in the ribonucleolytic activity of human angiogenin. Proc Natl Acad Sci U S A 1994; 91(8):2920-4; PMID:8159680; http://dx.doi.org/10.1073/pnas.91.8.2920
- Shapiro R, Vallee BL. Site-directed mutagenesis of histidine-13 and histidine-114 of human angiogenin. Alanine derivatives inhibit angiogenin-induced angiogenesis. Biochemistry 1989; 28(18):7401-8; PMID:2479414; http://dx.doi.org/10.1021/bi00444a038
- Curran TP, Shapiro R, Riordan JF. Alteration of the enzymatic specificity of human angiogenin by site-directed mutagenesis. Biochemistry 1993; 32(9):2307-13; PMID:8095159; http://dx.doi.org/10.1021/bi00060a023
- Shapiro R, Riordan JF, Vallee BL. Characteristic ribonucleolytic activity of human angiogenin. Biochemistry 1986; 25(12):3527-32; PMID:2424496; http://dx.doi.org/10.1021/bi00360a008
- Gao X, Xu Z. Mechanisms of action of angiogenin. Acta Biochim Biophys Sin (Shanghai) 2008; 40(7):619-24. Epub 2008/07/08; PMID:18604453; http://dx.doi.org/10.1111/j.1745-7270.2008.00442.x
- Goo SM, Cho S. The expansion and functional diversification of the mammalian ribonuclease a superfamily epitomizes the efficiency of multigene families at generating biological novelty. Genome Biol Evol 2013; 5(11):2124-40; PMID:24162010; http://dx.doi.org/10.1093/gbe/evt161
- Sorrentino S. The 8 human “canonical” ribonucleases: molecular diversity, catalytic properties, and special biological actions of the enzyme proteins. FEBS Lett 2010; 584(11):2194-200; PMID:20388512; http://dx.doi.org/10.1016/j.febslet.2010.04.018
- Moenner M, Gusse M, Hatzi E, Badet J. The widespread expression of angiogenin in different human cells suggests a biological function not only related to angiogenesis. Eur J Biochem 1994; 226(2):483-90; PMID:7528139; http://dx.doi.org/10.1111/j.1432-1033.1994.tb20073.x
- Olson KA, Verselis SJ, Fett JW. Angiogenin is regulated in vivo as an acute phase protein. Biochem Biophys Res Commun 1998; 242(3):480-3. Epub 1998/02/17; PMID:9464241; http://dx.doi.org/10.1006/bbrc.1997.7990
- Skorupa A, King MA, Aparicio IM, Dussmann H, Coughlan K, Breen B, Kieran D, Concannon CG, Marin P, Prehn JH. Motoneurons secrete angiogenin to induce RNA cleavage in astroglia. J Neurosci 2012; 32(15):5024-38; PMID:22496549; http://dx.doi.org/10.1523/JNEUROSCI.6366-11.2012
- Hu GF, Riordan JF, Vallee BL. A putative angiogenin receptor in angiogenin-responsive human endothelial cells. Proc Natl Acad Sci U S A 1997; 94(6):2204-9; PMID:9122172; http://dx.doi.org/10.1073/pnas.94.6.2204
- Harper JW, Vallee BL. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bond generated by regional mutagenesis. Biochemistry 1989; 28(4):1875-84; PMID:2719939; http://dx.doi.org/10.1021/bi00430a067
- Shapiro R, Vallee BL. Human placental ribonuclease inhibitor abolishes both angiogenic and ribonucleolytic activities of angiogenin. Proc Natl Acad Sci U S A 1987; 84(8):2238-41; PMID:3470787; http://dx.doi.org/10.1073/pnas.84.8.2238
- Rybak SM, Vallee BL. Base cleavage specificity of angiogenin with Saccharomyces cerevisiae and Escherichia coli 5S RNAs. Biochemistry 1988; 27(7):2288-94; PMID:3289612; http://dx.doi.org/10.1021/bi00407a007
- Li Z, Ender C, Meister G, Moore PS, Chang Y, John B. Extensive terminal and asymmetric processing of small RNAs from rRNAs, snoRNAs, snRNAs, and tRNAs. Nucleic Acids Res 2012; 40(14):6787-99; PMID:22492706; http://dx.doi.org/10.1093/nar/gks307
- Pereira ER, Liao N, Neale GA, Hendershot LM. Transcriptional and post-transcriptional regulation of proangiogenic factors by the unfolded protein response. PLoS One 2010; 5(9); PMID:20824063; http://dx.doi.org/10.1371/journal.pone.0012521
- Kishimoto K, Yoshida S, Ibaragi S, Yoshioka N, Okui T, Hu GF, Sasaki A. Hypoxia-induced up-regulation of angiogenin, besides VEGF, is related to progression of oral cancer. Oral Oncol 2012; 48(11):1120-7; PMID:22694909; http://dx.doi.org/10.1016/j.oraloncology.2012.05.009
- Ibaragi S, Yoshioka N, Kishikawa H, Hu JK, Sadow PM, Li M, Hu GF. Angiogenin-stimulated rRNA transcription is essential for initiation and survival of AKT-induced prostate intraepithelial neoplasia. Mol Cancer Res 2009; 7(3):415-24; PMID:19258415; http://dx.doi.org/10.1158/1541-7786.MCR-08-0137
- Yamasaki S, Ivanov P, Hu GF, Anderson P. Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 2009; 185(1):35-42; PMID:19332886; http://dx.doi.org/10.1083/jcb.200811106
- Fu H, Feng J, Liu Q, Sun F, Tie Y, Zhu J, Xing R, Sun Z, Zheng X. Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 2009; 583(2):437-42. Epub 2008/12/31; PMID:19114040; http://dx.doi.org/10.1016/j.febslet.2008.12.043
- Czech A, Wende S, Morl M, Pan T, Ignatova Z. Reversible and rapid transfer-RNA deactivation as a mechanism of translational repression in stress. PLoS Genet 2013; 9(8):e1003767; PMID:24009533; http://dx.doi.org/10.1371/journal.pgen.1003767
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 2015; 12(9):835-7; PMID:26214130; http://dx.doi.org/10.1038/nmeth.3478
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 2015; 12(9):879-84; PMID:26237225; http://dx.doi.org/10.1038/nmeth.3508
- Klungland A, Dahl JA. Dynamic RNA modifications in disease. Curr Opin Genet Dev 2014; 26:47-52; PMID:25005745; http://dx.doi.org/10.1016/j.gde.2014.05.006
- Wang X, He C. Dynamic RNA modifications in posttranscriptional regulation. Mol Cell 2014; 56(1):5-12; PMID:25280100; http://dx.doi.org/10.1016/j.molcel.2014.09.001
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev 2010; 24(15):1590-5. Epub 2010/08/04; PMID:20679393; http://dx.doi.org/10.1101/gad.586710
- Tuorto F, Liebers R, Musch T, Schaefer M, Hofmann S, Kellner S, Frye M, Helm M, Stoecklin G, Lyko F. RNA cytosine methylation by Dnmt2 and NSun2 promotes tRNA stability and protein synthesis. Nat Struct Mol Biol 2012; 19(9):900-5; PMID:22885326; http://dx.doi.org/10.1038/nsmb.2357
- Blanco S, Dietmann S, Flores JV, Hussain S, Kutter C, Humphreys P, Lukk M, Lombard P, Treps L, Popis M, et al. Aberrant methylation of tRNAs links cellular stress to neuro-developmental disorders. EMBO J 2014; 33(18):2020-39; PMID:25063673
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Cortés-Garrido R, Gkatza N, et al. Stem cell function and stress response are controlled by protein synthesis. Nature 2016; 534(7607):335-40; PMID:27306184; http://dx.doi.org/10.1038/nature18282
- Emara M, Ivanov P, Hickman T, Dawra N, Tisdale S, Kedersha N, Hu GF, Anderson P. Angiogenin-induced tiRNAs promote stress-induced stress granule assembly. J Biol Chem 2010; 285(14):10959-68; PMID:20129916; http://dx.doi.org/10.1074/jbc.M109.077560
- Panas MD, Ivanov P, Anderson P. Mechanistic insights into mammalian stress granule dynamics. J Cell Biol 2016; 215(3):313-23; PMID:27821493; http://dx.doi.org/10.1083/jcb.201609081
- Ivanov P, Emara MM, Villen J, Gygi SP, Anderson P. Angiogenin-Induced tRNA Fragments Inhibit Translation Initiation. Mol Cell 2011; 43(4):613-23; PMID:21855800; http://dx.doi.org/10.1016/j.molcel.2011.06.022
- Lyons SM, Achorn C, Kedersha NL, Anderson PJ, Ivanov P. YB-1 regulates tiRNA-induced Stress Granule formation but not translational repression. Nucleic Acids Res 2016; 44(14):6949-60; PMID:27174937; http://dx.doi.org/10.1093/nar/gkw418
- Saikia M, Jobava R, Parisien M, Putnam A, Krokowski D, Gao XH, Guan BJ, Yuan Y, Jankowsky E, Feng Z, et al. Angiogenin-Cleaved tRNA Halves Interact with Cytochrome c, Protecting Cells from Apoptosis during Osmotic Stress. Mol Cell Biol 2014; 34(13):2450-63; PMID:24752898; http://dx.doi.org/10.1128/MCB.00136-14
- Yuan S, Akey CW. Apoptosome structure, assembly, and procaspase activation. Structure 2013; 21(4):501-15; PMID:23561633; http://dx.doi.org/10.1016/j.str.2013.02.024
- Deng J, Ptashkin RN, Chen Y, Cheng Z, Liu G, Phan T, Deng X, Zhou J, Lee I, Lee YS, et al. Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol Ther 2015; 23(10):1622-9; PMID:26156244; http://dx.doi.org/10.1038/mt.2015.124
- Wang Q, Lee I, Ren J, Ajay SS, Lee YS, Bao X. Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol Ther 2013; 21(2):368-79; PMID:23183536; http://dx.doi.org/10.1038/mt.2012.237
- Selitsky SR, Baran-Gale J, Honda M, Yamane D, Masaki T, Fannin EE, Guerra B, Shirasaki T, Shimakami T, Kaneko S, et al. Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep 2015; 5:7675; PMID:25567797; http://dx.doi.org/10.1038/srep07675
- Anderson P, Ivanov P. tRNA fragments in human health and disease. FEBS Lett 2014; 588(23):4297-304; PMID:25220675; http://dx.doi.org/10.1016/j.febslet.2014.09.001
- Moroianu J, Riordan JF. Nuclear translocation of angiogenin in proliferating endothelial cells is essential to its angiogenic activity. Proc Natl Acad Sci U S A 1994; 91(5):1677-81; PMID:8127865; http://dx.doi.org/10.1073/pnas.91.5.1677
- Moroianu J, Riordan JF. Identification of the nucleolar targeting signal of human angiogenin. Biochem Biophys Res Commun 1994; 203(3):1765-72; PMID:7945327; http://dx.doi.org/10.1006/bbrc.1994.2391
- Henras AK, Plisson-Chastang C, O'Donohue MF, Chakraborty A, Gleizes PE. An overview of pre-ribosomal RNA processing in eukaryotes. Wiley interdisciplinary reviews RNA 2015; 6(2):225-42; PMID:25346433; http://dx.doi.org/10.1002/wrna.1269
- Mullineux ST, Lafontaine DL. Mapping the cleavage sites on mammalian pre-rRNAs: where do we stand? Biochimie 2012; 94(7):1521-32; PMID:22342225; http://dx.doi.org/10.1016/j.biochi.2012.02.001
- Goodfellow SJ, Zomerdijk JC. Basic mechanisms in RNA polymerase I transcription of the ribosomal RNA genes. Subcell Biochem 2013; 61:211-36; PMID:23150253; http://dx.doi.org/10.1007/978-94-007-4525-4_10
- Schneider DA, French SL, Osheim YN, Bailey AO, Vu L, Dodd J, Yates JR, Beyer AL, Nomura M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc Natl Acad Sci U S A 2006; 103(34):12707-12; PMID:16908835; http://dx.doi.org/10.1073/pnas.0605686103
- Schneider DA, Michel A, Sikes ML, Vu L, Dodd JA, Salgia S, Osheim YN, Beyer AL, Nomura M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol Cell 2007; 26(2):217-29; PMID:17466624; http://dx.doi.org/10.1016/j.molcel.2007.04.007
- Xu ZP, Tsuji T, Riordan JF, Hu GF. The nuclear function of angiogenin in endothelial cells is related to rRNA production. Biochem Biophys Res Commun 2002; 294(2):287-92; PMID:12051708; http://dx.doi.org/10.1016/S0006-291X(02)00479-5
- Xu ZP, Tsuji T, Riordan JF, Hu GF. Identification and characterization of an angiogenin-binding DNA sequence that stimulates luciferase reporter gene expression. Biochemistry 2003; 42(1):121-8; PMID:12515546; http://dx.doi.org/10.1021/bi020465x
- Li S, Hu MG, Sun Y, Yoshioka N, Ibaragi S, Sheng J, Sun G, Kishimoto K, Hu GF. Angiogenin mediates androgen-stimulated prostate cancer growth and enables castration resistance. Mol Cancer Res 2013; 11(10):1203-14; PMID:23851444; http://dx.doi.org/10.1158/1541-7786.MCR-13-0072
- Sheng J, Yu W, Gao X, Xu Z, Hu GF. Angiogenin stimulates ribosomal RNA transcription by epigenetic activation of the ribosomal DNA promoter. J Cell Physiol 2014; 229(4):521-9; PMID:24122807; http://dx.doi.org/10.1002/jcp.24477
- Ruggero D, Pandolfi PP. Does the ribosome translate cancer? Nat Rev Cancer 2003; 3(3):179-92; PMID:12612653; http://dx.doi.org/10.1038/nrc1015
- Ibaragi S, Yoshioka N, Li S, Hu MG, Hirukawa S, Sadow PM, Hu GF. Neamine inhibits prostate cancer growth by suppressing angiogenin-mediated rRNA transcription. Clin Cancer Res 2009; 15(6):1981-8; PMID:19276260; http://dx.doi.org/10.1158/1078-0432.CCR-08-2593
- Barcaroli D, Bongiorno-Borbone L, Terrinoni A, Hofmann TG, Rossi M, Knight RA, Matera AG, Melino G, De Laurenzi V. FLASH is required for histone transcription and S-phase progression. ProcNatlAcadSciUSA 2006; 103(40):14808-12; PMID:17003125; http://dx.doi.org/10.1073/pnas.0604227103
- Yang XC, Burch BD, Yan Y, Marzluff WF, Dominski Z. FLASH, a proapoptotic protein involved in activation of caspase-8, is essential for 3′ end processing of histone pre-mRNAs. Mol Cell 2009; 36:267-78; PMID:19854135; http://dx.doi.org/10.1016/j.molcel.2009.08.016
- Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Phosphorylation of tristetraprolin, a potential zinc finger transcription factor, by mitogen stimulation in intact cells and by mitogen-activated protein kinase in vitro. J Biol Chem 1995; 270(22):13341-7; PMID:7768935; http://dx.doi.org/10.1074/jbc.270.22.13341
- Taylor GA, Thompson MJ, Lai WS, Blackshear PJ. Mitogens stimulate the rapid nuclear to cytosolic translocation of tristetraprolin, a potential zinc-finger transcription factor. Mol Endocrinol 1996; 10(2):140-6; PMID:8825554; http://dx.doi.org/10.1210/mend.10.2.8825554
- Lai WS, Carballo E, Strum JR, Kennington EA, Phillips RS, Blackshear PJ. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol 1999; 19(6):4311-23; PMID:10330172; http://dx.doi.org/10.1128/MCB.19.6.4311
- Kass S, Craig N, Sollner-Webb B. Primary processing of mammalian rRNA involves 2 adjacent cleavages and is not species specific. Mol Cell Biol 1987; 7(8):2891-8; PMID:3670298; http://dx.doi.org/10.1128/MCB.7.8.2891
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell 2006; 22(3):351-61; PMID:16678107; http://dx.doi.org/10.1016/j.molcel.2006.03.028
- Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep 2008; 9(8):774-80; PMID:18600236; http://dx.doi.org/10.1038/embor.2008.109
- Saxena SK, Rybak SM, Davey RT, Jr., Youle RJ, Ackerman EJ. Angiogenin is a cytotoxic, tRNA-specific ribonuclease in the RNase A superfamily. J Biol Chem 1992; 267(30):21982-6; PMID:1400510
- Lee FS, Vallee BL. Binding of placental ribonuclease inhibitor to the active site of angiogenin. Biochemistry 1989; 28(8):3556-61; PMID:2742853
- Saikia M, Krokowski D, Guan BJ, Ivanov P, Parisien M, Hu GF, et al. Genome-wide identification and quantitative analysis of cleaved tRNA fragments induced by cellular stress. J Biol Chem 2012; 287(51):42708-25; PMID:23086926; http://dx.doi.org/10.1074/jbc.M112.371799
- Gebetsberger J, Polacek N. Slicing tRNAs to boost functional ncRNA diversity. RNA biology. 2013; 10(12):1798-806; PMID:24351723; http://dx.doi.org/10.4161/rna.27177
- Haussecker D, Huang Y, Lau A, Parameswaran P, Fire AZ, Kay MA. Human tRNA-derived small RNAs in the global regulation of RNA silencing. RNA 2010; 16(4):673-95. Epub 2010/02/26; PMID: 20181738; http://dx.doi.org/rna.2000810[pii]10.1261/rna.2000810
- Honda S, Loher P, Shigematsu M, Palazzo JP, Suzuki R, Imoto I, et al. Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc Natl Acad Sci U S A. 2015; 112(29):E3816-25; PMID:26124144; http://dx.doi.org/10.1073/pnas.1510077112
- Anderson P, Kedersha N, Ivanov P. Stress granules, P-bodies and cancer. Biochimica et biophysica acta 2015; 1849(7):861-70; PMID:25482014; http://dx.doi.org/ 10.1016/j.bbagrm.2014.11.009
- Morgan GT, Reeder RH, Bakken AH. Transcription in cloned spacers of Xenopus laevis ribosomal DNA. Proc Natl Acad Sci U S A. 1983; 80(21):6490-4; PMID:6579535
- Hoang TT, Raines RT. Molecular basis for the autonomous promotion of cell proliferation by angiogenin. Nucleic Acids Res 2016; PMID:27915233; http://dx.doi.org/10.1093/nar/gkw1192
