ABSTRACT
The relationship between epitranscriptomics and malignant tumours has become a popular research topic in recent years. N6-methyladenosine (m6A), the most common post-transcriptional modification in mammals, is involved in various physiological processes in different cancer types, including gastric cancer (GC). The incidence and mortality of GC have been increasing annually, especially in developing countries. Insights into the epitranscriptomic mechanisms of gastric carcinogenesis could provide potential strategies for the prevention, diagnosis, and treatment of GC. In this review, we describe the mechanisms of RNA m6A modification; the functions of m6A regulators in GC; the functional crosstalk among m6A, messenger RNA, and noncoding RNA; and the promising application of m6A in the diagnosis and treatment of GC.
1. Background
Epigenetic modifications occur in various macromolecules in vivo, such as DNA, nucleosomes, and histones. These modifications change the molecular phenotype while ensuring the invariance of molecular genotypes, resulting in changes in molecular gene functions. Epigenetics is the science that recognizes and explains this phenomenon [Citation1]. The study of epigenetic modification of RNA molecules is known as epitranscriptomics. There are more than 160 post-transcriptional chemical modifications of RNA [Citation2], which affect the metabolic function and interaction of RNA with other molecules without changing the RNA sequence. The term ‘epitranscriptome’ was first used by Saletore et al. [Citation3] in 2012. In the last decade, epitranscriptomics has developed vigorously in various fields owing to the development and application of high-resolution mass spectrometry and whole-transcriptome sequencing technology.
The N6-methyladenosine (m6A) modification is the most common post-transcriptional modification in mammals. Approximately 0.1–0.4% of adenosines in RNA are m6A, accounting for approximately 50% of all methylated ribonucleosides [Citation4]. m6A modifications are most common in messenger RNA (mRNA), accounting for up to 80% of RNA methylation. These methyl marks occur at the consensus sequence RRACH (R = A or G and H = A, C, or U) close to the transcript termination codons and the 3’ untranslated regions (3’ UTRs) [Citation5,Citation6].
The initial challenge in mechanistic studies of m6A was the lack of methods to determine the stoichiometry of m6A modifications; therefore, the role of epitranscriptomics in regulating the fate and function of mRNA transcripts was not completely accepted for many years. Although liquid chromatography-tandem mass spectrometry (LC-MS/MS) and colorimetry, which are common methods for micro-molecular quantification, can detect the overall m6A level of RNA, neither can distinguish differential m6A levels in individual RNA regions, rendering them incapable of analysing m6A changes in specific locations [Citation7]. The development of methylated RNA immunoprecipitation sequencing (meRIP-seq) in 2012 partly overcame this problem [Citation5,Citation8]. MeRIP-seq first uses an anti-m6A antibody to recognize and bind to RNA fragments for RNA immunoprecipitation (RIP). Then, the enriched m6A-positive RNA fragments are eluted and sequenced by RNA-seq. Finally, bioinformatic analysis is performed using a previously obtained library and another library as background controls to identify areas of m6A enrichment. Recently, other m6A mapping and measuring techniques have been developed, such as LAIC-seq, which utilizes differences in tandem alternative polyadenylation sites bound to methylated and non-methylated transcripts to quantify m6A levels on a transcriptome-wide scale [Citation9]; miCLIP-seq, which maps m6A residues at single-nucleotide resolution and distinguishes m6A from N6,2-O-dimethyladenosine (m6Am) by creating antibody-RNA crosslinks through UV light [Citation10]; and MAZTER-seq, which relies on the bacterial RNase MazF to cleave RNA and identify m6A sites without using anti-m6A antibodies [Citation11].
Owing to the development of high-throughput sequencing techniques, m6A has also been found in many types of noncoding RNAs (ncRNAs), including long ncRNAs (lncRNAs), microRNAs (miRNAs), circular RNAs (circRNAs), and ribosomal RNAs (rRNAs) [Citation2]. Moreover, three types of key regulators involved in the regulation of m6A have been confirmed: ‘writers’ (methyltransferases [MTases] or MTase complexes), ‘readers’ (m6A RNA binding proteins), and ‘erasers’ (demethylases) [Citation12]. Writers and erasers regulate m6A levels, whereas readers regulate the target RNA after m6A modification. In particular, the ability of m6A erasers to demethylate m6A in RNA confirms that m6A methylation is a dynamic and reversible process [Citation13,Citation14], which was another important factor in recognizing the field of epitranscriptomics, in addition to the development of mapping and measuring techniques.
Gastric cancer (GC) is one of the most common malignant tumours. In 2020, GC was the fifth (5.6%) most common tumour and the fourth (7.7%) deadliest among 36 types of tumours [Citation15], indicating that GC is a serious threat to global human health. Identifying the underlying molecular mechanisms of GC will promote the advancement of effective detection methods and novel therapeutic strategies for GC patients with poor prognoses. The dysregulation of RNA m6A modification has been demonstrated to play an important role in development, metastasis, and other biological processes in various malignant tumours [Citation16]. Therefore, this review describes the biological mechanisms and functions of RNA m6A modification in GC and the application of m6A modification in the diagnosis and treatment of GC, aiming to better understand the research status of epitranscriptomics in GC and forecast future research directions.
2. m6A modification system
2.1. m6A writers
Intracellular post-transcriptional modification of m6A methylation in eukaryotes, including humans, is catalysed by a multi-component m6A MTase complex in the nucleus. The MTase complex was initially isolated into three proteins approximately 30 kDa, 200 kDa, and 875 kDa in size, although only two had MTase activity [Citation17,Citation18]. It is now known that the ~200 kDa complex contains methyltransferase like 3 (METTL3) and methyltransferase like 14 (METTL14), whereas the ~875 kDa complex appears to facilitate the recruitment of METTL3 to RNA. Bokar et al [Citation19] purified the 70-kDa subunit MT-A70 from the ~200 kDa portion, now known as METTL3, in 1997 using conventional chromatography and electrophoresis. They found that METTL3 contains sites that bind to S-adenosyl methionine, the methyl donor, and proved its high conservation in eukaryotes from yeast to humans. Later, Bujnicki et al [Citation20] identified METTL14, the homolog of METTL3, through phylogenetic analysis. METTL14 shares 43% identity with METTL3. In 2014, Liu et al [Citation21] revealed that METTL3 and METTL14 form a complex at a stoichiometric ratio of 1:1 by analysing the co-expression of these two proteins through 2D native SDS-PAGE analysis and confirmed the stability of the heterodimer. METTL3 and METTL14 co-localize in nuclear speckles in mammalian cells, where their heterodimers methylate adenines in nuclear RNA [Citation21]. Functionally, METTL3 is primarily responsible for catalysing the transfer of the methyl group, whereas METTL14 assists in the localization of the MTase complex by recognizing histone H3 Lys36 trimethylation in a cotranscriptional manner and provides a structural scaffold that enhances the catalytic activity of METTL3 [Citation21–23]. Wilms tumour 1-associated protein (WTAP), a regulator involved in mammalian pre-mRNA splicing, is also a component of the m6A MTase complex [Citation24–26]. WTAP helps the METTL3-METTL14 dimer localize to the nuclear speckle, thereby promoting adenine methylation in mRNA [Citation27]. Although METTL14 and WTAP are catalytically inactive, their presence is essential for promoting the catalytic activity of the MTase complex and regulating m6A levels. If METTL3, METTL14, or WTAP is knocked down, m6A levels in eukaryotic cells are decreased to varying degrees, and the effects of METTL14 and WTAP knockdown on m6A levels is significantly greater than that of METTL3 knockdown [Citation21]. Therefore, METTL3/METTL14/WTAP is recognized as the core component of the m6A writer complex.
Other components of the m6A writer complex, such as VIRMA, RBM15/RBM15b, and ZC3H13, appear to regulate RNA methylation through a close association with WTAP. VIRMA-mediated adenosine methylation occurs preferentially in the vicinity of the 3’ UTR and stop codon and influences variable polyadenylation via polyadenylation cleavage factor and the m6A writer complex [Citation28]. The RNA-binding protein RBM15 and its homolog RBM15b interact with METTL3 in a WTAP-dependent manner, and RBM15 is a key component of the m6A writer complex in Drosophila [Citation29]. Recently, the zinc finger protein ZC3H13 was shown to be a bridging component between WTAP and RBM15 in the m6A writer complex in Drosophila, promoting efficient functional adenosine methylation and successful localization of the complex in the nucleus [Citation30]. Other proteins or enzymes, such as CBLL1 and PRMT1, interact directly or indirectly with the core catalytic component METTL3/METTL14/WTAP [Citation31,Citation32].
Recent studies indicate that although the METTL3/METTL14/WTAP writer complex is mainly responsible for the vast majority of m6A modifications in mRNAs, a significant portion of m6A modifications in ncRNAs are methylated by other m6A writers [Citation33]. For example, methyltransferase like 16 (METTL16), which is homologous to the Escherichia coli YbiN protein, is responsible for the N6-methylation of A1618 in the 23S rRNA [Citation34], and has been shown to methylate methionine adenosyl transferase 2A mRNA and the small nuclear RNA (snRNA) U6 [Citation35,Citation36]. Recently, Dai et al [Citation37] reported that METTL16 could downregulate the lncRNA RAB11B-AS1 in an m6A-dependent manner, promoting hepatocellular carcinoma progression. In addition, unlike METTL3/MELLT14/WTAP-mediated m6A modification, which occurs close to the stop codon, most METTL16 binding sites are in introns, suggesting that the mechanism of target recognition by METTL16 may be different [Citation36]. rRNA is the most abundant RNA in human cells and contains the majority of m6A. A single m6A modification in human 28S rRNA is generated by ZCCHC4, whereas a single m6A modification in 18S rRNA is generated by the METTL5-TRMT112 complex, in which METTL5 acts as a catalytic subunit and TRMT112 acts as an allosteric adapter [Citation38,Citation39]. These atypical m6A MTase complexes may provide novel insights into the mechanism of m6A modification.
2.2. m6A readers
m6A-binding proteins read the m6A RNA modification to exert m6A-mediated biological activity. Dozens of readers have been discovered, some of which have similar amino acid sequences, such as the YT521-B homology (YTH) domain family proteins YTHDC1, YTHDC2, YTHDF1, YTHDF2, and YTHDF3, and proteins without the YTH domain, including heterogeneous nuclear ribonucleoprotein (hnRNP) family proteins hnRNPA2B1, hnRNPC, and hnRNPG and insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs) IGF2BP1, IGF2BP2, and IGF2BP3. Other unique types of readers include eukaryotic initiation factor (eIF) 3 and fragile X retardation protein [Citation5,Citation12].
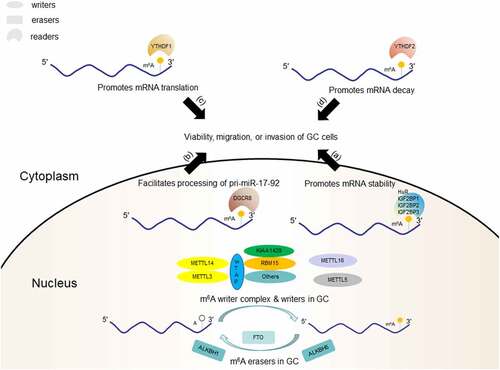
Readers have different downstream effects based on their subcellular localization (nucleus or cytoplasm) and recognition mechanisms (direct or indirect). In the cytoplasm, YTHDF2 accelerates the decay of m6A-modified transcripts and reduces their stability by directly recruiting the CCR4-NOT adenylate complex [Citation40], whereas YTHDF1 interacts with eIFs and ribosomes to improve the translation efficiency of target m6A-modified mRNAs [Citation41]. Intracellular YTHDF3 not only cooperates with YTHDF1 to promote the translation of m6A-modified mRNA but also directly interacts with YTHDF2 to accelerate the degradation of mRNA [Citation42,Citation43]. In addition, YTHDF3 and eIF4G2 participate in the initiation of m6A-modified circRNA translation [Citation44]. However, a recent study suggests that all DF paralogs have similar functions. Through structural analysis, motif analysis, CLIP studies, and immunofluorescence, Zaccara et al [Citation45] demonstrated that all DF proteins have highly similar sequences, functional domains, protein binding partners, and intracellular localizations. Therefore, like YTHDF2, the major effect of YTHDF1 and YTHDF3 is to promote mRNA degradation rather than translation. Hence, there is controversy remains over the function of YTHDF proteins, which needs to be verified by more studies in the future.
The functions of YTHDC1 in the nucleus include regulation of the exportation of m6A-modified mRNA, participation in mRNA splicing by recruiting the pre-mRNA splicing factor SRSF3 and inhibiting the binding of SRSF10 to mRNA, and priority identification of m6A residues on the lncRNA XIST, which mediates its silencing [Citation46–48]. hnRNPC and hnPNPG in the nucleus bind to pre-mRNAs adjacent to the m6A modification and indirectly affect their splicing [Citation49,Citation50].
The functions and mechanisms of some m6A readers are not clear. They exist in both the nucleus and cytoplasm, and their affinity for m6A is weaker than that of other readers. They directly or indirectly affect cellular activities through their effect on m6A-modified RNAs. IG2BPs and YTHDC2 are representative of these readers. The former enhances the stability of m6A-modified mRNA, whereas the latter is associated with mRNA degradation and regulation of translation initiation [Citation51,Citation52]. m6A readers are essential for the biological functions of m6A and the best entry point to study how m6A regulates the occurrence, development, and prognosis of diseases at the molecular level. The discovery of more potential m6A readers and clarification of their related mechanisms will be helpful for the prevention and treatment of diseases in clinical practice.
2.3. m6A erasers
m6A erasers catalyse demethylation in a dynamic, rapid, signal-dependent manner, which is critical for the functional regulation of m6A modifications [Citation53]. Fat mass and obesity-associated protein (FTO) was the first confirmed m6A eraser and is involved in the oxidative demethylation of 3-methylthymidine and 3-methyluracil in single-stranded DNA and RNA [Citation13,Citation54]. In mammals, FTO oxidizes m6A to N6-hydroxymethyladenosine (hm6A) as an intermediate modification and further oxidizes hm6A to N6-formyladenosine. These relatively stable intermediates eliminate the possibility of re-methylation mediated by the m6A writer complex at the same site and can be used as another modification to recruit different readers, which differentially regulate subsequent RNA-related pathways. Moreover, they can be used as markers for newly transcribed RNA by exploiting their intrinsic degradation kinetic [Citation55].
FTO belongs to the α-ketoglutarate-dependent dioxygenase homolog (ALBKH) family, and several other family members are also m6A erasers. ALKBH5 was the second m6A eraser discovered in mammals. ALKBH5 catalyzes the demethylation of m6A in an Fe(II)- and α-ketoglutarate-dependent manner [Citation14]. ALKBH3 mediates the demethylation of m6A-modified tRNA, which significantly improves the translation efficiency of certain mRNAs, a process closely related to tumour cell proliferation [Citation56]. ALKBH1, an m6A demethylase, is upregulated in lung cancer cells and reduces the m6A level, thereby promoting the metastasis and invasion of lung cancer cells [Citation57].
There are still many questions regarding how m6A erasers regulate post-transcriptional m6A demethylation and select demethylation sites in target RNAs. snRNAs may have a stronger affinity for m6A erasers than mRNAs, and erasers may be more closely associated with post-transcriptional epigenetic modifications other than m6A. For example, Mauer et al. [Citation58] suggested that the catalytic efficiency of FTO was approximately 100-fold higher towards m6Am than m6A based on the results of in vitro biochemical assays, which confirmed that m6Am was the preferred cellular substrate for FTO in vitro. However, another study showed that m6Am and m6A in the same RNA sequence had the same affinity for FTO, suggesting that the interaction of residues in the catalytic pocket with the nucleobase determines the activity and substrate specificity of erasers [Citation59]. Moreover, the catalytic activity of FTO can be influenced by the sequence and tertiary structure of the RNA. Recent studies have demonstrated that some potential eraser-interacting proteins can influence the selection of target RNAs by erasers. Song et al. [Citation60] reported that FTO interacts with the RNA-binding protein splicing factor, proline- and glutamine-rich (SFPQ) via its C-terminal domain and mediates the demethylation of SFPQ-bound RNA. More potential eraser regulators and demethylation mechanisms need to be investigated in the future.
2.4. m6A dysregulation in GC
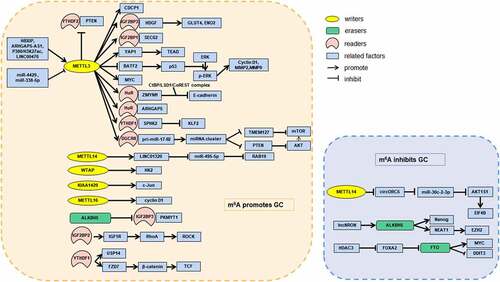
Many studies have shown epitranscriptomic dysregulation of m6A in GC tissues, with most showing increased m6A compared to that in paired adjacent normal tissues ( highlights the results of some of these studies). However, the role of m6A in carcinogenesis and the development of GC is unclear. Multiple studies have also validated abnormal expression of some m6A regulators, which is associated with the malignant biological properties of GC. These findings can help us understand the potential mechanism of m6A in the occurrence, development, and metastasis of GC [Citation61].
Table 1. Changes in total RNA m6A in GC.
Writers are currently the most widely studied m6A regulators in GC. Yue et al. [Citation62] found that GC patients with high METTL3 expression had a shorter median survival and more advanced TNM stage and were more likely to have diffuse-type disease compared to those with low METTL3 expression, and this corresponded to elevated m6A levels. By analysing big datasets acquired from The Cancer Genome Atlas (TCGA) and exploring METTL3 expression in GC cell lines and tumour tissues, Liu et al. [Citation63] also reached the conclusion that METTL3 was significantly increased in GC tissues and correlated with poor prognosis. Many other studies suggest that MELLT3 is an oncogene in GC, and transcription factors such as P300 and HBXIP might be involved in inducing METTL3 transcription [Citation64,Citation65]. Two other components of the m6A writer complex, WTAP and RBM15, are both highly expressed in GC; WTAP expression is associated with worse outcomes, and RBM15 expression is associated with better survival [Citation66,Citation67]. By contrast, METTL14 is downregulated in GC cells and tissues [Citation68]. This may be because METTL14 can suppress the progression of GC by deactivating the PI3K/AKT/mTOR pathway and epithelial-mesenchymal transition (EMT) through regulating m6A modification. Through gene set enrichment analysis and in vitro experiments, Zhang et al. [Citation69] demonstrated that METTL14 could be a potential tumour suppressor by blocking Wnt and PI3K/AKT signalling. In addition, they demonstrated that reduced m6A (represented by METTL14 knockdown) promotes the proliferation and invasiveness of GC cells, which could be reversed by m6A elevation (represented by FTO knockdown). m6A regulators seem to directly control the PI3K/AKT/mTOR signalling pathway, suggesting their potential as targets for antitumor therapy. The writer METTL5 was also proved to be significantly decreased in GC in a study of 168 GC patients [Citation70]. As a tumour suppressor gene, high METTL5 protein expression is closely related to better prognosis (P = 0.0051 based on 168 GC samples, P = 0.048 based on TCGA, and P = 0.00042 based on the Gene Expression Omnibus [GEO]). However, the pathways regulated by METTL5 in GC remain to be elucidated [Citation70].
Increased expression of m6A erasers, such as FTO, is associated with the tumorigenesis and progression of GC. Xu et al. [Citation71] confirmed that FTO mRNA (P < 0.001) and protein levels (IHC: P = 0.023; WB: P = 0.024) were significantly elevated in GC tissues compared to corresponding adjacent non-tumour tissues. Moreover, high FTO expression was closely related to poor differentiation (P < 0.001), lymph node metastasis (P = 0.029), TNM stage (P < 0.001), and poor prognosis (P < 0.001). Li et al. [Citation72] reported that high FTO (P = 0.0017) and ALKBH1 (P = 0.0241) levels were closely related to worse overall survival in GC patients; however, FTO and ALKBH1 were significantly downregulated at the protein level in GC tissues and closely correlated with clinicopathological features and prognosis of GC patients. The expression pattern of ALKBH5 in GC remains unclear. Shimura et al. [Citation73] and Hu et al. [Citation74] reported decreased ALKBH5 levels in GC samples, which were correlated with worse survival (both P < 0.001), but Jing et al. [Citation75] and Zhang et al. [Citation76] showed that GC patients with ALKBH5 overexpression had poor prognosis. Similar arguments exist regarding the expression of other m6A regulators. Therefore, multi-omics and multicenter studies are needed to further clarify the differential expression of m6A regulators in GC.
Readers are also dysregulated in GC, which leads to biological changes in an m6A-dependent manner. According to one analysis based on datasets from TCGA, the levels of YTHDF1/2/3, YTHDC1, hnRNPC, and hnRNPA2B1 were significantly increased in GC [Citation77]. GEO dataset analysis showed that YTHDF1 was significantly upregulated in three of five GEO datasets; hnRNPC, hnRNPA2B1, YTHDC1, and YTHDF3 were significantly upregulated in one of five GEO datasets, but YTHDF2 was not significantly altered in any of the datasets examined [Citation77]. IHC analysis verified that hnRNPC and YTHDF1/2/3 were overexpressed at the protein level, whereas hnRNPA2B1 and YTHDC1 were not differentially expressed in GC. Moreover, there was no significant relationship between the expression of m6A readers and prognosis in GC [Citation77]. Different readers have been reported to have different biological functions in GC, which makes it difficult to determine the clinical value of readers as biomarkers. The functions of GC-related m6A readers, as well as their target RNAs, are explained in detail in Chapter 3 and shown in .
Figure 1. m6A regulators in GC cell
As mentioned above, the increase in total RNA m6A levels in GC is a complex result of differential expression of writers, readers, and erasers. It is not possible to explain the oncological mechanisms of m6A in GC by studying the abnormal expression of one or two m6A regulators because of the complexity and heterogeneity of GC. Various m6A writers, erasers, and readers are involved in the biological progression of GC and maintain a relationship with each other. In addition, although abnormal m6A levels in the transcriptome are known to be related to GC, more work is needed for m6A levels to be a viable biomarker because their increase and decrease are affected by many different factors, which makes them unreliable and inaccurate. To advance the study of m6A regulators in GC, we should pay attention to the specific downstream molecules regulated by abnormally expressed enzymes and their functions.
3. Biological functions of m6A RNA modifications in GC
3.1. Regulation of mRNA
The epigenetic modification of oncogenic and tumour suppressor genes is considered a novel mechanism related to the occurrence, development, and outcome of GC. The role of m6A modification in the regulation of gene expression depends on the mRNA, the locations of m6A modification, and the involved m6A regulators [Citation78]. Myelocytomatosis viral oncogene homolog (MYC) is an oncogene associated with advanced tumour stage, diffuse type, deeper tumour extension, and lymph node metastasis in GC [Citation79]. A recent study showed that the m6A eraser FTO could stabilize MYC mRNA through m6A demethylation in GC cells, which enhanced their ability to proliferate, migrate, and invade [Citation80]. Upstream of the FTO/m6A/MYC axis, another oncogene, histone deacetylase 3, promotes GC progression by degrading forkhead box transcription factor A2 (FOXA2), and FOXA2 directly mediates FTO transcriptional repression. In contrast, MYC methylation by METTL3 in the 3 UTR enhances MYC translation and stability, which promotes proliferation, migration, and invasion in GC cells [Citation64]. Therefore, different m6A modification sites in MYC mRNA exhibit different functions [Citation81]. Identifying precise m6A positions on a transcriptome-wide level is important for exploring the functions of different m6A sites on the same mRNA. Fortunately, novel detection techniques, such as miCLIP-seq and MAZTER-seq, can map m6A residues at single-nucleotide resolution [Citation10].
Many other m6A-modified mRNAs participate in the biological processes of GC. The methylation of zinc finger MYM-type containing 1 by METTL3 enhances its stability, which facilitates GC EMT and metastasis [Citation62]. Additionally, METTL3 methylates hepatoma-derived growth factor, which enhances its stability and recognition by IGF2BP3, which leads to tumour angiogenesis [Citation65]. Conversely, METTL3 knockdown leads to increased basic leucine zipper ATF-like transcription factor 2 (BATF2) expression in GC cells, because m6A modification suppresses BATF2 translation [Citation82]. High expression of YTHDF1 leads to an increase in the translation of frizzled 7 (FZD7), a key Wnt receptor, in an m6A-dependent manner [Citation83]. As the expression of FZD7 increases, the Wnt/β-catenin pathway becomes hyperactivated, which contributes to gastric carcinogenesis. PKMYT1, an oncogene that plays a role in the growth of multiple tumours, is a target of ALKBH5 in GC [Citation74]. With the significant downregulation of ALKBH5 in GC, the m6A levels of PKMYT1 are increased, increasing its recognition by IGF2BP3 and enhanced stability. This increased PKMYT1 promotes invasion and migration in GC. Furthermore, one study revealed a special function of mRNA m6A modification in GC, indicating that methylation of E2F transcription factor 3 (E2F3) at the 3 UTR mediates the inhibitory effect of miR-660 on E2F3 [Citation84]. This m6A modification is required for the interaction of miR-660 and E2F3. The mechanisms by which GC-related mRNAs are regulated by m6A modification are summarized in .
Table 2. m6A-modified mRNAs in GC.
3.2. Regulation of ncRNA
High-throughput technology has verified that ncRNAs, such as lncRNAs, miRNAs, and circRNAs [Citation85–87], are involved in tumorigenesis, progression, and metastasis in GC. Indeed, methylation of RNAs, including ncRNAs, has been recognized as a unique ‘epitranscriptomic language’ in oncology, although its mechanisms are not completely understood [Citation88]. Recently, Hu et al. [Citation89]
found that m6A could upregulate the lncRNA LINC01320, which promotes the proliferation, migration, and invasion of GC cells, implying that GC-related ncRNAs can be regulated by m6A modification. Pulldown assays and RIP experiments verified that LINC01320 is methylated by METTL14. Zhang et al. [Citation76] determined that demethylation of the lncRNA NEAT1 by ALKBH5 increases NEAT1 levels and promotes GC invasion and metastasis. Sun et al. [Citation97] showed that the methylation of pri-miR-17-92 by METTL3 facilitates its processing into the miR-17-92 cluster in an m6A/DGCR8-dependent manner and that the miR-17-92 cluster inhibits PTEN/TMEM127 expression and activates the AKT/mTOR pathway. Moreover, they demonstrated that everolimus, an mTOR inhibitor, suppressed the viability of METTL3-high GC cells significantly better than that of METTL3-low GC cells (P < 0.05 on days 2 and 3), which was consistent with the results of an in vivo experiment, which measured tumour volume of mice carrying METTL3-high or control tumours (P = 0.0465). Therefore, everolimus has the potential to treat patients with m6A/METTL3-high GC. Zhang et al. [Citation98] analysed differentially expressed circRNAs in poorly differentiated GC and found that most differentially expressed circRNAs had m6A modifications, which was consistent with the circRNA expression levels. Recently, circORC5 was identified as a target of METTL14 in GC102]. Low expression of METTL14 in GC reduced the m6A level of circORC5, which resulted in increased circORC5 expression and poor survival of patients with GC. In addition, m6A in circRNAs mediates cap-independent translation, and proteins or peptides encoded by these circRNAs can play an important role in human cancers [Citation99]. However, there has been little research on the mechanisms and functions of proteins translated from m6A-modified circRNAs in GC. All recent studies on m6A-modified ncRNAs in GC are summarized in .
Table 3. m6A-modified ncRNAs in GC.
ncRNAs can also interact with RNA-binding proteins, including m6A regulators, to regulate cancer cell phenotypes [Citation101]. Zhu et al. [Citation102] reported that the lncRNA ARHGAP5-AS1 can recruit METTL3 to facilitate methylation of ARHGAP5, which stabilizes ARHGAP5 in the cytoplasm. The upregulation of ARHGAP5 promotes chemoresistance of GC cells, whereas its downregulation significantly reverses chemoresistance. Their study provided a potential strategy for addressing chemotherapy resistance in GC. The lncRNA LINC00470, which is correlated with distant metastasis, TNM stage, and poor prognosis in GC, promotes PTEN methylation via association with METTL3 [Citation103]. Then, the m6A reader YTHDF2 is involved in the degradation of PTEN, and the half-life of PTEN is increased after YTHDF2 knockdown. The lncRNA lncNRON was recently suggested to exert oncogenic functions in GC by recruiting ALKHB5 to inhibit Nanog mRNA decay [Citation104]. With regard to miRNAs, miR-338-5p can inhibit the methylation of CDCP1 by suppressing METTL3, which is important because m6A modification stimulates CDCP1 translation and accelerates the proliferation and invasion of GC cells [Citation105]. Upstream of the miR-338-5p/METTL3/CDCP1 axis, embryonic ectoderm development protein promotes GC progression by downregulating miR-338-5p through histone methylation. METTL3 is also suppressed by miR-4429, and reduced METTL3 prevents GC progression by attenuating m6A-induced stabilization of SEC62 [Citation106]. These ncRNAs may serve as promising targets for molecular therapy of GC. Overall, interactions between RNAs (mRNAs and ncRNAs) and m6A regulators are indeed related to the biological behaviours of GC, which are potential targets for molecular diagnosis and treatment ().
4. Application of RNA m6A modification in GC
Since increasing evidence demonstrates that RNA m6A modification is correlated with tumorigenesis and the progression of GC, the detection of dysregulated m6A levels may provide a promising method for diagnosing, predicting, and assessing GC. For example, Zhang et al. [Citation107] constructed an m6A scoring system to quantify the m6A modification pattern in individual GC patients and characterize immune cell infiltration. The m6A scoring system was divided into three distinct patterns that were highly consistent with the three types of immune cell-infiltrating characteristics. In this system, GC patients with a low m6A score tend to have better survival and more effective immune infiltration than those with a high m6A score. Moreover, analysis of two cohorts receiving immunotherapy demonstrated that patients with a lower m6A score exhibited significantly more clinical benefits and better prognosis after treatment with anti-PD-1/L1 immunotherapy (P = 0.002 and 0.013, respectively), implying the ability of m6A analysis to guide which patients should receive immunotherapy. Ge et al. [Citation108] evaluated the dysregulation of m6A modification in the peripheral blood of GC patients and found that m6A levels were related to the progression and metastasis of GC. As a novel biomarker, m6A levels in peripheral blood RNA (AUC = 0.929) had a better ability to distinguish GC patients from healthy individuals than the conventional tumour markers CEA and CA19-9 (AUC = 0.694 and 0.603, respectively). In addition, the combination of m6A, CEA, and CA19-9 improved the AUC to 0.955 (95% CI, 0.91–0.98; P < 0.0001). Subsequently, a serum diagnostic signature based on m6A target miRNAs was developed for the mass detection of cancer [Citation109]. The m6A-miRNAs signature showed a promising ability to distinguish GC patients from non-cancer controls and from mixed samples with other cancers (AUC = 0.989 and 0.791, respectively). When the signature was used to diagnose early GC, its AUC reached 0.989 (95% CI, 0.987–0.990), which was higher than those of CEA and CA19-9. This study provides a new direction for further investigation of the diagnostic value of m6A.
Aberrant expression or genetic variants of m6A regulators can also serve as valuable sources for screening and prognosis assessment. By analysing the expression profiles of m6A regulator genes in TCGA cohorts, Liu et al. [Citation110] constructed an m6A-gene based diagnostic signature for GC. The signature showed high diagnostic accuracy (AUC = 0.986) in distinguishing between 375 tumour samples and 32 normal samples; however, there was no external validation cohort. Wang et al. [Citation111] reported a significant increase in the m6A readers IGF2BP1, IGF2BP2, and IGF2BP3 at the RNA level (P < 0.05) in GC tissues and identified the single nucleotide polymorphism rs9906944 in IGF2BP1, which is associated with GC risk (P < 0.05). Analysis of 2,900 GC cases and 3,536 controls indicated that the T allele of rs9906944 was significantly associated with decreased GC risk (odds ratio = 0.75, 95% CI 0.64–0.88, P= 5.76 × 10−4). Furthermore, elevated IGF2BP1 levels are associated with worse overall survival in GC patients (HR = 1.49, 95% CI 1.16–1.91, P= 1.50 × 10−3). In addition, Li et al. [Citation112] established a risk ratio model of potential m6A regulators through differential analysis based on TCGA datasets and least absolute shrinkage and selection operator Cox regression analysis. After adding another risk model, patients with GC were divided into high- and low-risk groups. The OS in the low-risk group was significantly higher than that in the high-risk group (P < 0.05), and the AUC of this risk model was 0.667. Another recent study demonstrated the presence of multiple differentially expressed m6A regulators in GC tissues by analysing data from UALCAN and Oncomine web [Citation75]. High expression of FTO and YTHDC2 could be used as unfavourable prognostic markers in patients with GC. Interestingly, omeprazole, a proton pump inhibitor, enhances m6A levels by decreasing FTO and promotes chemosensitivity in GC cells [Citation113]. Mechanistically, FTO inhibition increases DDIT3 expression and activates mTORC1, which promotes pro-survival autophagy [Citation114]. Further studies on the dysregulation of m6A regulators in GC may provide more promising biomarkers and treatment strategies.
5. Conclusions
With the recent development of epitranscriptomic research, the mechanism of RNA m6A modification, which is the most common post-transcriptional modification, has been largely clarified. Epitranscriptomic research has subsequently been extended to various types of malignant tumours, including GC. GC-related mRNAs and ncRNAs are regulated by m6A regulators, and these m6A-modified RNAs exert pro- and anti-tumorigenic downstream functions. Moreover, ncRNAs can affect the methylation of downstream RNAs by regulating the expression of m6A regulators. Clinically, abnormal changes in RNA m6A modification levels or m6A regulators have an important impact on tumorigenesis, development, metastasis, and prognosis in GC. By detecting and evaluating specific m6A-related molecules, clinicians can diagnose GC at an early stage, develop appropriate treatment strategies, and accurately predict individual prognoses. Future research should focus on a comprehensive understanding of the interaction of various m6A regulators, m6A-modified RNAs, and m6A-modified sites in the biological process of GC to identify better m6A markers with high sensitivity and specificity and develop novel molecular therapies targeting the pathways of m6A modification.
Abbreviations
| (m6A) | = | N6-methyladenosine |
| (GC) | = | gastric cancer |
| (mRNA) | = | messenger RNA |
| (ncRNA) | = | non-coding RNA |
| (3’ UTRs) | = | 3’ untranslated regions |
| (LC-MS/MS) | = | liquid chromatography-tandem mass spectrometry |
| (meRIP-seq) | = | methylated RNA immunoprecipitation sequencing |
| (LAIC-seq) | = | m6A-level and isoform-characterization sequencing |
| (miCLIP-seq) | = | m6A individual-nucleotide-resolution cross-linking and immunoprecipitation sequencing |
| (RIP) | = | RNA immunoprecipitation |
| (APA) | = | alternative polyadenylation |
| (m6Am) | = | N6, 2’-O-dimethyladenosine |
| (lncRNAs) | = | long non-coding RNAs |
| (miRNAs) | = | microRNAs |
| (circRNAs) | = | circular RNAs |
| (rRNAs) | = | ribosomal RNAs |
| (MTase) | = | methyltransferase |
| (METTL3) | = | methyltransferase like 3 |
| (SAM) | = | S-adenosyl methionine |
| (METTL14) | = | methyltransferase like 14 |
| (H3K36me3) | = | histone H3 on Lys36 |
| (WTAP) | = | Wilms Tumour 1-associated protein |
| (METTL16) | = | Methyltransferase like 16 |
| (YTH) | = | YT521-B homology |
| (HNRNPs) | = | heterogeneous nuclear ribonucleoproteins |
| (IGF2BPs) | = | insulin-like growth factor 2 mRNA-binding proteins |
| (eIF3) | = | eukaryotic Initiation factor 3 |
| (FMRP) | = | fragile X retardation protein |
| (FTO) | = | Fat mass and obesity-associated protein |
| (hm6A) | = | n6-hydroxymethyladenosine |
| (f6A) | = | N6-formyladenosine |
| (ALKBH5) | = | α-ketoglutarate-dependent dioxygenase homolog 5 |
| (snRNA) | = | small nuclear RNAs |
| (SFPQ) | = | splicing factor, proline- and glutamine-rich |
| (TCGA) | = | The Cancer Genome Atlas |
| (EMT) | = | epithelial-mesenchymal transition |
| (GSEA) | = | gene set enrichment analysis |
| (IHC) | = | immunohistochemistry |
| (MYC) | = | myelocytomatosis viral oncogene homolog |
| (HDAC3) | = | histone deacetylase 3 |
| (FOXA2) | = | forkhead box transcription factor A2 |
| (ZMYM1) | = | zinc finger MYM-type containing 1 |
| (HDGF) | = | hepatoma-derived growth factor |
| (BATF2) | = | basic leucine zipper ATF-like transcription factor 2 |
| (FZD7) | = | frizzled 7 |
| (E2F3) | = | E2F transcription factor 3 |
| (EED) | = | embryonic ectoderm development protein |
| (TME) | = | tumour microenvironment |
| (SNP) | = | single-nucleotide polymorphism |
| (LASSO) | = | least absolute shrinkage and selection operator |
Author contributions
Yitian Xu: Data curation, Writing - Original draft preparation, Writing- reviewing and editing. Chen Huang: Conceptualization, Supervision, Writing- reviewing and editing.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27.
- Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways 2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307.
- Saletore Y, Meyer K, Korlach J, et al. The birth of the Epitranscriptome: deciphering the function of RNA modifications. Genome Biol. 2012;13(10):175.
- Wei CM, Gershowitz A, Moss B. Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell. 1975;4(4):379–386.
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485(7397):201–206.
- Schwartz S, Mumbach MR, Jovanovic M, et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014;8(1):284–296.
- Thüring K, Schmid K, Keller P, et al. LC-MS analysis of methylated RNA. Methods Mol Biol. 2017;1562:3–18.
- Meyer KD, Saletore Y, Zumbo P, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell. 2012;149(7):1635–1646.
- Molinie B, Wang J, Lim KS, et al. m(6)A-LAIC-seq reveals the census and complexity of the m(6)A epitranscriptome. Nat Methods. 2016;13(8):692–698.
- Linder B, Grozhik AV, Olarerin-George AO, et al. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat Methods. 2015;12(8):767–772.
- Garcia-Campos MA, Edelheit S, Toth U, et al. Deciphering the “m6A Code” via Antibody-Independent quantitative profiling. Cell. 2019;178(3):731–747.e16.
- Zaccara S, Ries RJ, Jaffrey SR. Reading, writing and erasing mRNA methylation. Nat Rev Mol Cell Biol. 2019;20(10):608–624.
- Jia G, Fu Y, Zhao X, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7(12):885–887.
- Zheng G, Dahl JA, Niu Y, et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol Cell. 2013;49(1):18–29.
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249.
- Jaffrey SR, Kharas MG. Emerging links between m6A and misregulated mRNA methylation in cancer. Genome Med. 2017;9(1):2.
- Bokar JA, Rath-Shambaugh ME, Ludwiczak R, et al. Characterization and partial purification of mRNA N6-adenosine methyltransferase from HeLa cell nuclei. Internal mRNA methylation requires a multisubunit complex. J Biol Chem. 1994;269(26):17697–17704.
- Rottman FM, Bokar JA, Narayan P, et al. N6-adenosine methylation in mRNA: substrate specificity and enzyme complexity. Biochimie. 1994;76(12):1109–1114.
- Bokar JA, Shambaugh ME, Polayes D, et al. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA. 1997;3(11):1233–1247.
- Bujnicki JM, Feder M, Radlinska M, et al. Structure prediction and phylogenetic analysis of a functionally diverse family of proteins homologous to the MT-A70 subunit of the human mRNA: m(6)Amethyltransferase. J Mol Evol. 2002;55(4):431–444.
- Liu J, Yue Y, Han D, et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol. 2014;10(2):93–95.
- Wang Y, Li Y, Toth JI, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16(2):191–198.
- Śledź P, Jinek M. Structural insights into the molecular mechanism of the m(6)A writer complex. Elife. 2016;5:e18434.
- Ortega A, Niksic M, Bachi A, et al. Biochemical function of female-lethal (2)D/Wilms’ tumor suppressor-1-associated proteins in alternative pre-mRNA splicing. J Biol Chem. 2003;278(5):3040–3047.
- Zhong S, Li H, Bodi Z, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. Plant Cell. 2008;20(5):1278–1288.
- Agarwala SD, Blitzblau HG, Hochwagen A, et al. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8(6):e1002732.
- XL P, BF S, Wang L, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24(2):177–189.
- Yue Y, Liu J, Cui X, et al. VIRMA mediates preferential m6A mRNA methylation in 3ʹUTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4(1):10.
- Lence T, Akhtar J, Bayer M, et al. m6A modulates neuronal functions and sex determination in drosophila. Nature. 2016;540(7632):242–247.
- Knuckles P, Lence T, Haussmann IU, et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018;32(5–6):415–429.
- Růžička K, Zhang M, Campilho A, et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215(1):157–172.
- Wang Z, Pan Z, Adhikari S, et al. m6A deposition is regulated by PRMT1-mediated arginine methylation of METTL14 in its disordered C-terminal region. EMBO J. 2021;40(5):e106309.
- Oerum S, Meynier V, Catala M, et al. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021;49(13):7239–7255.
- Sergiev PV, Serebryakova MV, Bogdanov AA, et al. The ybiN gene of Escherichia coli encodes adenine-N6 methyltransferase specific for modification of A1618 of 23S ribosomal RNA, a methylated residue located close to the ribosomal exit tunnel. J Mol Biol. 2008;375(1):291–300.
- Pendleton KE, Chen B, Liu K, et al. The U6 snRNA m6A methyltransferase METTL16 regulates SAM synthetase intron retention. Cell. 2017;169(5):824–835.e14.
- Warda AS, Kretschmer J, Hackert P, et al. Human METTL16 is a N6-methyladenosine (m6A) methyltransferase that targets pre-mRNAs and various non-coding RNAs. EMBO Rep. 2017;18(11):2004–2014.
- Dai YZ, Liu YD, Li J, et al. METTL16 promotes hepatocellular carcinoma progression through downregulating RAB11B-AS1 in an m6A-dependent manner. Cell Mol Biol Lett. 2022;27(1):41.
- Ma H, Wang X, Cai J, et al. N6-Methyladenosine methyltransferase ZCCHC4 mediates ribosomal RNA methylation. Nat Chem Biol. 2019;15(1):88–94.
- van Tran N, Ernst FGM, Hawley BR, et al. The human 18S rRNA m6A methyltransferase METTL5 is stabilized by TRMT112. Nucleic Acids Res. 2019;47(15):7719–7733.
- Du H, Zhao Y, He J, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7(1):12626.
- Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161(6):1388–1399.
- Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–328.
- Li A, YS C, XL P, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res. 2017;27(3):444–447.
- Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–641.
- Zaccara S, Jaffrey SR. A unified model for the function of YTHDF proteins in regulating m6A-Modified mRNA. Cell. 2020;181(7):1582–1595.e18.
- Roundtree IA, Luo GZ, Zhang Z, et al. YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. Elife. 2017;6:e31311.
- Xiao W, Adhikari S, Dahal U, et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol Cell. 2016;61(4):507–519.
- Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373.
- Liu N, Dai Q, Zheng G, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518(7540):560–564.
- Liu N, Zhou KI, Parisien M, et al. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051–6063.
- Huang H, Weng H, Sun W, et al. Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat Cell Biol. 2018;20(3):285–295.
- Kretschmer J, Rao H, Hackert P, et al. The m6A reader protein YTHDC2 interacts with the small ribosomal subunit and the 5’-3’ exoribonuclease XRN1. RNA. 2018;24(10):1339–1350.
- Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2017;18(1):31–42.
- Jia G, Yang CG, Yang S, et al. Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 2008;582(23–24):3313–3319.
- Fu Y, Jia G, Pang X, et al. FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat Commun. 2013;4(1):1798.
- Ueda Y, Ooshio I, Fusamae Y, et al. AlkB homolog 3-mediated tRNA demethylation promotes protein synthesis in cancer cells. Sci Rep. 2017;7(1):42271.
- Li H, Zhang Y, Guo Y, et al. ALKBH1 promotes lung cancer by regulating m6A RNA demethylation. Biochem Pharmacol. 2021;189:114284.
- Mauer J, Luo X, Blanjoie A, et al. Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature. 2017;541(7637):371–375.
- Zhang X, Wei LH, Wang Y, et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc Natl Acad Sci U S A. 2019;116(8):2919–2924.
- Song H , Wang Y, Wang R, et al. SFPQ Is an FTO-Binding Protein that Facilitates the Demethylation Substrate Preference. Cell Chem Biol. 2020;27(3):283–291.
- He L, Li H, Wu A, et al. Functions of N6-methyladenosine and its role in cancer. Mol Cancer. 2019;18(1):176.
- Yue B, Song C, Yang L, et al. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol Cancer. 2019;18(1):142.
- Liu T, Yang S, Sui J, et al. Dysregulated N6-methyladenosine methylation writer METTL3 contributes to the proliferation and migration of gastric cancer. J Cell Physiol. 2020;235(1):548–562.
- Yang Z, Jiang X, Li D, et al. HBXIP promotes gastric cancer via METTL3-mediated MYC mRNA m6A modification. Aging (Albany NY). 2020;12(24):24967–24982.
- Wang Q, Chen C, Ding Q, et al. METTL3-mediated m6A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut. 2020;69(7):1193–1205.
- Li H, Su Q, Li B, et al. High expression of WTAP leads to poor prognosis of gastric cancer by influencing tumour-associated T lymphocyte infiltration. J Cell Mol Med. 2020;24(8):4452–4465.
- Su Y, Huang J, Hu J. m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gastric cancer. Front Oncol. 2019;9:1038.
- Liu X, Xiao M, Zhang L, et al. The m6A methyltransferase METTL14 inhibits the proliferation, migration, and invasion of gastric cancer by regulating the PI3K/AKT/mTOR signaling pathway. J Clin Lab Anal. 2021;35(3):e23655.
- Zhang C, Zhang M, Ge S, et al. Reduced m6A modification predicts malignant phenotypes and augmented Wnt/PI3K-Akt signaling in gastric cancer. Cancer Med. 2019;8(10):4766–4781.
- Wang Z, Liu J, Yang Y, et al. Expression and prognostic potential of ribosome 18S RNA m6A methyltransferase METTL5 in gastric cancer. Cancer Cell Int. 2021;21(1):569.
- Xu D, Shao W, Jiang Y, et al. FTO expression is associated with the occurrence of gastric cancer and prognosis. Oncol Rep. 2017;38(4):2285–2292.
- Li Y, Zheng D, Wang F, et al. Expression of demethylase genes, FTO and ALKBH1, Is associated with prognosis of gastric cancer. Dig Dis Sci. 2019;64(6):1503–1513.
- Shimura T, Kandimalla R, Okugawa Y, et al. Novel evidence for m6A methylation regulators as prognostic biomarkers and FTO as a potential therapeutic target in gastric cancer. Br J Cancer. 2022;126(2):228–237.
- Hu Y, Gong C, Li Z, et al. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol Cancer. 2022;21(1):34.
- Jing JJ, Zhao X, Li H, et al. Expression profiles and prognostic roles of m6A writers, erasers and readers in gastric cancer. Future Oncol. 2021;17(20):2605–2620.
- Zhang J, Guo S, Piao HY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem. 2019;75(3):379–389.
- Guan K, Liu X, Li J, et al. Expression status and prognostic value of M6A-associated genes in gastric cancer. J Cancer. 2020;11(10):3027–3040.
- Kobayashi M, Ohsugi M, Sasako T, et al. The RNA methyltransferase Complex of WTAP, METTL3, and METTL14 regulates mitotic clonal expansion in Adipogenesis. Mol Cell Biol. 2018;38(16):e00116–18.
- De Souza CR, Leal MF, Calcagno DQ, et al. MYC deregulation in gastric cancer and its clinicopathological implications. PLoS One. 2013;8(5):e64420.
- Yang Z, Jiang X, Zhang Z, et al. HDAC3-dependent transcriptional repression of FOXA2 regulates FTO/m6A/MYC signaling to contribute to the development of gastric cancer. Cancer Gene Ther. 2021;28(1–2):141–155.
- Deng X, Su R, Weng H, et al. RNA N6-methyladenosine modification in cancers: current status and perspectives. Cell Res. 2018;28(5):507–517.
- Xie JW, Huang XB, Chen QY, et al. m6A modification-mediated BATF2 acts as a tumor suppressor in gastric cancer through inhibition of ERK signaling. Mol Cancer. 2020;19(1):114.
- Pi J, Wang W, Ji M, et al. YTHDF1 promotes gastric carcinogenesis by controlling translation of FZD7. Cancer Res. 2021;81(10):2651–2665.
- He X, Shu Y. RNA N6-methyladenosine modification participates in miR-660/E2F3 axis-mediated inhibition of cell proliferation in gastric cancer. Pathol Res Pract. 2019;215(6):152393.
- Xie S, Chang Y, Jin H, et al. Non-coding RNAs in gastric cancer. Cancer Lett. 2020;493:55–70.
- Luo Z, Rong Z, Zhang J, et al. Circular RNA circCCDC9 acts as a miR-6792-3p sponge to suppress the progression of gastric cancer through regulating CAV1 expression. Mol Cancer. 2020;19(1):86.
- Zhu Z, Rong Z, Luo Z, et al. Circular RNA circNHSL1 promotes gastric cancer progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer. 2019;18(1):126.
- Coker H, Wei G, Brockdorff N. m6A modification of non-coding RNA and the control of mammalian gene expression. Biochim Biophys Acta Gene Regul Mech. 2019;1862(3):310–318.
- Hu N, Ji H. N6-methyladenosine (m6A)-mediated up-regulation of long noncoding RNA LINC01320 promotes the proliferation, migration, and invasion of gastric cancer via miR495-5p/RAB19 axis. Bioengineered. 2021;12(1):4081–4091.
- Liu D, Xia AD, Wu LP, et al. IGF2BP2 promotes gastric cancer progression by regulating the IGF1R-RhoA-ROCK signaling pathway. Cell Signal. 2022;94:110313.
- Zhou W, Xian Q, Wang Q, et al. m6A methyltransferase 3 promotes the proliferation and migration of gastric cancer cells through the m6A modification of YAP1. J Oncol. 2021;2021:8875424.
- Chen XY, Liang R, Yi YC, et al. The m6A reader YTHDF1 facilitates the tumorigenesis and metastasis of gastric cancer via USP14 translation in an m6A-Dependent manner. Front Cell Dev Biol. 2021;9:647702.
- Huo FC, Zhu ZM, Zhu WT, et al. METTL3-mediated m6A methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene. 2021;40(16):2968–2981.
- Yu H, Zhao K, Zeng H, et al. N6-methyladenosine (m6A) methyltransferase WTAP accelerates the Warburg effect of gastric cancer through regulating HK2 stability. Biomed Pharmacother. 2021;133:111075.
- Miao R, Dai CC, Mei L, et al. KIAA1429 regulates cell proliferation by targeting c-Jun messenger RNA directly in gastric cancer. J Cell Physiol. 2020;235(10):7420–7432.
- Wang XK, Zhang YW, Wang CM, et al. METTL16 promotes cell proliferation by up-regulating cyclin D1 expression in gastric cancer. J Cell Mol Med. 2021;25(14):6602–6617.
- Sun Y, Li S, Yu W, et al. N6-methyladenosine-dependent pri-miR-17-92 maturation suppresses PTEN/TMEM127 and promotes sensitivity to everolimus in gastric cancer. Cell Death Dis. 2020;11(10):836.
- Zhang C, Wang J, Geng X, et al. Circular RNA expression profile and m6A modification analysis in poorly differentiated adenocarcinoma of the stomach. Epigenomics. 2020;12(12):1027–1040.
- Lu Y, Li Z, Lin C, et al. Translation role of circRNAs in cancers. J Clin Lab Anal. 2021;35(7):e23866.
- Fan HN, Chen ZY, Chen XY, et al. METTL14-mediated m6A modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol Cancer. 2022;21(1):51.
- Yao ZT, Yang YM, Sun MM, et al. New insights into the interplay between long non-coding RNAs and RNA-binding proteins in cancer. Cancer Commun. 2022;42(2):117–140.
- Zhu L, Zhu Y, Han S, et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019;10(6):383.
- Yan J, Huang X, Zhang X, et al. LncRNA LINC00470 promotes the degradation of PTEN mRNA to facilitate malignant behavior in gastric cancer cells. Biochem Biophys Res Commun. 2020;521(4):887–893.
- Wang S, Wang Y, Zhang Z, et al. Long Non-Coding RNA NRON promotes tumor proliferation by regulating ALKBH5 and nanog in gastric cancer. J Cancer. 2021;12(22):6861–6872.
- Zhang F, Yan Y, Cao X, et al. Methylation of microRNA-338-5p by EED promotes METTL3-mediated translation of oncogene CDCP1 in gastric cancer. Aging (Albany NY). 2021;13(8):12224–12238.
- He H, Wu W, Sun Z, et al. MiR-4429 prevented gastric cancer progression through targeting METTL3 to inhibit m6A-caused stabilization of SEC62. Biochem Biophys Res Commun. 2019;517(4):581–587.
- Zhang B, Wu Q, Li B, et al. m6A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer. 2020;19(1):53.
- Ge L, Zhang N, Chen Z, et al. Level of N6-Methyladenosine in peripheral blood RNA: a novel predictive biomarker for gastric cancer. Clin Chem. 2020;66(2):342–351.
- Zhang B, Chen Z, Tao B, et al. m6A target microRNAs in serum for cancer detection. Mol Cancer. 2021;20(1):170.
- Liu T, Yang S, Cheng YP, et al. The N6-Methyladenosine (m6A) methylation gene YTHDF1 reveals a potential diagnostic role for gastric cancer. Cancer Manag Res. 2020;12:11953–11964.
- Wang X, Guan D, Wang D, et al. Genetic variants in m6A regulators are associated with gastric cancer risk. Arch Toxicol. 2021;95(3):1081–1088.
- Li J, Zuo Z, Lai S, et al. Differential analysis of RNA methylation regulators in gastric cancer based on TCGA data set and construction of a prognostic model. J Gastrointest Oncol. 2021;12(4):1384–1397.
- Feng S, Qiu G, Yang L, et al. Omeprazole improves chemosensitivity of gastric cancer cells by m6A demethylase FTO-mediated activation of mTORC1 and DDIT3 up-regulation. Biosci Rep. 2021;41(1):BSR20200842.
- Yang H, Wen Y, Zhang M, et al. MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint. Autophagy. 2020;16(2):271–288.