ABSTRACT
Understanding how cells sense temperature is a fundamental question in biology and is pivotal for the evolution of life. In numerous organisms, temperature is not only sensed but also generated due to cellular processes. Consequently, the mechanisms governing temperature sensation in various organisms have been experimentally elucidated. Extending upon others’ proposals and demonstration of protein- and nucleic acid-based thermosensors, and utilizing a colonial India ‘punkah-wallahs’ analogy, I present my rationale for the necessity of temperature sensing in every organelle in a cell. Finally, I propose temperature-sensing riboceptors (ribonucleic acid receptors) to integrate all the RNA molecules (mRNA, non-coding RNA, and so forth) capable of sensing temperature and triggering a signaling event, which I call as thermocrine signaling. This approach could enable the identification of riboceptors in every cell of almost every organism, not only for temperature but also for other classes of ligands, including gaseous solutes, and water.
Have we identified all the temperature-sensing protein-based receptors?
Temperature is one of the pivotal regulators of the evolution of molecular machinery and life. The 600 million years of evolutionary conservation of temperature regulation capacity has been suggested to provide a survival benefit in organisms [Citation1]. Temperature can affect growth rates across various cells and organisms [Citation2–6]. Apart from environmental changes in temperature, pathogen infections and toxin exposure in plants and animals can also increase organismal temperatures [Citation1,Citation7–10]. The mechanisms that allow organisms to survive high temperatures have been attributed to heat shock proteins, temperature-sensitive gene cluster architecture, differential osmotic activity, differential antioxidant defense mechanisms, differential sequences that allow the formation of a relatively higher number of protein disulfide bonds, and protein stabilization [Citation11–13].
The mechanisms that allow cells to sense membranal temperature include the temperature-sensing TRP channels (transient receptor potential) that are referred to as sensory thermoreceptors [Citation14–16]. Such temperature-sensing TRP channel proteins have relatively high Q10 (temperature coefficient: the fold-increase in rate per 10°C increase) in comparison to other non-temperature sensing channel proteins [Citation17]. In Drosophila melanogaster, dTrpA1 in the warmth-activated anterior cell neurons and GR28b.d in the antennal arista are membranal temperature-sensing proteins [Citation18–20]. In the insect triatomine (Rhodnius prolixus), TRPA5B is also a temperature-sensing protein that exhibit a high temperature coefficient (Q10 = 25) [Citation21]. In addition to TRP channels, some GPCRs (G protein-coupled receptors) also exhibit temperature-sensing properties. For instance, light-sensitive rhodopsin in mammals and Drosophila, as well as mouse melanopsin (Opn4), have been identified as membranal temperature-sensing proteins (thermosensors) [Citation22–24].
In addition to membranal proteins, temperature can also be sensed internally via other intracellular proteins (thermosensors or thermometers) that are intrinsically temperature-sensitive. Such proteins either have temperature-sensitive domains or lose their dimerization capabilities at high temperatures. For instance, bacterial GrpE, sigma 32, and DesK are temperature-sensing proteins that exhibit nucleotide exchange factor, transcriptional factor, and kinase activity, respectively [Citation25–27]. In E. coli, the dimeric GrpE, at heat shock temperatures, undergoes unfolding of the long N-terminal helix pair, resulting in the loss of its nucleotide exchange factor activity [Citation28]. In Salmonella, the temperature-sensing activity of the DNA-binding autoregulator TlpA depends on the monomer-to-coiled-coil equilibrium [Citation29]. At high temperatures, TlpA loses its DNA binding and tlpA repressor activity. Another example is TdcA (thermosensory diguanylate cyclase), a temperature-sensing protein that exhibits a 100-fold increase in the activity of c-di-GMP generation upon a 10°C increase [Citation30]. TdcA diguanylate cyclase is inactive at 22°C and highly active at 37°C. TdcA and some TdcA homologs are thermosensitive via a conserved temperature-sensitive PAS (Per-Arnt-SIM) domain. In yeast, nucleotide exchange factor Mge1, a GrpE homolog, was identified as a thermosensor. At heat-shock temperatures, it loses its dimerization, interaction with Hsp70, and the capability to regulate its ATPase activity [Citation31,Citation32]. Human HSF1 (heat shock factor 1) is also a temperature-sensing protein [Citation33]. HSF1 is a transcription factor that trimerizes at increased temperature, binds to promoter sites with heat shock elements, induces gene expression, and confers proteostasis and stress response. In Arabidopsis, TWA1 (Thermo-with abscisic acid-response 1), an intrinsically disordered protein, is a temperature-sensing transcriptional co-regulator that undergoes a conformational change at high temperature, accumulates in the nucleus, and binds to other transcriptional factors that promote the expression of genes that confer thermotolerance [Citation34]. Overall, similar to the gas-sensing gasoreceptor proteins that have diverse signaling domains and activity (kinase, phosphodiesterase, transcription factor, guanylate cyclase, adenylate cyclase, etc.), temperature-sensing proteins or receptors also seem to have diverse signaling domains [Citation35–37]. But are the above-listed proteins the only temperature-sensing proteins in cells? [Citation38,Citation39]
Temperature differences exist internally across different regions and cellular organelles. For instance, the nucleus is marginally hotter than the cytoplasm [Citation40]. In neuron-like cells in vitro, cell body temperature is higher than in neurite-like structures [Citation41]. Temperatures in highly energy-active organelles such as mitochondria is around 50°C, and ATP synthesis perturbations in mitochondria can also cause temperature differences [Citation42–44]. Therefore, temperature-sensing must occur in almost all organelles and regions within the cell.
To illustrate the need for region/organelle-specific temperature-sensing mechanisms, I draw an analogy to colonial India. The British, struggling to acclimate to the Indian heat while maintaining their aristocratic attire, employed ‘punkah wallahs’ – servants or slaves who manually operated ceiling fans called punkahs. These fans, mounted on rectangular wooden frames with cloth, were pulled via a pulley system by the punkah wallahs, cooling the British as they moved between rooms [Citation45,Citation46]. In some cases, the punkah wallahs were completely isolated from the room, as a small hole in the room allowed the rope to be pulled via the pulley. Similar to the punkah wallahs adjusting the cooling in each room, temperature-sensing mechanisms must exist in each cellular organelle and region. The primary function of these mechanisms must be to ensure that the temperature in the cellular microenvironment is tightly regulated probably also by acting along with aquareceptors [Citation47].
Despite the advent of electricity and air conditioners, which require temperature sensors to sense room temperature, cells similarly need internal temperature sensors. Hence, we must identify all temperature-sensing receptors through a systematic approach, akin to research that identifies specific receptors for ligands or transcription factor targets. Although enzyme activity generally decreases at increased temperatures (16-fold per 25°C), proteins with extreme temperature-dependent activity variations are likely candidates for either heat or cold-sensing receptors [Citation48]. Systemically identifying temperature-sensing receptors with unusually high temperature-coefficient will allow us to identify all temperature-sensing receptors. I propose the term ‘agnireceptors’ (‘Agni’ in Sanskrit for ‘God of fire’) for all temperature-sensing proteins (thermoreceptors, thermosensors, and cold-sensing proteins), regardless of their localization and signaling domains and ‘thermocrine signaling’ for all temperature-dependent signaling events that are sensed and triggered via temperature-sensing receptors [Citation14,Citation49–51]. A unified terminology may facilitate the systemic study of temperature-sensing proteins’ functions, especially with advanced deep-learning algorithms that aim to replace animal models in drug discovery [Citation49].
Have we identified all the temperature-sensing nucleic acid-based receptors?
During evolution, temperature sensing was likely crucial for proto-organisms that lacked proteins or with limited protein-based cellular machinery [Citation52,Citation53]. To understand how temperature sensing might have occurred in such organisms, we can study RNA-based temperature-sensing mechanisms observed in microorganisms. These mechanisms often rely on stem-loop-based secondary structures [Citation54,Citation55]. For instance, certain mRNAs contain temperature-sensing elements that regulate the translation of downstream genes, permitting translation only at higher temperatures when ribosome-binding sites are accessible [Citation56–59]. An example is the temperature-sensitive 5’ mRNA coding sequence of the bacterial rpoH gene, which codes for the heat shock transcription factor sigma 32 [Citation60]. The stability of mRNA secondary structures is temperature-dependent, destabilizing under conditions like heat shock (shift from 30°C to 42°C). Another example is found in Yersinia pestis where the lcrF Shine-Dalgarno sequence is sequestered in a temperature-sensitive stem-loop. This structure allows ribosome access and initiates translation only after temperature-induced conformational changes [Citation61,Citation62]. Similarly, cold temperature-induced ncRNA (non-coding RNA) COOLAIR, COLDWRAP, and COLDAIR in plants have also been reported but whether these are intrinsically temperature-sensitive is unclear [Citation63,Citation64]. In my view, ‘RNA thermometers’ or ‘RNA thermosensors’ can be considered as RNA-based temperature-sensing receptors or proto-receptors. A discussion is warranted on when to categorize these structures as receptors. Riboswitches, for instance, have been proposed as receptors for metabolites and hormones, and temperature-sensing riboswitches have been identified as well [Citation65–67]. The determination of when an RNA thermometer or thermosensor qualifies as a receptor depends on the extent of downstream signaling events it triggers. Analogous to ‘ribozymes’ for RNA-based enzymes, it would be beneficial to adopt a unifying term for RNA-based receptors [Citation68].
To encompass all RNA molecules (mRNA, non-coding RNA, etc.) capable of directly sensing various stimuli within a cell – such as temperature, gravity, gaseous solutes, water, metal ions, metal clusters, amino acids, pH, and other biological molecules – and triggering a signal, I propose the term ‘riboceptors’ (ribonucleic acid receptors). Examples of riboceptors also include RNA-based evolutionarily old proto-receptors, such as the riboswitches sensing lithium, sodium, thiamine, lysine, magnesium, manganese, nickel, cobalt, and others [Citation69–73]. For temperature-sensing riboceptors, temperature per se serves as the ligand, with hot and cold temperatures acting as agonists and antagonists, or vice versa ().
Figure 1. Model of putative temperature-sensing riboceptors.
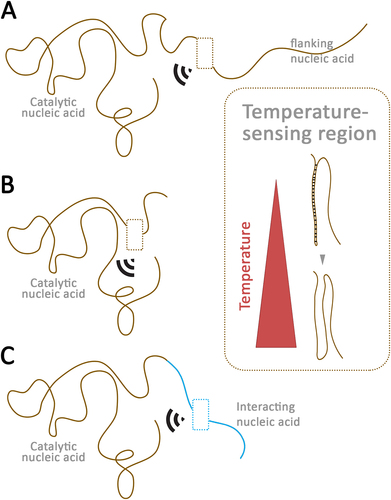
How to identify temperature-sensing riboceptors?
To identify temperature-sensing riboceptors, we first need to systematically identify all RNA structures with catalytic (or even transcription factor) activity in vivo. In such functional RNA molecules, we need to identify which contain or are flanked with highly temperature-sensitive sequences and structures (). Several computational tools have been developed and applied to identify such temperature-sensitive structures. However, the predictions made by these tools do not always align closely with in vivo temperature sensitivity. This discrepancy is not surprising, as even the binding of ribosomes to specific temperature-sensitive structures can promote RNA unwinding/melting at high temperatures [Citation74–80]. Next, we should determine which ribozymes exhibit an unusually high temperature coefficient (Q10) in vivo. Such highly temperature-sensitive ribozymes are likely to be temperature-sensing riboceptors. Mutational studies disrupting temperature-sensitive regions and structural stability should confirm the loss of temperature-sensing activity and modulation of ribozyme activity. We must also debate how distant the temperature-sensitive flanking sequence can be from the ribozyme to still be considered a temperature-sensing riboceptor (). Another possibility is that a ribozyme lacking temperature-sensitive regions may interact with a temperature-sensitive nucleic acid, and the resulting complex may act as a temperature-sensing riboceptor ().
Is it temperature sensing per se, or is it loss of binding partners?
One potential caveat when identifying temperature-sensing riboceptors is determining whether the changes in temperature affect only the nucleic acid structure or also impact the binding of other components necessary for catalytic or signaling activity. However, the question arises: are there any proteins or nucleic acids that function entirely independently without interacting with other molecules? Even the temperature-sensing TRPV1 channel can bind to various classes of endogenous and exogenous molecules [Citation81–83]. Both TRPV1 and TRPV3 channels seem to require lipid binding or ejection for temperature sensitivity [Citation84,Citation85]. Yet, they are still considered temperature-sensing receptors and not lipid-sensing receptors. Similarly, an adenine-sensing riboswitch was identified as temperature-sensitive due to its 35-nucleotide temperature-sensitive module and has been proposed as a riboswitch-thermostat [Citation67,Citation86,Citation87]. In the case of the RNA thermometer MiniROSE (Repression Of heat Shock gene Expression) RNA and agsA mRNA-based thermometer, the temperature-based ‘melting’ of RNA is facilitated by ribosome binding [Citation55,Citation59,Citation80]. Overall, temperature-sensing riboceptor activity could be modulated by its binding factors, similar to the above-mentioned examples of temperature-sensing proteins and RNA. Additionally, besides RNA, DNA also contains temperature-sensitive structures, and DNA structures (deoxyribozymes or DNAzyme) can also exhibit catalytic activity [Citation88–94]. Therefore, it is theoretically plausible that temperature-sensing deoxyriboceptors may exist as well.
Overall, until all the putative temperature-sensitive nucleic acid structures have been systemically characterized for their ability to sense temperature in vivo, it remains challenging to dismiss the possibility of temperature-sensing riboceptors. Identifying temperature-sensing agnireceptor proteins and riboceptors may help us better understand processes such as RNA editing and the cross-talk with gas-sensing gasoreceptors, water-sensing aquareceptors, and organism physiology [Citation30,Citation35,Citation95–97]. It may also help us understand the evolutionary origins of receptors, cellular signaling, and environmental sensing. With temperature and gases likely preceding RNA and protein world, thermocrine and gasocrine signaling via riboceptors are likely one of the earliest cellular signaling mechanisms during the evolution of life [Citation98].
Author contributions
Savani Anbalagan: conceptualization, writing of the original draft, and review and editing.
Disclosure statement
The funding agency and institution S.A. is affiliated with was not involved in the contents of the manuscript.
Correction Statement
This article has been corrected with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Evans SS, Repasky EA, Fisher DT. Fever and the thermal regulation of immunity: the immune system feels the heat. Nat Rev Immunol. 2015;15(6):335–349. doi: 10.1038/nri3843
- Ben-Haj-Salah H, Tardieu F. Temperature affects expansion rate of maize leaves without change in spatial distribution of cell length (analysis of the coordination between cell division and cell expansion). Plant Physiol. 1995;109(3):861–870. doi: 10.1104/pp.109.3.861
- Delomas TA, Dabrowski K. Larval rearing of zebrafish at suboptimal temperatures. J Therm Biol. 2018;74:170–173. doi: 10.1016/j.jtherbio.2018.03.017
- Howard DH. Effect of temperature on the intracellular growth of Histoplasma capsulatum. J Bacteriol. 1967;93(1):438–444. doi: 10.1128/jb.93.1.438-444.1967
- Watanabe I, Okada S. Effects of temperature on growth rate of cultured mammalian cells (L5178Y). J Cell Biol. 1967;32(2):309–323. doi: 10.1083/jcb.32.2.309
- Barnett SA, Neil AC. Growth and reproduction of mice cross-fostered between parents reared at different temperatures. J Physiol. 1971;215(3):665–678. doi: 10.1113/jphysiol.1971.sp009490
- Inada N. A Guide to plant intracellular temperature imaging using fluorescent thermometers. Plant Cell Physiol. 2023;64(1):7–18. doi: 10.1093/pcp/pcac123
- Walter EJ, Hanna-Jumma S, Carraretto M, et al. The pathophysiological basis and consequences of fever. Crit Care. 2016;20(1):200. doi: 10.1186/s13054-016-1375-5
- Haddad F, Soliman AM, Wong ME, et al. Fever integrates antimicrobial defences, inflammation control, and tissue repair in a cold-blooded vertebrate. Elife. 2023;12:e83644. doi: 10.7554/eLife.83644
- Gross L. Anatomy of a fever. PLOS Biol. 2006;4(9):e305. doi: 10.1371/journal.pbio.0040305
- Wang Q, Cen Z, Zhao J. The survival mechanisms of thermophiles at high temperatures: an angle of Omics. Physiol (Bethesda). 2015;30(2):97–106. doi: 10.1152/physiol.00066.2013
- Sezgin Muslu A, Kadıoğlu A. Role of abscisic acid, osmolytes and heat shock factors in high temperature thermotolerance of heliotropium thermophilum. Physiol Mol Biol Plants. 2021;27(4):861–871. doi: 10.1007/s12298-021-00975-7
- Seth P, Sebastian J. Plants and global warming: challenges and strategies for a warming world. Plant Cell Rep. 2024;43(1):27. doi: 10.1007/s00299-023-03083-w
- Dhaka A, Viswanath V, Patapoutian A. Trp ion channels and temperature sensation. Annu Rev Neurosci. 2006;29(1):135–161. doi: 10.1146/annurev.neuro.29.051605.112958
- Zhang M, Ma Y, Ye X, et al. TRP (transient receptor potential) ion channel family: structures, biological functions and therapeutic interventions for diseases. Sig Transduct Target Ther. 2023;8(1):1–38. doi: 10.1038/s41392-023-01464-x
- Rosenbaum T, Islas LD. Molecular physiology of TRPV channels: controversies and future challenges. Annu Rev Physiol. 2023;85(1):293–316. doi: 10.1146/annurev-physiol-030222-012349
- Baez-Nieto D, Castillo JP, Dragicevic C, et al. Thermo-TRP channels: biophysics of polymodal receptors. Adv Exp Med Biol. 2011;704:469–490.
- Yamamoto T, Vukelic J, Hertzberg EL, et al. Differential anatomical and cellular patterns of connexin43 expression during postnatal development of rat brain. Brain Res Dev Brain Res. 1992;66(2):165–180. doi: 10.1016/0165-3806(92)90077-A
- Ni L, Bronk P, Chang EC, et al. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature. 2013;500(7464):580–584. doi: 10.1038/nature12390
- Hamada FN, Rosenzweig M, Kang K, et al. An internal thermal sensor controlling temperature preference in Drosophila. Nature. 2008;454(7201):217–220. doi: 10.1038/nature07001
- Liénard MA, Baez-Nieto D, Tsai C-C, et al. TRPA5 encodes a thermosensitive ankyrin ion channel receptor in a triatomine insect. iScience. 2024;27(4):109541. doi: 10.1016/j.isci.2024.109541
- Pérez-Cerezales S, Boryshpolets S, Afanzar O, et al. Involvement of opsins in mammalian sperm thermotaxis. Sci Rep. 2015;5(1):16146. doi: 10.1038/srep16146
- Sokabe T, Chen H-C, Luo J, et al. A switch in thermal preference in drosophila larvae depends on multiple rhodopsins. Cell Rep. 2016;17(2):336–344. doi: 10.1016/j.celrep.2016.09.028
- Shen WL, Kwon Y, Adegbola AA, et al. Function of rhodopsin in temperature discrimination in drosophila. Science. 2011;331(6022):1333–1336. doi: 10.1126/science.1198904
- Groemping Y, Reinstein J. Folding properties of the nucleotide exchange factor GrpE from thermus thermophilus: GrpE is a thermosensor that mediates heat shock response 1. J Mol Biol. 2001;314(1):167–178. doi: 10.1006/jmbi.2001.5116
- Cybulski LE, Ballering J, Moussatova A, et al. Activation of the bacterial thermosensor DesK involves a serine zipper dimerization motif that is modulated by bilayer thickness. Proc Natl Acad Sci U S A. 2015;112(20):6353–6358. doi: 10.1073/pnas.1422446112
- Chattopadhyay R, Roy S. DnaK-sigma 32 interaction is temperature-dependent: implication for the mechanism of heat shock response *. J Biol Chem. 2002;277(37):33641–33647. doi: 10.1074/jbc.M203197200
- Gelinas AD, Langsetmo K, Toth J, et al. A structure-based interpretation of E. coli GrpE thermodynamic properties. J Mol Biol. 2002;323(1):131–142. doi: 10.1016/S0022-2836(02)00915-4
- Hurme R, Berndt KD, Normark SJ, et al. A proteinaceous gene regulatory thermometer in Salmonella. Cell. 1997;90(1):55–64. doi: 10.1016/S0092-8674(00)80313-X
- Almblad H, Randall TE, Liu F, et al. Bacterial cyclic diguanylate signaling networks sense temperature. Nat Commun. 2021;12(1):1986. doi: 10.1038/s41467-021-22176-2
- Moro F, Muga A. Thermal adaptation of the yeast mitochondrial Hsp70 system is regulated by the reversible unfolding of its nucleotide exchange factor. J Mol Biol. 2006;358(5):1367–1377. doi: 10.1016/j.jmb.2006.03.027
- Marada A, Karri S, Singh S, et al. A single point mutation in mitochondrial Hsp70 cochaperone Mge1 gains thermal stability and resistance. Biochemistry. 2016;55(51):7065–7072. doi: 10.1021/acs.biochem.6b00232
- Hentze N, Le Breton L, Wiesner J, et al. Molecular mechanism of thermosensory function of human heat shock transcription factor Hsf1. Elife. 2016;5:e11576. doi: 10.7554/eLife.11576
- Bohn L, Huang J, Weidig S, et al. The temperature sensor TWA1 is required for thermotolerance in arabidopsis. Nature. 2024;629(8014):1126–1132. doi: 10.1038/s41586-024-07424-x
- Anbalagan S. Heme-based oxygen gasoreceptors. Am J Physiol Endocrinol Metab. 2024;326(2):E178–81. doi: 10.1152/ajpendo.00004.2024
- Anbalagan S. Oxygen is an essential gasotransmitter directly sensed via protein gasoreceptors. Anim Model Exp Med. 2024;7(2):189–193. doi: 10.1002/ame2.12400
- Aono S. Gas sensing in cells. United Kingdom: Royal Society of Chemistry; 2017.
- Zhou J, Del Rosal B, Jaque D, et al. Advances and challenges for fluorescence nanothermometry. Nat Methods. 2020;17(10):967–980. doi: 10.1038/s41592-020-0957-y
- Di X, Wang D, Su QP, et al. Spatiotemporally mapping temperature dynamics of lysosomes and mitochondria using cascade organelle-targeting upconversion nanoparticles. Proc Natl Acad Sci USA. 2022;119(45):e2207402119. doi: 10.1073/pnas.2207402119
- Okabe K, Uchiyama S. Intracellular thermometry uncovers spontaneous thermogenesis and associated thermal signaling. Commun Biol. 2021;4(1):1–7. doi: 10.1038/s42003-021-02908-2
- Tanimoto R, Hiraiwa T, Nakai Y, et al. Detection of temperature difference in neuronal cells. Sci Rep. 2016;6(1):22071. doi: 10.1038/srep22071
- Terzioglu M, Veeroja K, Montonen T, et al. Mitochondrial temperature homeostasis resists external metabolic stresses. Elife. 2023;12: RP89232.10.7554/eLife.89232.3
- Chrétien D, Bénit P, H-H H, et al. Mitochondria are physiologically maintained at close to 50 °C. PLOS Biol. 2018;16(1):e2003992. doi: 10.1371/journal.pbio.2003992
- Hayashi T, Fukuda N, Uchiyama S, et al. A cell-permeable fluorescent polymeric thermometer for intracellular temperature mapping in mammalian cell lines. PLOS ONE. 2015;10(2):e0117677. doi: 10.1371/journal.pone.0117677
- Parsons RC. On the working of Punkahs in India; as at present carried out by coolie labour, and the same operation effected by machinery. London-New York. 1878.
- Sengupta R. (2022). Keeping the master cool, every day, all day: Punkah-pulling in colonial India. The Indian Economic & Social History Review, 59(1), 37–73. doi: 10.1177/00194646211064592
- Anbalagan S. 2024. Heme-based aquareceptors. Postepy Biochem. doi: 10.18388/pb.2021_551
- Elias M, Wieczorek G, Rosenne S, et al. The universality of enzymatic rate–temperature dependency. Trends Biochem Sci. 2014;39(1):1–7. doi: 10.1016/j.tibs.2013.11.001
- Anbalagan S. “Blind men and an elephant”: the need for animals in research, drug safety studies, and understanding civilizational diseases. Anim Model Exp Med. 2023;6(6):627–633. doi: 10.1002/ame2.12364
- Vu LD, Gevaert K, De Smet I. Feeling the heat: searching for plant thermosensors. Trends Plant Sci. 2019;24(3):210–219. doi: 10.1016/j.tplants.2018.11.004
- Jung J-H, Seo PJ, Oh E, et al. Temperature perception by plants. Trends Plant Sci. 2023;28(8):924–940. doi: 10.1016/j.tplants.2023.03.006
- Lancet D, Zidovetzki R, Markovitch O. Systems protobiology: origin of life in lipid catalytic networks. J R Soc Interface. 2018;15(144):20180159. doi: 10.1098/rsif.2018.0159
- Cavalier-Smith T. Obcells as proto-organisms: membrane heredity, lithophosphorylation, and the origins of the genetic code, the first cells, and photosynthesis. J Mol Evol. 2001;53(4–5):555–595. doi: 10.1007/s002390010245
- Stephenson W, Keller S, Santiago R, et al. Combining temperature and force to study folding of an RNA hairpin. Phys Chem Chem Phys. 2013;16(3):906–917. doi: 10.1039/C3CP52042K
- Meyer S, Carlson PD, Lucks JB. Characterizing the structure–function relationship of a naturally occurring RNA thermometer. Biochemistry. 2017;56(51):6629–6638. doi: 10.1021/acs.biochem.7b01170
- Loh E, Righetti F, Eichner H, et al. RNA thermometers in bacterial pathogens. Microbiol Spectr. 2018;6(2):6. doi: 10.1128/microbiolspec.RWR-0012-2017
- Mandin P, Johansson J. Feeling the heat at the millennium: thermosensors playing with fire. Mol Microbiol. 2020;113(3):588–592. doi: 10.1111/mmi.14468
- Somero GN. RNA thermosensors: how might animals exploit their regulatory potential? J Exp Biol. 2018;221(4):jeb162842. doi: 10.1242/jeb.162842
- Chowdhury S, Maris C, Allain FH, et al. Molecular basis for temperature sensing by an RNA thermometer. Embo J. 2006;25(11):2487–2497. doi: 10.1038/sj.emboj.7601128
- Morita MT, Tanaka Y, Kodama TS, et al. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13(6):655–665. doi: 10.1101/gad.13.6.655
- Hoe NP, Goguen JD. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993;175(24):7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993
- Böhme K, Steinmann R, Kortmann J, et al. Concerted actions of a thermo-labile regulator and a unique intergenic RNA thermosensor control Yersinia virulence. PlOS Pathog. 2012;8(2):e1002518. doi: 10.1371/journal.ppat.1002518
- Jha UC, Nayyar H, Roychowdhury R, et al. Non-coding RNAs (ncRNAs) in plant: master regulators for adapting to extreme temperature conditions. Plant Physiol Biochem. 2023;205:108164. doi: 10.1016/j.plaphy.2023.108164
- Yuan C, He R, Zhao W, et al. Insights into the roles of long noncoding RNAs in the communication between plants and the environment. Plant Genome. 2023;16(4):e20277. doi: 10.1002/tpg2.20277
- Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5(6):451–463. doi: 10.1038/nrm1403
- Grojean J, Downes B. Riboswitches as hormone receptors: hypothetical cytokinin-binding riboswitches in Arabidopsis thaliana. Biol Direct. 2010;5(1):60. doi: 10.1186/1745-6150-5-60
- Reining A, Nozinovic S, Schlepckow K, et al. Three-state mechanism couples ligand and temperature sensing in riboswitches. Nature. 2013;499(7458):355–359. doi: 10.1038/nature12378
- Walter NG, Engelke DR. Ribozymes: catalytic RNAs that cut things, make things, and do odd and useful jobs. Biologist (Lond). 2002;49(5):199–203.
- White N, Sadeeshkumar H, Sun A, et al. Lithium-sensing riboswitch classes regulate expression of bacterial cation transporter genes. Sci Rep. 2022;12(1):19145. doi: 10.1038/s41598-022-20695-6
- White N, Sadeeshkumar H, Sun A, et al. Na+ riboswitches regulate genes for diverse physiological processes in bacteria. Nat Chem Biol. 2022;18(8):878–885. doi: 10.1038/s41589-022-01086-4
- Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419(6910):952–956. doi: 10.1038/nature01145
- Sudarsan N, Wickiser JK, Nakamura S, et al. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes Dev. 2003;17(21):2688–2697. doi: 10.1101/gad.1140003
- Wedekind JE, Dutta D, Belashov IA, et al. Metalloriboswitches: RNA-based inorganic ion sensors that regulate genes. J Biol Chem. 2017;292(23):9441–9450. doi: 10.1074/jbc.R117.787713
- Shah P, Gilchrist MA, Spirin AS. Is thermosensing property of RNA thermometers unique? PLOS ONE. 2010;5(7):e11308. doi: 10.1371/journal.pone.0011308
- Churkin A, Avihoo A, Shapira M, et al. RNAthermsw: direct temperature simulations for predicting the location of RNA thermometers. PLOS ONE. 2014;9(4):e94340. doi: 10.1371/journal.pone.0094340
- Waldminghaus T, Gaubig LC, Narberhaus F. Genome-wide bioinformatic prediction and experimental evaluation of potential RNA thermometers. Mol Genet Genomics. 2007;278(5):555–564. doi: 10.1007/s00438-007-0272-7
- Chursov A, Kopetzky SJ, Bocharov G, et al. RNAtips: analysis of temperature-induced changes of RNA secondary structure. Nucleic Acids Res. 2013;41(W1):W486–491. doi: 10.1093/nar/gkt486
- Sato K, Akiyama M, Sakakibara Y. RNA secondary structure prediction using deep learning with thermodynamic integration. Nat Commun. 2021;12(1):941. doi: 10.1038/s41467-021-21194-4
- Zhang J, Lang M, Zhou Y, et al. Predicting RNA structures and functions by artificial intelligence. Trends Genet. 2024;40(1):94–107. doi: 10.1016/j.tig.2023.10.001
- Narayan S, Kombrabail MH, Das S, et al. Site-specific fluorescence dynamics in an RNA ‘thermometer’ reveals the role of ribosome binding in its temperature-sensitive switch function. Nucleic Acids Res. 2015;43(1):493–503. doi: 10.1093/nar/gku1264
- Riera CE, Vogel H, Simon SA, et al. Artificial sweeteners and salts producing a metallic taste sensation activate TRPV1 receptors. Am J Physiol Regul Integr Comp Physiol. 2007;293(2):R626–634. doi: 10.1152/ajpregu.00286.2007
- Oláh Z, Jósvay K, Pecze L, et al. Anti-calmodulins and tricyclic adjuvants in pain therapy block the TRPV1 channel. PLOS ONE. 2007;2(6):e545. doi: 10.1371/journal.pone.0000545
- Ahern GP, Brooks IM, Miyares RL, et al. Extracellular cations sensitize and gate capsaicin receptor TRPV1 modulating pain signaling. J Neurosci. 2005;25(21):5109–5116. doi: 10.1523/JNEUROSCI.0237-05.2005
- Singh AK, McGoldrick LL, Demirkhanyan L, et al. Structural basis of temperature sensation by the TRP channel TRPV3. Nat Struct Mol Biol. 2019;26(11):994–998. doi: 10.1038/s41594-019-0318-7
- Gao Y, Cao E, Julius D, et al. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534(7607):347–351. doi: 10.1038/nature17964
- Fürtig B, Oberhauser EM, Zetzsche H, et al. Refolding through a linear transition state enables fast temperature adaptation of a translational Riboswitch. Biochemistry. 2020;59(10):1081–1086. doi: 10.1021/acs.biochem.9b01044
- Wu L, Liu Z, Liu Y. Thermal adaptation of structural dynamics and regulatory function of adenine riboswitch. RNA Biol. 2021;18(11):2007–2015. doi: 10.1080/15476286.2021.1886755
- Ponce-Salvatierra A, Wawrzyniak-Turek K, Steuerwald U, et al. Crystal structure of a DNA catalyst. Nature. 2016;529(7585):231–234. doi: 10.1038/nature16471
- Dong X, Qiu Z, Wang Z, et al. Efficient Silver(I)-containing i-motif DNA hybrid catalyst for Enantioselective Diels-Alder reactions. Angew Chem Int Ed Engl. 2024:e202407838. doi: 10.1002/anie.202407838
- Cozma I, Em M, Brennan JD, et al. DNAzymes as key components of biosensing systems for the detection of biological targets. Biosens Bioelectron. 2021;177:112972. doi: 10.1016/j.bios.2021.112972
- Okamoto C, Momotake A, Yamamoto Y. Structural and functional characterization of complexes between heme and dimeric parallel G-quadruplex DNAs. J Inorg Biochem. 2021;216:111336. doi: 10.1016/j.jinorgbio.2020.111336
- Driessen RPC, Sitters G, Laurens N, et al. Effect of temperature on the intrinsic flexibility of DNA and its interaction with architectural proteins. Biochemistry. 2014;53(41):6430–6438. doi: 10.1021/bi500344j
- Brunet A, Salomé L, Rousseau P, et al. How does temperature impact the conformation of single DNA molecules below melting temperature? Nucleic Acids Res. 2018;46(4):2074–2081. doi: 10.1093/nar/gkx1285
- Mizushima T, Kataoka K, Ogata Y, et al. Increase in negative supercoiling of plasmid DNA in Escherichia coli exposed to cold shock. Mol Microbiol. 1997;23(2):381–386. doi: 10.1046/j.1365-2958.1997.2181582.x
- Birk MA, Liscovitch-Brauer N, Dominguez MJ, et al. Temperature-dependent RNA editing in octopus extensively recodes the neural proteome. Cell. 2023;186(12):2544–2555.e13. doi: 10.1016/j.cell.2023.05.004
- Li W, Bu M, Hu R, et al. Tissue-specific temperature dependence of RNA editing levels in zebrafish. BMC Biol. 2023;21(1):262. doi: 10.1186/s12915-023-01738-4
- Su R, Zhou M, Lin J, et al. A circular RNA-gawky-chromatin regulatory axis modulates stress-induced transcription. Nucleic Acids Res. 2024;52(7):3702–3721. doi: 10.1093/nar/gkae157
- Anbalagan S. Gas-sensing riboceptors. RNA Biol. 2024 doi: 10.1080/15476286.2024.2379607
