 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
TUT4 and the closely related TUT7 are non-templated poly(U) polymerases required at different stages of development, and their mis-regulation or mutation has been linked to important cancer pathologies. While TUT4(7) interaction with its pre-miRNA targets has been characterized in detail, the molecular bases of the broader target recognition process are unclear. Here, we examine RNA binding by the ZnF domains of the protein. We show that TUT4(7) ZnF2 contains two distinct RNA binding surfaces that are used in the interaction with different RNA nucleobases in different targets, i.e that this small domain encodes diversity in TUT4(7) selectivity and molecular function. Interestingly and unlike other well-characterized CCHC ZnFs, ZnF2 is not physically coupled to the flanking ZnF3 and acts independently in miRNA recognition, while the remaining CCHC ZnF of TUT4(7), ZnF1, has lost its intrinsic RNA binding capability. Together, our data suggest that the ZnFs of TUT4(7) are independent units for RNA and, possibly, protein-protein interactions that underlay the protein’s functional flexibility and are likely to play an important role in building its interaction network.
Introduction
TUT4/Zcchc11 (zinc-finger, CCHC domain-containing protein 11) is a non-templated U-specific polymerase that, together with the related and partially redundant protein TUT7, plays a key role in organismal development. TUT4(7) is essential for oocyte development [Citation1], the maternal-to-zygotic transition [Citation2] and the later development of the embryo, among others [Citation3,Citation4]. Not surprisingly, dis-regulation of TUT4(7) is linked to a large set of cancer pathologies [Citation3,Citation5,Citation6].
TUT4(7) plays important roles in different stages in development and in adults these are linked to its uridylation of a broad range of RNA targets that differ in structure and sequence, including mRNAs [Citation7–9] but also miRNA precursors [Citation10–15] and mature miRNAs [Citation16–20]. In some settings, TUT4(7) binding leads to 3ʹ poly(U)ridylation and subsequent RNA degradation. In others, TUT4(7) mediates the addition of a single U that stabilizes the target RNA.
The best studied function of TUT4(7) is the regulation of the biogenesis of the tumour suppressor Let-7 miRNA. TUT4(7) acts in combination with the protein adaptor Lin28 to recognize and poly(U)ridylate the Let-7 miRNA precursor, leading to its degradation by the exonuclease DIS3L2 [Citation10,(Citation11,Citation12]. TUT4-mediated poly(U)ridylation of pre-Let-7 requires the formation of a three-component pre-let-7-Lin28-TUT4(7) complex, where TUT4(7) interacts with both the Lin28 protein and the pre-Let-7 miRNA. Recent structural and molecular work on the TUT4 and TUT7 proteins has provided an insight into this interaction but has also suggested that one of the ZnF domains of TUT4(7) makes direct contact with the polyU, and facilitates processive poly-uridylation [Citation21]. hTUT4(7) contains an amino-terminal domain, also known as the Lin-28 interacting module (LIM), and a carboxy terminal domain or catalytic module (CM) (). The CM module comprises a catalytically active domain (NTD2) sandwiched between two CCHC ZnFs (ZnF1 and 2) followed by a low sequence complexity region that includes a third ZnF (ZnF3). The CM interacts with the terminal base pair and the 3’single stranded end of the RNA molecule (7). Importantly, the second ZnF of the CM (ZnF2) makes contacts with the O2 and O4 moieties of a Uridine in position U2 of a single stranded poly U RNA. This specific contact has been proposed to stabilize the nascent poly(U) chain and to promote efficient poly(U) tailing [Citation21,Citation22].
Figure 1. A) Domain structure of hTUT4. The three CCHC ZnF domains are coloured in Orange, blue and green. the boundaries of the ZnF1, ZnF2 and ZnF3 constructs used in this study are reported using both the full-length hTUT4 amino acid count (black) and, for clarity, the amino acid count of that we use in this manuscript (colours). please note that the ZnF2-3 construct starts at the N-terminal boundary of ZnF2 and ends and the C-terminal boundary of ZnF3. b) [15N 1H] HSQC spectra of CCHC-ZnF2. c) O verlay of the [15N 1H] HSQC spectra of ZnF2 with NNNN RNA at ratios from 1 to 0 (light blue) to 1 to 10 (dark blue). d) S caffold Independent Analysis (SIA) of the hTUT4 ZnF2. left: SIA determines the nucleobase preference of a domain in any wanted position of the bound sequence by comparing the interaction of the domain with four quasi-degenerate RNA sequences, each with one of the four nucleobases in the position to be scanned. A workflow and representative data for this assay are found in Supplementary Figure 1. right: SIA scores for hTUT4 ZnF2. e) l.eft: overlay of [15N 1H] HSQC spectra recorded on ZnF2 with CGGA at protein to RNA ratios from 1 to 0 (light blue) to 1 to 10 (dark blue). I. nset: zoom of the Cys 3 amide resonance. In the inset, the corresponding peaks from equivalent titrations with the CGUA, CUGA RNAs are also displayed. please note that peaks from the other two titrations have been shifted in order to minimize overlap and better appreciate differences in saturation and linewidth. Top right: CSP measured in the CGGA titration plotted against the protein sequence
![Figure 1. A) Domain structure of hTUT4. The three CCHC ZnF domains are coloured in Orange, blue and green. the boundaries of the ZnF1, ZnF2 and ZnF3 constructs used in this study are reported using both the full-length hTUT4 amino acid count (black) and, for clarity, the amino acid count of that we use in this manuscript (colours). please note that the ZnF2-3 construct starts at the N-terminal boundary of ZnF2 and ends and the C-terminal boundary of ZnF3. b) [15N 1H] HSQC spectra of CCHC-ZnF2. c) O verlay of the [15N 1H] HSQC spectra of ZnF2 with NNNN RNA at ratios from 1 to 0 (light blue) to 1 to 10 (dark blue). d) S caffold Independent Analysis (SIA) of the hTUT4 ZnF2. left: SIA determines the nucleobase preference of a domain in any wanted position of the bound sequence by comparing the interaction of the domain with four quasi-degenerate RNA sequences, each with one of the four nucleobases in the position to be scanned. A workflow and representative data for this assay are found in Supplementary Figure 1. right: SIA scores for hTUT4 ZnF2. e) l.eft: overlay of [15N 1H] HSQC spectra recorded on ZnF2 with CGGA at protein to RNA ratios from 1 to 0 (light blue) to 1 to 10 (dark blue). I. nset: zoom of the Cys 3 amide resonance. In the inset, the corresponding peaks from equivalent titrations with the CGUA, CUGA RNAs are also displayed. please note that peaks from the other two titrations have been shifted in order to minimize overlap and better appreciate differences in saturation and linewidth. Top right: CSP measured in the CGGA titration plotted against the protein sequence](/cms/asset/3e63d3e6-c366-45d6-ac48-00e9fdafedfc/krnb_a_1991169_f0001_oc.jpg)
In addition to recognizing pre-miRNA, hTUT4(7) has been shown to interact with and regulate poly-uridylate mRNAs(7) [Citation1,Citation2,Citation8,Citation9], mature miRNAs [Citation16,Citation17,Citation18–20] and other ncRNAs [Citation23,Citation24]. Interestingly, one of the miRNAs regulated by TUT4(7) is the tumour suppressor Let-7i, and TUT4(7) has been shown to regulate Let-7i mono-uridylation and Hox gene regulation during development.
While structural and molecular work from a number of groups has provided an account of TUT4(7) interactions with the hLin28 protein and the Let-7 pre-miRNA target, we have a limited understanding of how hTUT4(7) recognizes the diverse set of cognate RNAs. It seems likely that the single-stranded, sequence specific ZnF domains play a key role in this recognition. ZnF2 has been shown to recognize RNA in the poly-Uridylated targets in a very specific structural setting, but how ZnF1 and ZnF3 interact with the RNA targets is unclear. In this work, we ask what is the role of TUT4(7) ZnF domains in RNA recognition.
Results
ZnF2 folds as an independent structural unit that recognizes specific RNA sequences
A recent crystal structure has shown that the second ZnF domain of hTUT7 makes specific contacts with a U within a UU dinucleotide, and it has been proposed that the domain stabilizes the poly(U) tail and increases processivity in pre-Let-7 poly-Uridylation. However, it is not clear whether this domain, which flanks the NTD2 catalytic domain, plays a wider role in recognizing and recruiting different RNA targets. Interestingly, ZnF2 directly engages in RNA recognition when NTD2 is bound to the UU di-nucleotide but not when a U1-derived RNA is bound instead. We therefore examined whether ZnF2 can fold and recognize RNA independently from NTD2, and what its base sequence specificity is. As a first step, we tested the fold and stability of the free ZnF2 using NMR. The 15N-correlation spectrum of the domain displays 29 well-dispersed peaks. These correspond to 29 backbone amide resonances, which is the expected number for ZnF2, and indicates that the construct folds into a single, structured, conformation in solution (). Then, we tested the sequence-independent RNA binding of ZnF2 using a randomized NNNN RNA tetramer and NMR spectroscopy. The comparison of 15N-correlation experiments recorded at different points of a titration showed selective changes in a set of ZnF2 amide resonances and confirmed RNA binding takes place on a specific surface ().
The resonance shifts in the NNNN titration indicate that the protein interacts with RNA with Kd values in the micromolar range. As NMR operates at micromolar concentrations of protein, it is ideally suited to report on the interactions. Next, we investigated de novo the nucleobase preference of hTUT4 ZnF2 using NMR Scaffold Independent Analysis (SIA), a method that can be used to define the nucleobase preference of isolated RNA-binding domains at each of the bound positions [Citation25] (Supplementary Figure 1). The concept of SIA is based on comparison assays where a domain is bound to quasi-degenerate sets of RNA, with only one specific nucleobase. The binding to the different sets is examined using the shift of peaks in the NMR spectra upon addition of the RNAs, which allows one to establish how RNA sets with different nucleobases in the same position bind to RNA. The data are then normalized and averaged across different RNAs and NMR peaks [Citation25] (Supplementary Figure 1), obtaining a series of comparative semi quantitative scores that report on the nucleobase preference of the ZnF domain (Supplementary Figure 1). In our SIA assays, we tested the potential of hTUT4 ZnF2 to recognize up to three nucleobases within a four-nucleotide RNA (we found it best to add a nucleotide at the 5ʹ to minimize boundary effects). Our analysis indicates that hTUT4 ZnF2 recognizes two Gs with significant specificity in the first and second scanned position (), while the third scanned position shows low nucleobase preference. We validated and quantified the nucleobase preference in positions one and two by comparing the binding of hTUT4 ZnF2 to the GG-containing CGGA sequence to the previously observed NNNN RNAs and to two sequences where one of the two Gs was mutated to U (CUGA and CGUA) ( and Supplementary Figure 2). In our NMR experiments, the resonances are in a fast exchange regime on a chemical shift timescale. Binding of CUGA and CGUA is too weak to obtain an accurate value for the Kd; however, comparison of the shift of peaks at equivalent protein to RNA ratios can be used as a proxy to evaluate the bound protein molar fraction. This indicated that CUGA and CGUA both interact with the protein, but with significantly lower affinity than CGGA (Supplementary Figure 2), in agreement with the SIA results (). Because the binding to CGGA RNA is tighter than to CUGA/CGUA saturation was reached in the titration. This allowed us to fit the shifts from the free to the bound position as a function of RNA concentration and obtain a Kd of 3.7 ± 0.4 µM for the ZnF2-CGGA complex (Supplementary Figure 3).
ZnF2 recognizes poly(U) RNA tails and G-rich target sequences using distinct surfaces
One important question is whether the TUT4(7) ZnF2 interacts with the CGGA RNA and the UU RNA in the TUT7 complex, using the same surface. In addition to providing information on affinity and specificity, our NMR Chemical Shift Perturbation (CSP) data allow us to define the ZnF2 surface that mediates the interaction with the CGGA RNA, provided we can assign the individual NMR cross-peaks to the amide resonances of specific amino acids. In order to do this, we assigned the backbone resonances of ZnF2 using a standard set of NMR experiments () and mapped the amide chemical shift changes of hTUT4 on the published structure of the close homologue hTUT7. This defined a continuous surface, where the largest changes are in the backbone amides of Cys 2, Cys 3, Met 13 and Lys 14 (). We then compared the ZnF2-CGGA and the ZnF2-UU interaction surfaces. The latter interaction involves the side chains of residues His 1355 and Lys 1352 (His 11 and Lys 8 in this work) (), which are highly conserved across species (Supplementary Figure 4). The comparison shows that ZnF2 uses opposite surfaces and very different residues to bind the two RNAs, with both sets of residues being completely conserved in TUT4 and TUT7. In order to validate that U recognition is context-dependent, we tested the binding of the isolated hTUT4 ZnF2 to a UUUU RNA and observed no significant binding (). The use of two alternative surfaces for recognizing RNA points to the ZnF2 domain playing an additional and much broader role in the recognition of the RNA targets which is dependent on the overall context.
Figure 2. The CCHC ZnF domain binds RNA using different surfaces in different protein-RNA complexes. a) Top: The protein-RNA interaction surfaces in the hTUT4 ZnF2-CGGA (NMR) and hTUT7 ZnF2-UU (X-ray) complexes are mapped in red (top) and yellow (bottom) respectively on the structure of hTUT7, which is here displayed using both a cartoon and surface representation, for clarity of visualization. bottom: the same colour coding is used to map the interactions on the sequence alignment of hTUT4 and hTUT7 ZFs2. b) O verlay of [15 N1 H] HSQC spectra of ZnF2 with UUUU at protein to RNA ratios 1 to 0 (green) and 1 to 10 (dark blue). The region containing the Cys3 and His11 residues has been enlarged. no significant chemical shifts perturbation is observed, indicating the lack of meaningful interactions. c) Top left, and right of this panel: the protein-RNA interaction surfaces in the hTUT4 ZnF2-CGGA and hLin28 ZnF2-AGGAGAU NMR complexes were mapped in red (top) and yellow (bottom) respectively on the structure of hLin28 ZnF2, which is here displayed using both a cartoon and surface representation, for clarity of visualization. Bottom left of the panel: the same colour coding is used to map the residues on the sequence alignment of the two domains. The two residues with the largest chemical shift changes upon RNA binding are the same in the hTUT4 and hLin28 complexes and they are highlighted with a box
![Figure 2. The CCHC ZnF domain binds RNA using different surfaces in different protein-RNA complexes. a) Top: The protein-RNA interaction surfaces in the hTUT4 ZnF2-CGGA (NMR) and hTUT7 ZnF2-UU (X-ray) complexes are mapped in red (top) and yellow (bottom) respectively on the structure of hTUT7, which is here displayed using both a cartoon and surface representation, for clarity of visualization. bottom: the same colour coding is used to map the interactions on the sequence alignment of hTUT4 and hTUT7 ZFs2. b) O verlay of [15 N1 H] HSQC spectra of ZnF2 with UUUU at protein to RNA ratios 1 to 0 (green) and 1 to 10 (dark blue). The region containing the Cys3 and His11 residues has been enlarged. no significant chemical shifts perturbation is observed, indicating the lack of meaningful interactions. c) Top left, and right of this panel: the protein-RNA interaction surfaces in the hTUT4 ZnF2-CGGA and hLin28 ZnF2-AGGAGAU NMR complexes were mapped in red (top) and yellow (bottom) respectively on the structure of hLin28 ZnF2, which is here displayed using both a cartoon and surface representation, for clarity of visualization. Bottom left of the panel: the same colour coding is used to map the residues on the sequence alignment of the two domains. The two residues with the largest chemical shift changes upon RNA binding are the same in the hTUT4 and hLin28 complexes and they are highlighted with a box](/cms/asset/c5209096-3aae-4f09-b7d9-618f904f1d22/krnb_a_1991169_f0002_oc.jpg)
Interestingly, while we have no molecular data on the interaction of TUT4(7) (or other individual CCHC ZnFs) in complex with a G-rich sequence, high resolution structural data and biophysical data exists on the interaction of double CCHC ZnFs and short G-rich sequences. In particular, both high resolution structural information and CSP data are available on the interaction between the ZnF di-domain of hLin28 and the pre-Let-7 RNA target sequence. In this di-domain unit, the second ZnF interacts with a four nucleotide stretch within a larger RNA, with extensive contacts made with two central Gs whose individual mutation results in a drop in the affinity of the interaction [Citation26]. We therefore compared the ZnF2-CGGA interaction surface mapped by our CSP data with the surface mediating the interaction between hLin28 ZnF2 and GG-containing 4-nucleotide RNA sequence, mapped using public domain data [Citation26]. The comparison indicated that equivalent surfaces are used by hTUT4 ZnF2 and hLin28 ZnF2 (), although the residues mediating the interactions are not conserved between the two proteins, which implies the details of nucleobase recognition are different.
ZnF2 is uncoupled from ZnF3 and acts independently in miRNA recognition
Two important questions are whether ZnF2 interaction with a short G-rich sequence recapitulates the one with a physiological RNA target(s) and, relatedly, whether the domain cooperates with hTUT4 ZnF1 and ZnF3 domains in this interaction. First, we explored the affinity and kinetics of ZnF2 interaction with the well-characterized Let7i miRNA and, compared them with those obtained for the interaction with a short RNA containing the SIA isolated sequence () using Biolayer Interferometry (BLI). In our BLI assays, a biotinylated RNA is immobilized on a sensor and exposed to a series of protein concentrations () and, in the short RNA, the SIA sequence has been distanced from the 5ʹ to prevent steric hindrance. Our data show that ZnF2 binds to both RNAs with affinity in the low micromolar range (~4 μM), and very similar kinetics (). Interestingly, the Let-7i sequence includes a GG di-nucleotide and a number of isolated Gs (), and mutation of the Gs in the miRNA resulted in the cumulative loss of binding and function [Citation10,Citation16].
Figure 3. hTUT4 ZnF2-RNA binding using BioLayer Interferometry (BLI). a) Biotinylated RNAs used in the assays. b) Interferograms recorded using Streptavidin sensors coated with the biotinylated RNAs, in buffer and then exposed to increasing concentrations (1 µM, 2 µM, 4 µM and 8 µM) of hTUT4 ZnF2-3 domain. The interferograms are aligned using the buffer baseline. The baseline and the association and dissociation steps are displayed in the top panels. The region used to fit the association curves and to calculate kobs is displayed in the bottom panel. c) Plots of kobs at the different protein concentration. The plots were used to calculate kon and koff, and from those Kd, using the program Anabel
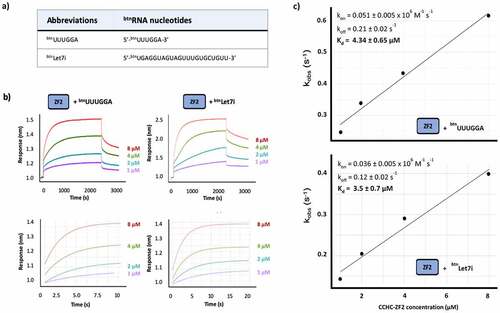
Next, we want to understand whether ZnF2 and ZnF3 cooperate to create a stronger or more long-lasting interaction. CCHC ZnF domains have been shown to recognize RNA as a di-domain unit. However, the linker between the two domains is typically of a few amino acids. TUT4(7) ZnF2 is separated from the neighbouring ZnF3 domain by a long (~ 40 amino acids) linker. Further, although we have previously shown that ZnF3 selectively binds G-containing RNAs [Citation25], the role of ZnF3 in promoting poly(U)ridylating pre-Let-7 miRNA is debated [Citation21,Citation22]. Whether the ZnF2 and ZnF3 domains cooperate in the recognition of the miRNA targets, and in general of TUT4 G-rich targets, is not known and requires direct investigation.
In order to assess whether the ZnF2 and ZnF3 domains are interacting and operating as an RNA binding unit, we initially compared the fingerprint 15N-correlation NMR spectra of the ZnF2, ZnF3 and ZnF2-3 constructs (). Comparison of the spectra indicated that only minor differences are observed between the resonances of the isolated domains and the same resonances in two-domain construct indicating the two domains are unlikely to interact (). Consistently, the resonance attributable to the ZnF2-ZnF3 interdomain linker are in the random coil region, indicating the linker is unfolded. In order to validate this initial conclusion, backbone amide 15N T1 and T2 NMR relaxation experiments were recorded on the two-domain construct ( and Supplementary Figure 5). The analysis of the relaxation series showed that ZnF2 and ZnF3 have rotational correlation times of τc = 4.2 ± 0.6ns, and τc = 5.2 ± 0.9ns, respectively. The domains’ correlation times confirm they are largely independent structural units that do not form a stable interaction. Next, we asked whether the two domains cooperate in binding the Let-7i miRNA. To answer this, we measured the strength and kinetics of the interaction between the ZnF2-3 di-domain unit and both the Let7i RNAs and the UUUGGA RNA, as a control where only one domain can bind (). We then compared the equilibrium and kinetic parameters with the ones obtained for the isolated ZnF2. Our experiments show that the ZnF2 and ZnF2-3 bind to both the miRNA and the short control RNA with very similar affinity and kinetics, although the ZnF2-3-miRNA complex has a slightly longer lifetime and slower association than the ZnF2 complex (~2-fold), and that binding of the two domains is, by and large, decoupled.
Figure 4. Structural and functional assessment of the coupling between hTUT4 ZnF2 and ZnF3 domains. a) Overlay of [15N 1H] HSQC spectra of ZnF2 (blue), ZnF3 (green) and ZnF2-3 di-domain (black). The organization of the two domains and the boundaries of the three constructs are displayed. b) 15N T1 and T2 and rotational correlation time (τc) values of the ZnF2-3 amides are plotted along the protein sequence. Values for the ZnF2 and for the ZnF3 amides are reported in two separate plots, to better compare the two domains. Residues number refers to the sequence of each domain. c) Plots of Kobs at the different protein concentration. The plots were used to calculate kon and koff, and from those Kd, using the program Anabel
![Figure 4. Structural and functional assessment of the coupling between hTUT4 ZnF2 and ZnF3 domains. a) Overlay of [15N 1H] HSQC spectra of ZnF2 (blue), ZnF3 (green) and ZnF2-3 di-domain (black). The organization of the two domains and the boundaries of the three constructs are displayed. b) 15N T1 and T2 and rotational correlation time (τc) values of the ZnF2-3 amides are plotted along the protein sequence. Values for the ZnF2 and for the ZnF3 amides are reported in two separate plots, to better compare the two domains. Residues number refers to the sequence of each domain. c) Plots of Kobs at the different protein concentration. The plots were used to calculate kon and koff, and from those Kd, using the program Anabel](/cms/asset/707699e2-39cf-4f53-ada2-3bee37ad647c/krnb_a_1991169_f0004_oc.jpg)
hTUT4 ZnF1 has lost its ssRNA binding capability
Finally, we wanted to assess whether the first CCHC ZnF1 of hTUT4 may also, in principle contribute to the recognition of the ssRNA regions/targets. The structural work by Daehnle and co-workers indicates this domain is likely to be decoupled by the NTD2 structure. We expressed and purified ZnF1, and recorded an NMR 15N-correlation spectrum that showed the domain’s amide resonance are well dispersed and the peaks have similar intensity. The number of peaks is consistent with the number of residues in the folded part of the domain confirming that ZnF1 is folded and assumes one stable conformation ().
Figure 5. A) [15N 1H] HSQC spectra of CCHC-ZnF1. b) Overlay of the [15N 1H] HSQC spectra of ZnF1 free and with a 1:4 protein:NNNN RNA ratio. Zoom region of one ZnF1 residue to confirm spectral alignment and spectra quality. c) Sequence alignment of CCHC-type zinc fingers from hTUT4, HIV-1 nucleocapsid and hLin28 proteins. The ClustalX colour scheme is used to highlight conservation and residue type. In hTUT4 CCHC-ZnF1, two serines substitute bulky residues which are involved in RNA binding in the Lin-28 and Nucleocapside structures. d) Overlay of the [15N 1H] HSQC spectra of CCHC-ZnF3 (left) and CCHC-ZnF3 F1360S/V1368S mutant (right) free (black) and with NNGN RNA at a 1 to 4 ratio (green), which shows how mutating the two serines above impairs RNA binding
![Figure 5. A) [15N 1H] HSQC spectra of CCHC-ZnF1. b) Overlay of the [15N 1H] HSQC spectra of ZnF1 free and with a 1:4 protein:NNNN RNA ratio. Zoom region of one ZnF1 residue to confirm spectral alignment and spectra quality. c) Sequence alignment of CCHC-type zinc fingers from hTUT4, HIV-1 nucleocapsid and hLin28 proteins. The ClustalX colour scheme is used to highlight conservation and residue type. In hTUT4 CCHC-ZnF1, two serines substitute bulky residues which are involved in RNA binding in the Lin-28 and Nucleocapside structures. d) Overlay of the [15N 1H] HSQC spectra of CCHC-ZnF3 (left) and CCHC-ZnF3 F1360S/V1368S mutant (right) free (black) and with NNGN RNA at a 1 to 4 ratio (green), which shows how mutating the two serines above impairs RNA binding](/cms/asset/35eb03c4-f9db-4aa5-bf71-b04fd6e4d63d/krnb_a_1991169_f0005_oc.jpg)
Next, we explored ZnF1 RNA binding properties by titrating a NNNN RNA tetramer and monitoring spectra changes using NMR. We used a fully degenerate sequence in order to capture the sequence specificity of the domain which is unknown. The comparison of 15N-correlation experiments recorded at different titration points showed that ZnF1 amide resonances did not change their position in the presence of RNA, indicating the domain does not bind to RNA at a µM protein and RNA concentration (). This was surprising and in order to rationalize the lack of ZnF1 RNA binding activity, we compared the sequence of hTUT4 ZnF1 with the one of other two CCHC ZnF domains (ZnF2 and ZnF3) and with the sequences of the hLin28 and NCP double ZnFs [Citation27], and mapped the residues important for the interaction in well-studied CCHC ZnF-RNA complexes. The sequence alignment highlights that a variable bulky hydrophobic residue that plays a key role in protein-RNA binding in the complexes above and is conserved in the ZnF2 and ZnF3 domains, is instead mutated to a Serine in ZnF1 (). A second, less conserved, bulky residue is also mutated to Serine. These differences provide a possible explanation for the loss of RNA binding by hTUT4 ZnF1. To validate our hypothesis, we generated a chimera in the ZnF3 domain where these residues are mutated to serine (F1360S/V1368S), to create a KH1 mimic. We then tested the mutant’s folding and RNA binding properties (). NMR 15N-correlation experiments showed that mutant ZnF3 maintains its fold but loses the capability to bind to a previously tested RNA oligo, NNGN [Citation28].
Discussion
TUT4, and the closely related TUT7 protein, are key factors in mammalian development that regulate the stability and processing of a range of RNA targets. The role of TUT4(7) in the uridylation of Let-7 miRNA precursor molecules has been studied in detail, but it is unclear how the protein(s) would recognize other ssRNA targets, including a set of mature ss miRNA molecules regulating immune response, growth hormone and development [Citation16–18]. Here, we examine RNA binding by TUT4 ZnF domains and report that, while ZnF1 does not interact with ssRNA, ZnF2 recognizes a G-rich sequence using a surface alternative to the one mediating poly(U) recognition during the uridylation of Let-7 miRNA precursor. We then discuss the role of ZnF2 in the TUT4 interaction with the Let-7i miRNA, as a paradigm for the mature miRNA targets [Citation21,Citation26,Citation29,Citation30], and explore its relation with the adjacent ZnF3.
Our data show that hTUT4 ZnF1 and ZnF2 are independent folding units. However, while ZnF1 does not bind RNA, ZnF2 recognizes ssRNA with significant specificity. Importantly, ZnF2 does not recognize the U nucleobase that has been shown to make specific contacts with two amino acid side chains in a larger hTUT7 protein construct [Citation21]. These contacts have been proposed to strengthen TUT4(7) processivity, leading to the synthesis of longer poly(U) chains [Citation21]. Instead, our SIA assays and follow-up validation indicate that ZnF2 recognizes a GG di-nucleotide. A mechanistically important question is whether the interaction with the G-rich targets would prevent hTUT4 engaging the poly(U) tail of the pre-miRNA molecules, acting as a general molecular switch between different functions. While we do not have a definitive answer for the specific Let-7 pre-miRNA complex, the comparison of the two U- and GG-recognition surfaces indicates that the domain uses two non-overlapped sets of residues on opposite sides of ZnF2 to recognize the different sequences, and binding of one RNA does not imply release of the other. In addition, we do not observe binding of poly(U) sequences in our model system, even in the absence of other RNA, hinting that U recognition is dependent on a specific structural context. Regardless, our data indicate that ZnF2 uses a G-specific RNA-binding surface to recognize and recruit single stranded G-containing miRNA targets for TUT4 mono-uridylation and that the domain plays a role which is broader than the support of processive poly-uridylation in the pre-miRNA targets.
Many of TUT4(7) G-rich targets, including its miRNA targets, are regulated by adding a single Uridine [Citation16]. In our model, the ZnF2 domain would help recognize those targets and recruit them to the polymerase. Therefore, we tested the interaction between ZnF2 and the best studied of these miRNA, Let-7i. Our data show that ZnF2 binds the full-length miRNA and the short SIA-derived G-rich target with very similar affinity and kinetics, indicating an interaction with a single site and an affinity which is not dependent on an avidity effect. This is confirmed by the association and dissociation kinetics which are similar to the ones we have observed for the interaction of single stranded, sequence specific RNA binding domains with the cognate RNAs [Citation31,Citation32]. This indicates that ZnF2 interacts with individual sites with low μM Kd, and a higher affinity in the cell is likely reached by multiple synergistic interactions.
CCHC ZnF domains have been shown to recognize RNA sequences as part of di-ZnF structural units [Citation26]. Although the linker between ZnF2 and ZnF3 is longer than the one observed in other di-domain units, ZnF2 could synergize with the downstream ZnF3, which we have shown also prefers G-rich sequences [Citation25], to achieve the required affinity for the RNA target. Our chemical shift and relaxation NMR data, indicate that the ZnF2-3 linker is flexible and that the two domains do not interact in the free protein. Our data also show that the affinity and kinetics of the two-domain ZnF2-3 interaction with the miRNA are similar to the ones of the isolated ZnF2, indicating the presence of ZnF3 does not increases the affinity of ZnF2 for the physiological RNA target. It is worth mentioning that, we do not take this to imply that the two ZnFs are not binding simultaneously to the RNA molecule in vivo. Such binding may take place and, for example, help orient the RNA in the larger TUT4 complex. Rather, the decoupling of the two domains indicates that, contrary to what is observed for hLin28, target recognition by hTUT4 ZnFs is mediated by binding of individual domains that recognize very short RNA sequences. This is consistent with the overall molecular context being more important for hTUT4-RNA than for hLin28-RNA recognition, and with hTUT4 recognizing a broad range of RNA targets.
Our data on hTUT4 ZnF domains, reveal a context-dependent RNA recognition mode for hTUT4(7) ZnF2 and lead to a target selection model where the individual CCHC ZnF domains of hTUT4(7) can play multiple roles by stabilizing the nascent poly(U) chain, recruiting G-rich targets and possibly orienting these targets in the larger protein complex. How the latter would take place will require high-resolution structural information on a complex between the large hTUT4(7) CM module and a G-rich RNA target. Importantly, ZnF2 is not coupled to ZnF3 in binding the miRNA target, and the affinity and kinetics of the interaction are typical of individual protein domain binding short RNA sequences, rather than of a CCHC ZnF di-domain. It seems likely that, as for many other RNA binding proteins, higher affinity is reached by multiple inter-molecular interactions and, in this context, the role of ZnF1, which does not bind RNA and yet has a conserved interaction surface, may be to mediate some of the complex’s protein-protein contacts. Overall, our data paint a picture of complexity, where the different CCHC ZnF domains of TUT4(7) play very different roles and contribute to creating a network of interactions that define the multi-faceted function of this protein. Dissecting the interaction network using functional and molecular studies in different settings will represent a key step of understanding this global role.
Material and methods
Cloning, expression and purification of the protein constructs
hTUT4 zinc finger constructs CCHC‐ZnF1 (903‐931), CCHC‐ZnF2 (1293‐1321), CCHC‐ZnF2-3 (1293‐1382) and CCHC-ZnF3 (1354–1382) (PubMed Accession number NP_001009881, sequence in Supplementary Figure 6) were cloned from human cDNA into a pET‐M11 vector [Citation33] to obtain protein constructs that include an N‐terminal hexa‐histidine tag (His6‐tag) followed by a cleavable tobacco etch virus (TEV) site and whose expression is under the control of a T7 promoter. Primers were designed to introduce the mutation F1360S/V1368V into CCHC-ZnF3. Point mutations were introduced into the construct by amplification of the plasmid using overlapping complementary primers with the mutation of interest inserted at the centre of the oligonucleotides. Following PCR amplification parent DNA was removed by DpnI digestion. The five constructs (ZnF1, ZnF2, ZnF2-3, ZnF3 and ZnF3 F1360S/V1368V) were expressed and purified using the protocols below.
In order to obtain 15N and, if required, 13C-labelled protein samples, the protein was expressed in M9 minimal media containing respectively (15NH4)2SO4 as the only nitrogen source and 13C6‐D‐glucose as the only carbon source. E. coli BL21(DE3) cells were transformed with the pET-M11 relevant plasmid and grown overnight at 37°C. The overnight culture was diluted to an OD600 of 0.1 in fresh media and cells were grown to an OD600 of 0.6 before the temperature was reduced to 22°C and protein expression induced with 0.5 mM IPTG. Protein expression was overnight at 22°C, and cells were harvested by centrifugation and stored at ‐80°C.
Cells were sonicated in denaturing buffer pH 8.0 (100 mM NaH2PO4, 10 mM Tris‐HCl pH 8.0, 8 M urea) and the lysate was centrifuged at ~7700 x g for 30 minutes. The supernatant was loaded on a Ni-NTA column and purified by immobilized metal ion affinity chromatography (IMAC). In more detail, 4 ml of Ni‐NTA resin per litre of cell culture was packed into a gravity flow column and equilibrated with 10 column volumes (CV) denaturing buffer. The clarified cell lysate was loaded onto the column and the flow through was collected and reloaded onto the column to enhance binding of the expressed protein. The protein was eluted with denaturing buffer pH 4.5 (100 mM NaH2PO4, 10 mM Tris‐HCl pH 4.5, 8 M urea). The eluted protein was dialysed against 4 litres of equilibration buffer (10 mM Tris pH 8.0, 10 mM Imidazole, 200 mM NaCl, 2 mM β‐mercaptoethanol, 10 μM ZnCl2) to reduce the urea content in the buffer. TEV protease was then added to a final concentration of 2.5 μM to cleave the His6‐tag. The reaction mixture was dialysed for a further 16 hours for cleavage to take place. Both protease and cleaved tag were separated from the protein by a reverse IMAC column (Ni‐NTA). The cleavage reaction mixture was loaded into a gravity flow column packed with Ni‐NTA resin equilibrated with 10 mM Tris pH 8.0, 10 mM Imidazole, 200 mM NaCl, 2 mM β‐mercaptoethanol, 10 μM ZnCl2. The flow through was collected and reloaded onto the column for a second time and the flow through from the second column was concentrated using a Vivaspin concentrator and further purified using a Hiload 16/60 Superdex 75 gel filtration column. Fractions containing pure protein as assessed by SDS-PAGE were pooled and concentrated before being dialysed into a final buffer of 10 mM Tris‐HCl pH 7.4, 100 mM NaCl, 0.5 mM TCEP and 10 μM ZnCl2 and concentrated in a Vivaspin to the final storage concentration. Protein concentration was determined from the absorbance at 280 nm and the molecular weights of the different constructs confirmed by mass spectrometry.
RNA sample preparation
RNA oligonucleotides (CGGA, CUGA, CGUA, UUUU, btnUUUGGA, btnUGAGGUAGUAGUUUGUGCUGUU and btnUCACCUACUACUUUCUCCUCUU) were purchased from Horizon Discovery, and deprotected as advised. RNA concentration was determined from the 260 nm absorbance of the sample.
NMR protein backbone assignment experiments
Labelled (15N or 13C15N) hTUT4 samples were prepared as described and concentrated to ~200 μM. NMR experiments were recorded at 25°C using a Bruker Avance NMR spectrometer operating at 600 MHz 1H frequency. Measurements were made in 90% H2O/10% D2O. A 13C15N labelled hTUT4 CCHC‐ZnF2 sample was used to acquire a HNCA, HN(CO)CA [Citation34], HNCACB [Citation35], HN(CO)CACB [Citation36] and HNCO at 10 mM Tris‐HCl pH 7.4, 100 mM NaCl, 0.5 mM TCEP and 10 μM ZnCl2 buffer conditions. Data were processed using Topspin 3.7 and assignment of protein resonances was done on CcpNmr analysis [Citation37].
NMR relaxation measurements
Relaxation experiments [Citation38] were recorded on a 15N‐labelled hTUT4 ZnF2-3 sample to obtain T1 and T2 values. Experiments were performed on a Varian Inova NMR spectrometer operating at 800 MHz 1H frequency. 20, 200, 400, 700, 1000, 1500 ms delays and 10, 20, 40, 70, 100, 150 ms delays were used for T1 and T2 measurements, respectively. Standard relaxation values were determined for each residue by fitting an exponential decay to the peak volume over the course of the data collected. Residues were excluded where overlap in the signals prevented accurate measurement on peak volume.
The hTUT4 ZnF2 peaks used in the di-domain relaxation to calculate the correlation time in that domain are C1295, R1296, V1297, G1299, G1302, H1303, Y1304, M1305, D1307, K1310 and R1311 (C3, R4, V5, G7, G10, H11, Y12, M13, D15, K18 and K19 in this work). In the hTUT4 ZnF3 case the residues used were C1360, F1361, I1362, G1364, D1365, A1366, G1367 and H1368 (C7, F8, I9, G11, D12, A13, G14, H15, in this work).
NMR titration experiments
NMR RNA titrations were performed in 10 mM Tris‐HCl pH 7.4, 100 mM NaCl, 0.5 mM TCEP and 10 μM ZnCl2 buffer conditions at 25°C. 15N HSQC NMR spectra were recorded during the titration of a 50 μM protein sample with increasing at each concentration of the different RNAs (at protein RNA ratios 1 to 0, 0.5, 1, 2, 4, 6, 8, 10, 12). Data were processed using Topspin 3.7 (Bruker) and a value for the 1H and 15N Chemical Shift changes was calculated for each residue applying the following equation: ( to the shifts in the two dimension.
The Kd for the hTUT4 ZnF2-CGGA interaction were calculated in the CcpNmr analysis program using the A(B + x-sqrt((B + x)^2-4x) equation. Kd were calculated for individual residues and then merged to obtain the global Kdan average of all individual Kd per residue [Citation39].
NMR scaffold independent analysis
70 μM hTUT4 ZnF2 stock was prepared in 100 mM NaCl, 10 mM Tris pH 7.4 buffer containing 0.5 mM TCEP, 10 μM ZnCl2, sodium azide and RNasin Plus. This was aliquoted out into 180 μM samples and RNA pools added where required at a protein to RNA ratio of 1 to 4. Samples were transferred to 3 mm NMR tubes and placed in a rack. Samples were automatically loaded into the spectrometer using the Bruker Sample Jet and 1H‐15N SOFAST‐HMQC spectra were recorded for each of the samples. NMR data were recorded on a Bruker Avance NMR spectrometer at 700 MHz. Spectra were processed using NmrPipe and data analysed using the weighted average chemical shift changes (for HN and N) with each RNA pool for a subset of shifting residues being measured and normalized with respect to the highest shift. The average normalized value across the subset of peaks is taken to give a comparative score of binding preference.
BioLayer Interferometry (BLI) measurements
BLI experiments were performed in 10 mM Tris pH 7.4, 100 mM NaCl, 0.5 mM TCEP, 10 μM ZnCl2, 0.5 mg/ml bovine serum albumin (BSA) on an Octet Red 96 instruments (ForteBio, Inc. Menlo Park, CA) operating at 25°C. 5ʹ-Biotinylated 5ʹ-btnUUUGGA-3ʹ and Let7i miRNA variations (5ʹ-btnUGAGGUAGUAGUUUGUGCUGUU-3ʹ, 5ʹ-btnUCACCUACUACUUUCUCCUCUU-3ʹ) (1 ng μL−1 solutions) were immobilized on streptavidin-coated biosensors and incubated with varying concentrations of hTUT4 ZnF2 and ZnF2-3 (1–8 μM). Equilibrium and kinetic parameters were extracted by the analysis of the observed association rate constant versus ligand concentration using the program Anabel, as described [Citation40].
Sequence alignment and structures used
All sequence alignments were carried out using the programs T-Coffe [Citation41] and EMBOSS Needle [Citation42] with default settings. Figures were generated using ClustalX [Citation43]. The hTUT7 coordinates used in our image building derive from the 5W0M PDB file, while the coordinates of the hLin28 ZnFs were extracted from the PDB file 2LI8.
Author Contributions
Cloning of the TUT4 ZnFs constructs was performed by KMC and EC. The protein samples were prepared by KMC and BCA. NMR experiments were recorded and analysed by KMC, BCA and GN. BLI experiments were recorded by BCA and analysed by BCA and SRM. The project was designed by A.R. and the manuscript was written by A.R. and KMC with contributions from all authors.
Data Sharing Statement
The plasmids expressing the constructs discussed in this study will be made available by the authors upon request. The resonances assignment of hTUT4 ZnF2 protein will be available the BMRB database (https://bmrb.io/) upon publication of the manuscript.
Supplemental Material
Download PDF (1.6 MB)Acknowledgments
NMR spectra were recorded at the MRC Biomedical NMR and UCL NMR facilities and we thank Geoff Kelly, Alain Oregioni and Angelo Pinto De Figueiredo for assistance.
Disclosure statement
The authors declare no competing financial interests.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Morgan M, Much C, DiGiacomo M, et al. MRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature. 2017;548(7667):347–351.
- Chang H, Yeo J, Kim JG, et al. Terminal uridylyltransferases execute programmed clearance of maternal transcriptome in vertebrate embryos. Mol Cell. 2018;70(1):72–82.e7.
- Menezes MR, Balzeau J, Hagan JP. 3ʹ RNA uridylation in epitranscriptomics, gene regulation, and disease. Front Mol Biosci. 2018;5. DOI:10.3389/fmolb.2018.00061
- Yu S, Kim VN. A tale of non-canonical tails: gene regulation by post-transcriptional RNA tailing. Nat Rev Mol Cell Biol. 2020;21(9):542–556.
- Barbieri IKouzarides T. Role of RNA modifications in cancer. Nat Rev Cancer. 2020;20(6:303-322.
- HallettRM, Hassell JA. E2F1 and KIAA0191 expression predicts breast cancer patient survival. BMC Res Notes. 2011;4(1). DOI:10.1186/1756-0500-4-95
- Lim J, Ha M, Chang H, et al. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell. 2014;159(6):1365–1376.
- Morgan M, Kabayama Y, Much C, et al. A programmed wave of uridylation-primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis. Cell Res. 2019;29(3):221–232.
- Thomas MP, Liu X, Whangbo J, et al. Apoptosis triggers specific, rapid, and global mRNA decay with 3ʹ uridylated intermediates degraded by DIS3L2. Cell Rep. 2015;11(7):1079–1089.
- Heo I, Joo C, Kim YK, et al. TUT4 in Concert with Lin28 Suppresses MicroRNA Biogenesis through Pre-MicroRNA Uridylation. Cell. 2009;138(4):696–708.
- Heo I, Ha M, Lim J, et al. Mono-uridylation of pre-microRNA as a key step in the biogenesis of group II let-7 microRNAs. Cell. 2012;151(3):521–532.
- Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16(10):1021–1025.
- Kim B, Ha M, Loeff L, et al. TUT 7 controls the fate of precursor micro RNAs by using three different uridylation mechanisms. EMBO J. 2015;34(13):1801–1815.
- Kim H, Kim J, Yu S, et al. A mechanism for microRNA arm switching regulated by uridylation. Mol Cell. 2020;78(6):1224–1236.e5.
- Kim H, Kim J, Kim K, et al. Bias-minimized quantification of microRNA reveals widespread alternative processing and 3 end modification. Nucleic Acids Res. 2019;47(5):2630–2640.
- Thornton JE, Du P, Jing L, et al. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res. 2014;42(18):11777–11791.
- Jones MR, Quinton LJ, Blahna MT, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11(9):1157–1163.
- Jones MR, Blahna MT, Kozlowski E, et al. Zcchc11 uridylates mature miRNAs to enhance neonatal IGF-1 expression, growth, and survival. PLoS Genet. 2012;8(11):e1003105.
- Yang A, Bofill-De Ros X, Shao TJ, et al. 3′ uridylation confers miRNAs with non-canonical target repertoires. Mol Cell. 2019;75(3):511–522.e4.
- Gutiérrez-Vázquez C, Enright AJ, Rodríguez-Galán A, et al. 3′ Uridylation controls mature microRNA turnover during CD4 T-cell activation. RNA. 2017;23(6):882–891.
- Faehnle CR, Walleshauser J, Joshua-Tor L. Multi-domain utilization by TUT4 and TUT7 in control of let-7 biogenesis. Nat Struct Mol Biol. 2017;24(8):658–665.
- Thornton JE, Chang HM, Piskounova E, et al. Lin28-mediated control of let-7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA. 2012;18(10):1875–1885.
- Pirouz M, Du P, Munafò M, et al. Dis3l2-mediated decay is a quality control pathway for noncoding RNAs. Cell Rep. 2016;16(7):1861–1873.
- łabno A, Warkocki Z, Kulínski T, et al. Perlman syndrome nuclease DIS3L2 controls cytoplasmic non-coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Res. 2016. Dec 1;44(21):10437-10453. DOI:10.1093/nar/gkw649
- Collins KM, Oregioni A, Robertson LE, et al. Protein-RNA specificity by high-throughput principal component analysis of NMR spectra. Nucleic Acids Res. 2015;43(6):e41.
- Loughlin FE, Gebert LFR, Towbin H, et al. Structural basis of pre-let-7 miRNA recognition by the zinc Knuckles of pluripotency factor Lin28. Nat Struct Mol Biol. 2012;19(1):84–91.
- Yamashita S, Nagaike T, Tomita K. Crystal structure of the Lin28-interacting module of human terminal uridylyltransferase that regulates let-7 expression. Nat Commun [Internet]. 2019;10(1):1960. Available from:
- Hollingworth D, Candel AM, Nicastro G, et al. KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res. 2012 Aug;40(14):6873–6886.
- Rybak A, Fuchs H, Smirnova L, et al. A feedback loop comprising lin-28 and let-7 controls pre-let-7 maturation during neural stem-cell commitment. Nat Cell Biol. 2008 Aug;10(8):882–891.
- Lee W, Revington MJ, Arrowsmith C, et al. A pulsed field gradient isotope-filtered 3D 13C HMQC-NOESY experiment for extracting intermolecular NOE contacts in molecular complexes. FEBS Lett. 1994 Aug;350(1):87–90.
- Dagil R, Ball NJ, Ogrodowicz RW, et al. IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties. Nucleic Acids Res. 2019;47(8):4334–4348.
- Nicastro G, Candel AM, Uhl M, et al. Mechanism of β-actin mRNA recognition by ZBP1. Cell Rep. 2017;18(5):1187–1199.
- Afitska K, Fucikova A, Shvadchak VV, et al. Modification of C terminus provides new insights into the mechanism of α-synuclein aggregation. Biophys J. 2017 Nov;113(10):2182–2191.
- Grzesiek S, Bax A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson. 1992;96(2):432–440.
- Grzesiek S, Bax A. An efficient experiment for sequential backbone assignment of medium-sized isotopically enriched proteins. J Magn Reson. 1992;99(1):201–207.
- Grzesiekt S, Bax A. Correlating backbone amide and side chain resonances in larger proteins by multiple relayed triple resonance NMR. J Am Chem Soc. 1992;114(16):6291–6293.
- Vranken WF, Boucher W, Stevens TJ, et al. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins Struct Funct Genet. 2005;59(4):687–696.
- Kay LE, Torchia DA, Bax A. backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28(23):8972–8979.
- Skinner SP, Fogh RH, Boucher W, et al. CcpNmr AnalysisAssign : a flexible platform for integrated NMR analysis. J Biomol NMR. 2016;66(2):111–124.
- Krämer SD, Wöhrle J, Rath C, et al. Anabel: an online tool for the real-time kinetic analysis of binding events. Bioinf Biol Insights [Internet]. 2019;13:1177932218821383. Available from: https://europepmc.org/articles/PMC6328958
- Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000 Sep;302(1):205–217.
- Madeira F, Park YM, Lee J, et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res [Internet]. 2019 Jul;47(W1):W636–W641. Available from: https://europepmc.org/articles/PMC6602479
- Waterhouse AM, Procter JB, Martin DMA, et al. Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009 May;25(9):1189–1191.
