 ?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.Abstract
Although numerous techniques have been developed to monitor autophagy and to probe its cellular functions, these methods cannot evaluate in sufficient detail the autophagy process, and suffer limitations from complex experimental setups and/or systematic errors. Here we developed a method to image, contextually, the number and pH of autophagic intermediates by using the probe mRFP-GFP-LC3B as a ratiometric pH sensor. This information is expressed functionally by AIPD, the pH distribution of the number of autophagic intermediates per cell. AIPD analysis reveals how intermediates are characterized by a continuous pH distribution, in the range 4.5–6.5, and therefore can be described by a more complex set of states rather than the usual biphasic one (autophagosomes and autolysosomes). AIPD shape and amplitude are sensitive to alterations in the autophagy pathway induced by drugs or environmental states, and allow a quantitative estimation of autophagic flux by retrieving the concentrations of autophagic intermediates.
Keywords:
- autophagic flux
- autophagy
- cathepsin B
- chloroquine
- fluorescence microscopy
- LysoSensor
- mechanistic target of rapamycin (serine/threonine kinase)
- microtubule-associated protein 1 light chain 3
- mRFP-GFP-LC3
- pH ratiometric sensor
- phosphatidylinositol 3-kinase catalytic subunit type 3
- pHrodo red
- rapamycin
- starvation
- 3-methyladenine
Abbreviations
| 3-MA | = | 3-methyladenine |
| AI | = | autophagic intermediates |
| AIPD | = | autophagic intermediates pH distribution |
| AL | = | autolysosomes |
| ALE | = | early autolysosomes |
| ALM | = | mature autolysosomes |
| AP | = | autophagosomes |
| CHL | = | chloroquine |
| CTSB | = | cathepsin B |
| DsRed | = | Discosoma sp. red fluorescent protein |
| EGFP | = | enhanced green fluorescent protein |
| FRET | = | fluorescence resonance energy transfer |
| HEK-293T | = | human embryonic kidney 293 |
| L | = | lysosomes |
| LPD | = | LysoSensor Yellow/Blue pH distribution |
| MAP1LC3B | = | microtubule-associated protein 1 light chain 3 β |
| mRFP | = | monomeric red fluorescent protein |
| mRFP-EGFP-LC3B | = | monomeric red fluorescent protein-enhanced green fluorescent protein-MAP1LC3B tandem construct |
| MTOR | = | mechanistic target of rapamycin (serine/threonine kinase) |
| PG | = | phagophores |
| RPS6 | = | ribosomal protein S6 |
| STX17 | = | syntaxin 17 |
| UV | = | ultraviolet radiation. |
Introduction
Autophagy is a fundamental cellular pathway whereby cytoplasmic material, including organelles, reach lysosomes for degradation.Citation1-3 Three primary types of autophagy have been classified (macroautophagy, microautophagy, and chaperone-mediated autophagy), and among these, macroautophagy (hereafter autophagy), the most extensively studied, is mediated by organelles known as autophagosomes. Upon induction, a vesicular sac called the phagophore, elongates, and encloses a portion of cytoplasm, which results in the formation of a double-membrane structure, the autophagosome (). Then, the outer membrane of the autophagosome fuses with a lysosome to form an autolysosome, leading to the degradation of the enclosed materials. Recovered amino acids and other small molecules are delivered back to the cytoplasm for recycling or for energy production. Increasing evidence suggests that the deregulation of autophagy may contribute to a broad spectrum of human disorders, including neurodegenerative diseases (Alzheimer, Parkinson, Huntington, and Creutzfeldt-Jakob), cardiomyopathy (Danon disease), tumorigenesis, aberrant antigen presentation, infectious diseases, cell death, type II diabetes, and fatty liver.Citation1,2,4 Consequently, there is a rapidly growing need for accurately monitoring autophagy in diverse biological processes.
Figure 1. mRFP-EGFP-LC3B is a ratiometric probe which allows monitoring autophagic flux. (A) Under autophagy induction, the phagophore (PG) engulfs a portion of the cytoplasm, forming a double-membrane autophagosome (AP). Subsequently, autophagosomes fuse with lysosomes (inner pH∼4.8), to become autolysosomes. During this process mRFP-EGFP-LC3B is lipidated at its C terminus and recruited to both the inner and outer membrane of the autophagosome. After or before fusion with lysosomes, mRFP-EGFP-LC3B bound to the outer membrane is delipidated and detached from the outer membrane of the autolysosomes. The signals from EGFP that were present within the inner autophagosome membrane are quenched in the autolysosomal environment (pK = 6.0), but mRFP signals are relatively stable (pK = 4.5). Three different possible states of the probe (n1, n2, n3) with different ratios and different pH are reported as an example. The pH of an autolysosome in an early state (ALE) is less acidic than the pH of an autolysosome in a mature state (ALM) (B) green and (C) red emission channel of mRFP-EGFP-LC3B. (D) The overlap of the 2 channels (E) Overlap of the 2 channels with different emission settings (green photomultiplier voltage higher than the red one) shows how an arbitrary regulation of photomultiplier tubes gain values can significantly alter, within the same image, the relative contributions from the 2 channels, thus converting arbitrarily to “red” the yellow spots or vice versa. The arrows are pointing to some intermediates in which such change is evident. Scale bar: 10 µm. (F) Fluorescence emission spectra of mRFP-EGFP-LC3B excited at 488 nm in function of pH. Purple line pH = 9.0, green line pH = 7.9, red line pH = 6.9, blue line pH = 5.8. (G) Ratio R vs. pH. Solid line is the fit to the data of Equation Equation1[1]
[1] .
![Figure 1. mRFP-EGFP-LC3B is a ratiometric probe which allows monitoring autophagic flux. (A) Under autophagy induction, the phagophore (PG) engulfs a portion of the cytoplasm, forming a double-membrane autophagosome (AP). Subsequently, autophagosomes fuse with lysosomes (inner pH∼4.8), to become autolysosomes. During this process mRFP-EGFP-LC3B is lipidated at its C terminus and recruited to both the inner and outer membrane of the autophagosome. After or before fusion with lysosomes, mRFP-EGFP-LC3B bound to the outer membrane is delipidated and detached from the outer membrane of the autolysosomes. The signals from EGFP that were present within the inner autophagosome membrane are quenched in the autolysosomal environment (pK = 6.0), but mRFP signals are relatively stable (pK = 4.5). Three different possible states of the probe (n1, n2, n3) with different ratios and different pH are reported as an example. The pH of an autolysosome in an early state (ALE) is less acidic than the pH of an autolysosome in a mature state (ALM) (B) green and (C) red emission channel of mRFP-EGFP-LC3B. (D) The overlap of the 2 channels (E) Overlap of the 2 channels with different emission settings (green photomultiplier voltage higher than the red one) shows how an arbitrary regulation of photomultiplier tubes gain values can significantly alter, within the same image, the relative contributions from the 2 channels, thus converting arbitrarily to “red” the yellow spots or vice versa. The arrows are pointing to some intermediates in which such change is evident. Scale bar: 10 µm. (F) Fluorescence emission spectra of mRFP-EGFP-LC3B excited at 488 nm in function of pH. Purple line pH = 9.0, green line pH = 7.9, red line pH = 6.9, blue line pH = 5.8. (G) Ratio R vs. pH. Solid line is the fit to the data of Equation Equation1[1] R=R0(Rf+10−(pH−pKA)1+10−(pH−pKA)).[1] .](/cms/asset/2f88c21e-fdfa-4f73-98fc-ea66efe65de5/kaup_a_1084455_f0001_oc.gif)
The term “autophagic flux” denotes the process of autophagosome synthesis, delivery of autophagic substrates to lysosomes, and degradation of autophagic substrates inside lysosomes. Ideally, an experimental method for the assessment of the autophagic flux should detect the different intermediates in the autophagy pathway (early autophagosome, autolysosome, autophagic degradation products) and their concentrations. This information is critical, for instance, to determine whether an increase in intermediates represents an increase in autophagic degradation, or rather an inhibition in some steps of the autophagic pathway. Microtubule-associated protein 1 light chain 3 β (MAP1LC3B), a mammalian ortholog of yeast Atg8, exists on autophagosomes,Citation5 and therefore, this protein serves as a widely used marker for these organelles ().Citation6,7 MAP1LC3B-based biochemical and microscopy assays, as well as the experimental manipulation of the autophagy pathway (through either knockout or knockdown of autophagy genes or the expression of dominant negative autophagy proteins) have facilitated the investigation of the autophagic system. However, these methods cannot monitor the autophagic flux as a whole, or require further cell manipulations that perturb the cellular state.Citation8
A useful assay to measure autophagic flux is based on the concept of lysosomal quenching of GFP in GFP-labeled autophagic substrates such as MAP1LC3B. GFP is a stably folded protein relatively resistant to lysosomal proteases. The low pH inside the autolysosome quenches the fluorescent signal of GFP.Citation6,9 In contrast, mRFP is not quenched in autolysosomes.Citation10 By exploiting the difference in the photostability of these 2 fluorescent proteins, autophagic flux can be morphologically traced with an mRFP-EGFP-MAP1LC3B tandem constructCitation11 (called mRFP-EGFP-LC3B; representative images of the EGFP and mRFP emission channels are shown in ). Indeed autophagosomes and autolysosomes, respectively labeled with yellow (mRFP and EGFP) and red (mRFP only) signals ("puncta"), can be counted to get a measure of the number of each intermediate, and if autophagic flux increases, both yellow and red puncta are increased. If, instead, autophagosome maturation into autolysosomes is blocked, only yellow puncta are increased. Although this assay can be used as an indicator of autophagic flux, it is not free from errors and biases, since, as shown in , active autolysosomes can assume all the color gradations between yellow and red, depending on the value of the endosomal pH that quenches the EGFP signal; furthermore, an arbitrary regulation of the detector's settings can significantly alter, within the same image, the relative contributions from the 2 channels, thus converting arbitrarily to “red” the yellow spots or vice versa (). Therefore, simple counting of yellow to red spot leads to a biased estimation of AP and AL number. To improve autophagic flux determination, an mTagRFP-mWasabi-LC3B reporter was engineered, with a monomeric green fluorescent protein that is more acid sensitive than EGFP and therefore completely quenched in autolysosomes.Citation12 However, this method only allows distinguishing between acidic and neutral compartments, with no contextual information on their pH. Rosado et al.Citation13 have constructed a pH-insensitive RFP combined with a pH-sensitive GFP. The resultant probe, “Rosella,” was used in live cells to follow autophagic engulfment of both cytosol and specific organelles. Unfortunately this probe requires dual excitation and dual emission; therefore it is generally inadequate to follow rapid events in highly motile structures. In this regard, the observation of autolysosome formation using Rosella may be limited by the poor spatiotemporal resolution of conventional confocal microscopes. Moreover, the red protein employed, DsRed-T3, has several drawbacks, including slow chromophore maturation and poor solubility.Citation14 Another pH-sensitive probe was employed to monitor autophagy, mKeima.Citation15 Unfortunately, mKeima is not a MAP1LC3B-associated probe, requiring therefore co-staining of autophagic organelles and dual-excitation, with the same temporal resolution limits of Rosella. Furthermore, since excitation wavelength of mKeima is in the ultraviolet (UV) spectrum, phototoxicity ensues as an unavoidable side effect. Moreover, autophagy can be induced by UV irradiation, perturbing unavoidably the autophagic assay.Citation16
We present in this paper a method that overcomes the above limitations. The method exploits the full potential of mRFP-EGFP-LC3B probe to image, contextually, the number and the pH of autophagy intermediates using the tandem fluorescent protein as a dual-emission ratiometric pH probe, in a fashion that is unbiased with respect to instrumental setup or observer subjectivity. With this technique, the autophagic flux is fully characterized by the pH distribution of the number of autophagic intermediates per cell, AIPD. The method reveals that intermediates are characterized by a broad pH distribution, compatible with a more complex set of states with respect to the expected biphasic one (autophagosomes and autolysosomes). Furthermore, differences in both shape and amplitude of AIPD reveal different kinetic properties for autophagy induced by drugs (rapamycin, chloroquine, 3-methyladenine) or environmental states (serum “starvation”), and allow a quantitative estimation of autophagic flux by the determination of the stoichiometry of intermediates.
Results
mRFP-EGFP-LC3B is a ratiometric pH-sensitive probe for acidic intracellular compartments
During autophagosome formation, mRFP-EGFP-LC3B is lipidated at its C terminus and recruited to both the inner and outer membrane of the forming autophagosome (MAP1LC3B-I to MAP1LC3B-II transition, ). After lysosome-autophagosome fusion, mRFP-GFP-LC3 bound to the outer membrane of the autolysosomes is delipidated and detached.Citation11 Although MAP1LC3B is currently thought to function primarily in the cytosol, where the formation of autophagosome occurs, EGFP-MAP1LC3B often appears to be enriched in the nucleoplasm relative to the cytoplasm ().Citation17 In order for autophagosomes and autolysosomes to be labeled with 2 distinct single colors (yellow and red), the pH of the intermediates should assume only 2 discrete values. Instead, active autolysosomes can assume all color gradations between yellow and red (), depending only on pH since the EGFP signal is instantly (<1 msec interval) quenched by the protonation of its fluorophore.Citation11,18 This complication, together with the arbitrary regulation of detector efficiencies, can bias the count of “red” and “yellow” spots (). Conversely, we speculated that the classification of autophagy intermediates by organelle pH, possibly a) using the same probe that marks autophagic intermediates to avoid dual transfections, and b) with a single excitation wavelength to avoid time lapses between acquisitions, would overcome the above systematic errors in number estimations.
EGFP is a sensitive pH indicator,Citation19 and it has been already used to monitor nonratiometrically pH variations in cell compartments.Citation19,20 EGFP is characterized by high emissivity of the anionic state in the green (ϵ488 = 60,000 M−1·cm−1, Φ = 0.7, 1e = 509nm21), although its pKa (5.8 to 6.2) makes it unsuitable for monitoring high pH subcellular components. For monitoring the pH of autophagic intermediates, however, this pKa value is optimal. Based on the different pH sensitivity of EGFP and mRFP (pK ≈ 4.5),Citation11 we measured the pH dependency of mRFP-EGFP-LC3B emission spectra excited at 488 nm, to examine whether a pH-ratiometric imaging is feasible (). The emission spectrum results in a superposition of EGFP emission (range 500 to 600nm) and mRFP emission (range 600 to 700 nm). The quenching effect of pH on EGFP fluorescence is visible from the drop off in fluorescence of 115%, when pH decreases from 9.0 to 5.8. The concurrent decrease in the range 650 to 700 (mRFP emission) is lower (about 40%), due to fluorescence resonance energy transfer (FRET) between EGFP and mRFP.Citation22 This contribution is small (low quantum efficiency of mRFP makes it disadvantageous as a FRET acceptorCitation23), and since the decrease is only one-third of that observed in EGFP window of emission, mRFP-EGFP-LC3 allows a ratiometric readout. Quenching of mRFP is negligible since it is present only at an extreme pH range (i.e. pH = 3.5–4.5). Lysosomal degradation of the whole tandem does not influence the readout, because variations in concentration are ruled out from the ratio; moreover, lysosomal degradation rate is conceivably the same for EGFP and mRFP, which have a very similar molecular mass (∼27 KdA) and conformation.Citation18,24,25
The ratio R between the emission intensity in the green range (FG) and the emission intensity in the red range (FR) reflects EGFP quenching by hydrogen ions. This ratio R = FG/FR can be modeled by the equationCitation26[1]
[1] Where Rf is the dynamic range of the probe, R0 is the ratiometric offset, pKA is the ratiometric pK. In , ratio R versus pH is plotted. A fit of the experimental points with Equation Equation1
[1]
[1] permits to determine the pKA of the probe equal to 6.0. By inverting Equation Equation[1]
[1]
[1] :
[2]
[2]
It is possible to link measured R values to pH, using as calibrators for R0 probe solutions of known pH in the range pKA+1, pKA-1, provided that during the experimental session detector's settings are kept unchanged. The dynamic range of the probe is comparable with other probes in the same pKA range () and is characterized by the lowest pKA, making it optimal to be used in acidic compartments. The reliability of pH read-out in live cells was further validated by using the pH-sensitive probe LysoSensor Yellow/Blue (see below).
Table 1. Dynamic range and pKA of principal redox sensitive fluorophores
Determination of AIPD
By using the pseudo-colored map of R we can simultaneously evaluate the number and pH of autophagic intermediates for untreated HEK-293T (CTRL) cells ( overlap of red and green channel, R-map; see Materials and Methods for details about the construction). AIPD (autophagy Intermediates pH distribution), which gives the number of intermediates per cell at an established pH, is retrieved by measuring directly the pH value on the R-map, for each intermediate. To construct the AIPD, the first step is to “bin” the range of pH values—that is, divide the entire range of pH values into a series of small intervals—and then count how many values fall into each interval. Each AIPD value is therefore proportional to the count of intermediates with the selected pH. According to this construction, the total area of the AIPD represents the number of puncta per cell n, equal to 10.2. AIPD () is characterized by a broad and continuous distribution in the range 4.5–6.5. The distribution is neither Gaussian, nor symmetric: the distribution presents a shoulder between pH = 4.5 and pH = 5.4, a peak at pH = 5.6 and then it damps at pH = 6.6.
Figure 2. AIPD as a tool for quantifying autophagic flux. (A) Overlaps of the 2 channels of mRFP-EGFP-LC3B for control (CTRL), serum-starved (STARV), rapamycin treated (RAP), rapamycin-treated and serum-starved cells (STARV + RAP), chloroquine-treated (CHL) and 3-MA-treated cells (3-MA). (B) R-maps of the cells with the above-mentioned treatments (16-color rainbow lookup table [ImageJ]), values from R0-black to Rf-white. (C) AIPDs for the cells with the above-mentioned treatments. Scale bar: 20 µm. A 3-Lorentzian deconvolution of the AIPD function allows measuring the relative contributions of the 3 populations: the first population represents autophagosomes (AP, pH = 6.0, orange line), whether the second and the third represent 2 different states of autolysosomes (AL), an early one (ALE, pH = 5.6, green line), and a mature, more acidic state (ALM, pH = 5.2, blue line). The black line represents the sum of the 3 populations.
![Figure 2. AIPD as a tool for quantifying autophagic flux. (A) Overlaps of the 2 channels of mRFP-EGFP-LC3B for control (CTRL), serum-starved (STARV), rapamycin treated (RAP), rapamycin-treated and serum-starved cells (STARV + RAP), chloroquine-treated (CHL) and 3-MA-treated cells (3-MA). (B) R-maps of the cells with the above-mentioned treatments (16-color rainbow lookup table [ImageJ]), values from R0-black to Rf-white. (C) AIPDs for the cells with the above-mentioned treatments. Scale bar: 20 µm. A 3-Lorentzian deconvolution of the AIPD function allows measuring the relative contributions of the 3 populations: the first population represents autophagosomes (AP, pH = 6.0, orange line), whether the second and the third represent 2 different states of autolysosomes (AL), an early one (ALE, pH = 5.6, green line), and a mature, more acidic state (ALM, pH = 5.2, blue line). The black line represents the sum of the 3 populations.](/cms/asset/983fd4c6-ce27-4d83-b060-14204fadae89/kaup_a_1084455_f0002_oc.gif)
Effects of autophagy inductors and inhibitors on autophagic intermediates
Blockade of the nutrient sensitive cascade triggered by MTOR (mechanistic target of rapamycin [serine/threonine kinase]), through either growth factor withdrawal or the antibiotic inhibitor rapamycin, represents a broadly used method to induce autophagy in mammalian cells.Citation27,28 Serum deprivation of HEK-293T cells for 16 h triggers autophagy, as biochemically revealed by a) the appearance of the PE-conjugated form of the fluorescent probe as a faster migrating band (LC3-II) in anti-MAP1LC3B western blots (Fig. S1A); b) almost complete disappearance of the endogenous unlipidated (LC3-I) MAP1LC3B in the same assay, suggestive of active autophagic degradation (Fig. S1B). These events are paralleled by a reduced activity of the nutrient-sensitive MTOR cascade, monitored by the (Ser235/236) phosphorylation of the downstream effector RPS6 (Fig. S1C). Of note, treatment of nutrient-replete HEK-293T cells with the lysosome inhibitor chloroquine allows the accumulation of the endogenous LC3B-II while the unlipidated form completely disappears. This pattern is consistent with chloroquine acting at multiple levels in the autophagic cascade: by blocking cargo degradation in lysosomes in the final step, and by accelerating the upstream events leading to autophagosome formation, through an inhibitory action on MTOR (Fig. S1C).Citation29
We investigated the effects of starvation on HEK-293T cells by retrieving R-map for serum-deprived cells (STARV, overlap in , R-map in ), and the relative AIPD (). Upon starvation, AIPD area increases from n = 10 (control distribution of the untreated sample CTRL, ) to n = 32. A shift of the peak to higher pH and a reduction of the distribution asymmetry were also observed. As expected, rapamycin treatment (RAP, overlap in , R-map in and AIPD in ) also leads to a similar increase of AIPD area (n = 36), only more pronounced at lower pH, with a concomitant increase of the distribution asymmetry. Treatment with both inducers (STARV+RAP, overlap in , R-map in and AIPD in ) leads to a shape more closely resembling that of untreated cells (n = 9).
Among autophagic inhibitors, we tested chloroquine (CHL) and 3-methyladenine (3-MA). Chloroquine is a lysosomotropic agent that prevents endosomal acidification. It accumulates inside the acidic parts of the cell, including endosomes and lysosomes, preventing both fusion of autophagosomes with lysosomes and lysosomal protein degradation.Citation8 Upon chloroquine treatment (CHL, overlap in , R-map in and AIPD in ), AIPD area increases up to n = 20 with a shift to higher pH values, and no obvious changes in the overall symmetry are observed. Chloroquine raised the number of intermediates by acting mainly on the ALM compartment (by a factor 9, ). In order to monitor an inhibitory agent that does not raise the pH of organelles, we used 3MA, which blocks autophagosome formation via the inhibition of type III phosphatidylinositol 3-kinases. 3MA (overlap in , R-map in and AIPD in ) had a weak effect on the overall number of intermediates, but a decrease of the right shoulder of AIPD and a concomitant increase of the left shoulders was observed.
pH and stoichiometric contribution of autophagic intermediates
To determine the relative stoichiometric contribution of the different intermediates, we further processed AIPD by Lorentzian deconvolution, which is more practical with respect to Gaussian deconvolution since the overlap area among the curves is greatly reduced. The same analytical method was used in deconvolution of histograms of redox images to separate the contributions of oxidized and reduced spots.Citation30,31
The 3 Lorentzian deconvolutionCitation30 turned out to be the most stable method, yielding 3 principal populations: the first peaked at pH = 6.0 (orange line), the second at pH = 5.6 (green line), the third at pH = 5.3 (blue line). The black line is therefore the sum of 3 contributions.
In the pH of the deconvolved populations are reported. It is conceivable that the first population represents autophagosomes (AP, pH = 6.0, orange), whether the second and the third represent, respectively, an early state (ALE, pH = 5.6, green), and a mature, more acidic state (ALM, pH = 5.2, blue) of autolysosomes. The number (and percentage) of organelles of each family was retrieved by the area fraction under the corresponding Lorentzian ().
Figure 3. pH, Absolute number and percentage of autophagic intermediates. (A) pH, weighted by the AIPD distributions, for autophagosomes (AP, pH = 6.0, orange line), early autolysosomes (ALE, pH = 5.6, green line), and mature autolysosomes (ALM, pH = 5.2, blue line). (B) Stacked column histogram of the number of puncta per cell for each population, calculated from AIPD area fractions. (C) Percentages of the number of puncta per cell for each population. Asterisks denote statistical significance of differences among percentage distributions (χ2-test). * P < 0.02; ***P < 0.002; ** P < 0.0001. Error bars represent the error associated with the fitting algorithm. (D) Overlap and (E) AIPD of untreated Caco cells. Scale bar: 10 µm. (F) Overlap and (G) AIPD of rapamycin-treated cells. (H) Stacked column histogram of the number of puncta per cell for each population, calculated from the AIPD.
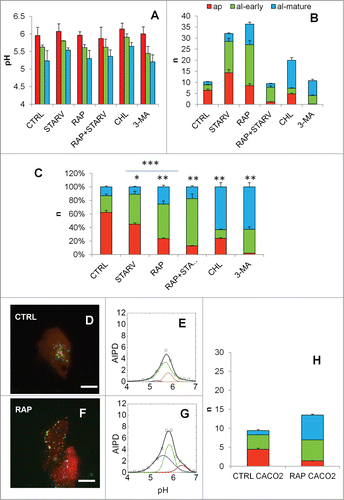
Absolute quantifications of autophagy intermediates revealed marked differences in cell response to different inducers. We observed a net increase of APs (by a factor 2.2) and ALs (by a factor 5.6 for ALE and 3 for ALM) in response to serum deprivation, whereas rapamycin increased ALs by a factor 7 in both of its forms but had a little effect on APs (20% increase). Combination of both stimuli led to normalization of total intermediates, with an 80% decrease of APs and a raise of ALE by a factor 2.6. Chloroquine raised the number of intermediates by acting mainly on the ALM compartment (by a factor 9, ). Upon 3-MA treatment, a severe reduction of AP, and a simultaneous raise in ALE and ALM were detected.
These changes were mirrored by important differences in percentage distributions among the different samples () with the fraction of APs dropping from 62% in control cells to 44%, 24%, and 13%, respectively, in cells exposed to starvation, rapamycin, or their combination. AP population decrease to 2% in 3-MA treated cells.
Use of mRFP-GFP-LC3 on the Caco2 cell line
HEK-293T cells usually demonstrate a significant induction of autophagy as determined by conversion of MAP1LC3B-I to MAP1LC3B-II detected by western blotting. Caco-2 cells, a heterogeneous human colorectal adenocarcinoma cell line, were transfected with mRFP-EGFP-LC3B. AIPD of untreated cells (overlap in , and AIPD in ) was very similar to that observed in HEK-293T cells, characterized by a broad and continuous distribution in the range 4.5–6.5. Rapamycin treatment (overlap in , and AIPD in ) led to an increase of AIPD area (n = 14) that was overall smaller with respect to HEK-293T cells, but more pronounced at lower pH, with a concomitant increase of the distribution asymmetry. In , absolute quantifications of autophagy intermediates are reported for Caco2 cells. The decrease of AP population was abrupt, passing from 4.5 to 1.4 puncta per cell, while the ALE population increased from 3.8 to 5.5 and ALM from 1.1 to 6.6 puncta per cell.
Figure 4. pH and absolute number of autophagic intermediates. (A) R-map ("cool" lookup table [imageJ], values from R0-black to Rf-white) of LysoSensor-marked cells of the control, serum-starved, rapamycin-treated, rapamycin+starvation-treated, chloroquine-treated and 3-MA-treated samples. (B) LPD of ctrl untreated cells serum-starved, rapamycin-treated, rapamycin+starvation-treated, chloroquine-treated, and 3-MA-treated samples. Scale bar: 20 µm. A 2-Lorentzian deconvolution of the AIPD function can be recovered to distinguish between the number of acidic organelles as lysosomes (L, pink line) and autophagic intermediates (AI = AP+AL, purple line). The black line is the sum of the 2 Lorentzians. (C) pH, weighted by the LPD distributions, for lysosomes (L, pH = 5.0, pink line), and autophagic intermediates (AI, pH = 5.4, purple line). (D) Stacked column histogram of the number of puncta per cell for L (pink) and AI (purple), calculated from LPD area fractions. Error bars represent the error associated to the fitting algorithm.
![Figure 4. pH and absolute number of autophagic intermediates. (A) R-map ("cool" lookup table [imageJ], values from R0-black to Rf-white) of LysoSensor-marked cells of the control, serum-starved, rapamycin-treated, rapamycin+starvation-treated, chloroquine-treated and 3-MA-treated samples. (B) LPD of ctrl untreated cells serum-starved, rapamycin-treated, rapamycin+starvation-treated, chloroquine-treated, and 3-MA-treated samples. Scale bar: 20 µm. A 2-Lorentzian deconvolution of the AIPD function can be recovered to distinguish between the number of acidic organelles as lysosomes (L, pink line) and autophagic intermediates (AI = AP+AL, purple line). The black line is the sum of the 2 Lorentzians. (C) pH, weighted by the LPD distributions, for lysosomes (L, pH = 5.0, pink line), and autophagic intermediates (AI, pH = 5.4, purple line). (D) Stacked column histogram of the number of puncta per cell for L (pink) and AI (purple), calculated from LPD area fractions. Error bars represent the error associated to the fitting algorithm.](/cms/asset/89b50f5d-71b1-43e3-9de6-855f5423ea19/kaup_a_1084455_f0004_oc.gif)
Effects of autophagy inducers and late suppressors on lysosomes
In order to test the reliability of the probe, and to get a contemporary assessment of lysosome stoichiometric weight, we analyzed cells marked with LysoSensor Yellow/Blue (pK = 4.3), which stains all the acidic organelles, in the same experimental conditions. From the ratio map of LysoSensor-stained cells () it is possible to extract the LysoSensor Yellow/Blue pH distribution (LPD), which gives number (nlys) and pH of all the acidic organelles. The control LPD, since it monitors also not-yet-fused lysosomes (), has a range of variation shifted toward lower pH values (∼0.8) with respect to AIPD. Upon starvation, distribution increases especially in its right shoulder, from nlys = 30 to 35, indicating a higher number of autophagic intermediates in the pH range 5.5–6.5, with respect to control distribution. Rapamycin treatment led to a doubling of LPD area (nlys = 65). Treatment with both inducers again led to a shape more similar to that of the control, but with a wider area (nlys = 43). As a main effect, chloroquine caused a marked reduction of stained acidic organelles (nlys = 20). Treatment with 3-MA (nlys = 30) led to a normalization to control values.
By a 2-Lorentzian deconvolution of LPD it is possible to recover and distinguish between the number of acidic organelles as lysosomes (L) and autophagic intermediates (AI = AP+AL). In this case the 3-Lorentzian deconvolution employed above does not give stable results, because the sensitivity of the probe is very low in the pH range higher than pK+1 = 5.3. The first Lorentzian (representing the contribution of Ls) has peaked at pHL = 5.0, whether the second (representing the AI contribution) at pHAI = 5.4. In the pH of the deconvolved populations are reported for each case. Significant pH shifts of AI population in the serum-starved and rapamycin-treated cells were in good agreement with that observed in AIPD (). Also in this case the number of organelles of each family was retrieved by the area fraction under each Lorentzian. In , the number of L and AI are reported. With respect to the control of untreated cells, there was a net increase upon starvation of AI (5.5 times) at the expenses of Ls, which dropped off abruptly (60% decrease), likely because of their enhanced fusion with newly formed APs. With rapamycin treatment an analog increase in both AI (4.7 times), and lysosomes (1.8 times) was detected. When cells were treated with both inducers, the increases in AI (16%) and L (52%) were less pronounced. Conversely, when cells were treated with chloroquine, a concomitant decrease of L (70%) and AI (46%) was detected. This can reflect a neutralization of acidic compartments, more than a change in their stoichiometry, and was consistent with biochemical evidence of lipidated MAP1LC3B (MAP1LC3BII) accumulation in this sample (Fig. S1). 3-MA treated cells showed a relative percentage of L and AI similar to that of chloroquine treated cells. In Fig. S2 a linear correlation between the number of AI retrieved by LysoSensor Yellow/Blue (nAI) and mRFP-GFP-LC3 (n), reveals a good agreement between the 2 methods. A systematic down-estimation of the number of autophagic intermediates by LysoSensor Yellow/Blue was caused by the worse sensitivity in the high (>5.3) pH region, and, to a lesser extent, by the intrinsic limit of the pH-based deconvolution method, which can include in the lysosomal fraction some autolysosomes (no more than 30%).
Distribution of CTSB content in AP, ALE, ALM
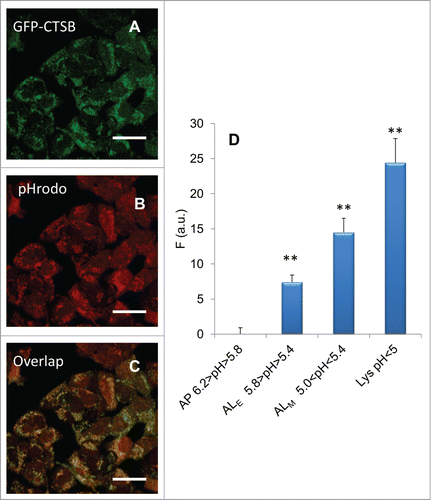
To establish if a functional difference exists among the 3 populations, we analyzed their CTSB (cathepsin B) content. CTSB is an enzymatic protein belonging to the peptidase (or protease) family. To this aim, we measured fluorescence by contextually labeling cells with EGFP-CTSB and pHrodo® Red AM. We could not use mRFP-GFP-LC3 for this peculiar characterization because this probe emits in the green thus masking EGFP-CTSB. By measuring the pH of the intermediates through pHrodo® Red AM we can discriminate, according to our classification, among lysosomes (5.0 > pH), ALM (5.0 < pH < 5.4), ALE (5.4 < pH < 5.8) and AP (5.8 < pH < 6.2). We can then measure the CTSB fluorescence intensity (proportional to CTSB concentration) from each region. In , the fluorescence image of GFP-CTSB construct for untreated cells is reported. In the pHrodo® image from the same sample is reported and in the overlap is reported. In the histogram in the concentrations of CTSB in the different compartments are reported, showing a continuous increase in CTSB, going from alkaline toward acidic compartments.
Figure 5. Differences in CTSB content in the different autophagic compartments. (A) Fluorescence image of EGFP-CTSB construct for untreated cells. (B) pHrodo® image from the same sample. (C) Overlap between EGFP-CTSB and pHrodo images. (D) Concentrations of CTSB in the different compartments. A continuous increase in CTSB going from alkaline toward acidic compartments is present. Error bars represent standard deviations of measured values (n = 100 cells). Asterisks denote statistical significance of differences among percentage distributions (χ2-test): **, P < 0.0001.
Discussion
In order to detect the different intermediates in the autophagy together with their concentrations, we developed a method to image, contextually, the number and the pH of autophagy intermediates using mRFP-EGFP-LC3B tandem employed in a ratiometric way, which allows the measurement of AIPD.
According to AIPD analysis, intermediates are characterized by a continuous pH distribution in the range 4.5–6.5. We have then identified a more complex set of states compared to the expected one, characterized by values clustered in 2 main populations (autophagosomes and autolysosomes). In our model, we dissected 3 major populations: AP, ALE and ALM. The pH distribution of each population is also a continuous rather than a discrete one: indeed, APs are characterized by a range of values going from 6.2 to 5.8, ALE are characterized by a range of values going from 5.8 to 5.4 and ALM are characterized by a range of values going from 5.4 to 5.0.
The reason for the pH heterogeneity of ALE and ALM populations is the acidification process triggered by the fusion of lysosomes with autophagosomes after the association of STX17 (syntaxin 17).Citation32 Indeed the pH of lysosomes is also heterogeneous and is not a prerequisite for fusion,Citation33 and the number of lysosomes fusing with an AP could vary, as shown by the continuous increase in the local concentration of CTSB, which is higher in ALM with respect to ALE and APs (). This observation confirms that the fusion with lysosome is a non-discrete process, and that a “2-state” based discrimination between APs and ALs could be an oversimplification. Moreover, the observation that CTSB content in the AP population is nearly zero, confirms that this organelle population has not encountered lysosomes yet. In this context, the reasons for the pH heterogeneity of the AP population (pH 5.8–6.2) should be independent from lysosomes: for example, differences in the organelle content and membrane protein composition, the balance of proton pumps and leaks,Citation34 differences in the organelles implicated in the origin of the phagophoreCitation35,36 may broaden the pH spectrum. Variations in the amplitude (variance) of the AP contribution on AIPD may detect these potential sources of heterogeneity.
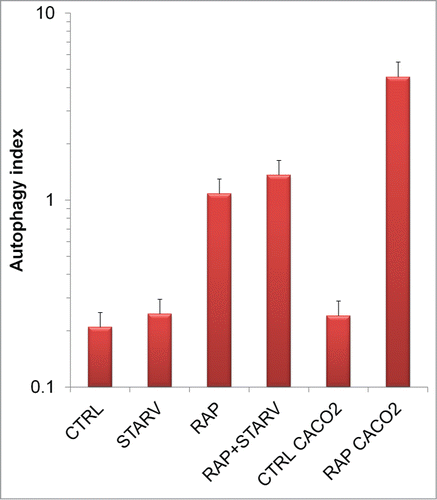
AIPD, in all cases examined, is sensible in shape and amplitude to alterations in the autophagy process and allows a quantitative estimation of the autophagic flux through the simultaneous measurements of the number of intermediates involved in the autophagic process, by taking the ratio Φ = ALM/AP.
The ratio [ALM]/[AP] is a reliable index for autophagic flux, because it corresponds to the reaction quotient, Q, which expresses the relative ratio of products to reactants at a given instant in the simple reaction model we adopted:[3]
[3]
Following serum deprivation, a prototypical trigger of autophagy, we observed a doubling in the autophagic flux from 0.2 (control value) to 0.25 (): indeed the doubling of AP is accompanied by an increase of AP-L fusion, as shown by the marked increase of AL (6 times) and the concomitant decrease of L(60%). In response to rapamycin, autophagic flux increase is even higher (ratio 1.1): the increase of AL in both of its forms by a factor 7 is this time accompanied by a decrease of AP. By LysoSensor Yellow/Blue an important increase of Ls is observed, which may be ascribed to augmented lysosome biogenesis: indeed inhibition of MTOR (pharmacological, or by starvation) activates TFEB (transcription factor EB), which promotes lysosome biogenesis.Citation37,38 This process can be an important factor to set a dose-response relationship and a temporal regulation of autophagy mechanisms. Under the combination of serum deprivation and rapamycin, the flux is even higher because, although the number of intermediates decreases, the ratio Φ is the highest with respect of all the observed cases. Of note, the synergistic effect of serum starvation and rapamycin—the latter a selective and potent MTOR complex 1 inhibitor—suggests that other pathways beside the MTOR cascade may contribute to autophagy triggering by serum deprivation. In Caco2 cells, where the nominal induction of autophagy seems less pronounced with respect to HEK-293T cells, rapamycin enhances the number of total organelles by 40% instead of the 200% observed for HEK-293T cells. Nonetheless, the method retrieves a more pronounced autophagic flux (from 0.2 to 5), despite a less total number of autophagic intermediates. Unfortunately the ratio [ALM]/[AP] does not make it possible to quantify the autophagic flux in processes in which the inhibition of AP formation is involved, as in 3-MA treated cells, since an assessment of the number of phagophores is needed in this case (the step [PG]→[AP] cannot be taken in account). Nevertheless a correct estimation of the different autophagic intermediates is possible. On the contrary, since the method relies on pH, the approach here presented is not useful in determining stoichiometry in conjunction with agents that neutralize lysosomal pH, as chloroquine. Additionally, conditions that alter lysosomal acidification may adversely affect results obtained from this assay.Citation39
Figure 6. Autophagic flux quantifications. AIPD sensibility to alterations in the autophagy process allows a quantitative estimation by the ratio ALM/AP of the autophagic flux. Error bars represent the error propagated on the indirect estimation.
A very accurate estimation of both puncta per cell and APs/ALs population is therefore possible with the dual-emission ratiometric use of mRFP-EGFP-LC3B assay, since the simultaneous measurement of the pH rules out systematic errors due to arbitrary detection and excitation settings and/or subjectivity of the observer in selection. The principal advantages of this probe are 1) the single, nonphototoxic, excitation wavelength; 2) it is targeted to autophagy intermediates, so it does not require double staining with an organelle-specific marker; 3) it is genetically encoded and ratiometric in emission, allowing real-time imaging. The exchange of m-CherryCitation40 with mRFP may constitute an improvement to the probe and could improve photostability, although enhanced FRET efficiency could limit the dynamic range of the probe. Similarly, substitution of EGFP with GfpH,Citation41 while extending dynamic range and photostability of the probe (), would also entail UV excitation, with phototoxicity as a side effect. For this kind of investigation, UV irradiation should be avoided even more, since it is itself conducive to autophagy.Citation16
The method outlined has a wide applicability: mRFP-EGFP can be a valuable tool to measure ratiometrically acidic pH in a cell, since it can be easily targeted to other kind of organelles. Moreover AIPD is, independently of the specific probe used to determine pH, a very useful tool to quantify the different intermediates, and to characterize analytically the process through the determination of autophagic flux.
In conclusion, the quantitative assessment of autophagic flux by the analytic measurement of AIPD through the ratiometric employment of mRFP-EGFP-LC3B allows investigating with high accuracy molecular mechanisms as the basis of a broad spectrum of autophagy-related diseases.
Materials and Methods
Cells
HEK-293T cells (E1A-transformed human embryonic kidney cells carrying a temperature sensitive T antigen), were obtained from ATCC/LGC Standards (ATCC, CRL-3216), stored in liquid nitrogen and propagated for a limited number of passages to avoid selection of clonal variants. Cells were routinely maintained in Dulbecco's modified Eagle's medium containing 4.5g/l glucose, 2 mM glutamine, 1 mM sodium pyruvate, nonessential amino acids, penicillin-streptomycin and 10% v/v fetal calf serum (EUROBIO, CVFSVF00–01). Cultures were passaged every 4 or 5 d by trypsinization.
Plasmids
Plasmid ptfMAP1LC3B encoding the monomeric red fluorescent protein (mRFP) and the enhanced green fluorescent protein (EGFP) fused in tandem with rat MAP1LC3B (microtubule-associated protein 1 light chain 3 β), originally generated by Kimura S. et al.,Citation11 were obtained from the Addgene repository (plasmid 21074, deposited by Tamotsu Yoshimori) under a Material Transfer Agreement. Plasmid amplification and purification was performed by standard procedures using the Geneaid™ Maxi Plasmid Kit (Geneaid Biotech Ltd., PM-025). Plasmids encoding EGFP fused with rat MAP1LC3B and CTSB were a kind gift of Dr. Toren Finkel (NIH, Bethesda).
Cell transfection
HEK-293T cells were seeded in 6-well plates (Corning Life Science, 3335) at 3 × 105 cells/well in 1600 microliters of complete medium. After 24 h cell were transfected with 1 µg plasmid DNA/well using the EFFECTENE® transfection reagent (QIAGEN, 301425) according to the manufacturer's recommendations. Successful transfection was verified 24 h later under an inverted fluorescence microscope (NIKON ECLIPSE TE2000-U, Chiyoda, Tokyo, Japan).
Determination of organelle pH by LysoSensor™ and pHrodo® probes
Lysosome staining with the fluorescent acidotropic probe LysoSensor™ Yellow/Blue DND-160 and pHrodo red AM® (Life Technologies, L-7545 and P35372, respectively) was performed according to the manufacturer's recommendations. Briefly, HEK-293T cells were seeded in glass-bottom dishes, left to adhere, and exposed to autophagic stimuli for 16 h as described above. LysoSensor Yellow/Blue (stock 1 mM in DMSO, pKa 4.2) or pHrodo (pKa 6.5) was then added directly to the medium at the final concentration of 5 μM and incubated for 2 h at 37°C, 5% CO2. At the end of the incubation cells were directly transferred to the confocal facility without further manipulation. LysoSensor Yellow/Blue fluorescence was excited at 760 nm (2-photon) and collected in the range 400 to 490 nm and 510 to 600 nm (emission) for ratiometric calculation. pHrodo red AM was excited at 543 nm and emission fluorescence was collected in the range 620 to 700 nm. pH was determined based on a calibration curve prepared with the same probe dissolved in buffers of different pH.
Cell stimulation
Twenty-four h after transfection cells were trypsinized counted and reseeded at 105/dish in matrix-coated 3.5-mm glass-bottom dishes (Ibitreat, IbiDI, 81156). Cells were left to adhere and spread for 4.5 h, then the medium was replaced with serum-free Dulbecco's modified Eagle's medium, and 5% fetal calf serum, 200 nM rapamycin (LC Laboratorier, R-5000) or 100 µM chloroquine (Sigma, C6628) added alone or in combination where required. Cells were exposed to the drug for 16 h. 3 MA (Sigma, M9281) was added at 5 mM, 30 min before autophagy induction (i.e., by serum deprivation) and kept for the entire incubation time (16 h).
Western blotting
Cells processed for imaging studies were lysed directly in the culture dishes with a lysis buffer (150 mM NaCl, 50 mM Tris–HCl, pH 8, 2 mM EDTA) containing 1% Triton X-100 (Sigma, 93443), protease and phosphatase inhibitors (Sigma, respectively I3911, P5726). After 20 min of passive lysis on ice, unlysed cells/nuclei were removed by centrifugation (22000g × 10 min) and supernatant fractions assayed for protein content (DC Protein Assay; Bio-Rad, 5000111) mixed with 6x Laemmli buffer and subjected to SDS-PAGE, followed by electro-transfer onto nitrocellulose. Membranes were blocked at room temperature in 5% skim milk in TBS-T (Sigma, 91414) and incubated overnight with rabbit anti-MAP1LC3B or rabbit anti p-RPS6 (Ser235/236) antisera (Cell Signaling Technology, respectively 3868 and 2211). Immunocomplexes were visualized by incubation with a HRP-conjugated goat anti-rabbit secondary reagent (Bio-Rad, 1706515) followed by ECL detection (GE Healthcare, RPN2232).
Confocal microscopy
Images were obtained by using an inverted confocal microscope (DMIRE2, Leica Microsystems, Germany) fitted with a 63X oil immersion objective (NA 1.4) and LCS 2.61 acquisition Software (Leica Microsystems, Wetzlar, Germany). Internal photon multiplier tubes collected 8 bit, unsigned images at a 400 Hz scan speed. mRFP-GFP-LC3 was excited by an argon-ion laser line (excitation wavelength: 488 nm; emission ranges: 500 to 540: 640 to 740 nm). Photomultiplier tube gain values were set to 600 V and 450 V, and kept fixed during the experiment. Pinhole was set to 1 A.U. Z-stacks were acquired every 500 nm, to ensure a count of the puncta over the whole cell volume. Analysis of images acquired was performed with ImageJ 1.41 (NIH).
Construction of R-images
The R index was obtained by calculating the ratio between fluorescence emissions in the 500 to 540 and 640 to 740 nm ranges, upon sample excitation at 488 nm. By mapping R over the entire microscope scanning field, R-images can be created with the homemade downloadable software Redox Maps Generator 1.0.Citation42
Determination of number and pH of intermediates
Green and red images were overlaid, and maxima of red and green channels, representing autophagy intermediates ("puncta"), were retrieved by the FIND MAXIMA plugin (ImageJ). Regions of interests, including whole organelles, were manually drawn in correspondence of the maxima, and fluorescence intensity values measuredCitation43,44 directly on the R-image through the SYNC WINDOWS plug-in (ImageJ). pH values are then obtained by Equation Equation2[2]
[2] . Puncta without detectable EGFP fluorescence were minimized to less than 5% of the total number by setting adequate values for photomultipliers. At least 100 cells per sample were analyzed to build the histogram (∼1000–4000 puncta).
Statistical analyses
Fluorescence intensities and intensity ratios data were presented as Mean±SD and differences were assessed by using χ2-test. values of p < 0.05 were considered significant.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary Material
Supplemental data for this article can be accessed on the publisher's website.
1084455_supplemental_files.zip
Download Zip (422.5 KB)Acknowledgments
The confocal analysis has been performed at Labcemi, UCSC, Rome. The authors acknowledge Sig. Mario Amici for his excellent technical support, Dr. Matteo Minasi for critically reading the manuscript, and the COST Action CM1201 “biomimetic radical chemistry” for useful discussions.
Funding
This work was supported by Fondi di Ateneo, UCSC Rome, Italy (Linea D1 to GM, GP and MDS, and linea 3.2–2013 to GP).
References
- Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell 2008; 132:27-42; http://dx.doi.org/10.1016/j.cell.2007.12.018
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature 2008; 451:1069-75; http://dx.doi.org/10.1038/nature06639
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 2006; 443:780-6; http://dx.doi.org/10.1038/nature05291
- Tesori V, Piscaglia AC, Samengo D, Barba M, Bernardini C, Scatena R, Pontoglio A, Castellini L, Spelbrink JN, Maulucci G, et al. The multikinase inhibitor Sorafenib enhances glycolysis and synergizes with glycolysis blockade for cancer cell killing. Sci Rep 2015; 5:9149; http://dx.doi.org/10.1038/srep09149
- Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J Cell Sci 2004; 117:2805-12; http://dx.doi.org/10.1242/jcs.01131
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19:5720-8; http://dx.doi.org/10.1093/emboj/19.21.5720
- Mizushima N. Methods for monitoring autophagy. Int J Biochem Cell Biol 2004; 36:2491-502; http://dx.doi.org/10.1016/j.biocel.2004.02.005
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140:313-26; http://dx.doi.org/10.1016/j.cell.2010.01.028
- Bampton ETW, Goemans CG, Niranjan D, Mizushima N, Tolkovsky AM. The dynamics of autophagy visualized in live cells: from autophagosome formation to fusion with endo/lysosomes. Autophagy 2005; 1:23-36; http://dx.doi.org/10.4161/auto.1.1.1495
- Katayama H, Yamamoto A, Mizushima N, Yoshimori T, Miyawaki A. GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct 2008; 33:1-12; http://dx.doi.org/10.1247/csf.07011
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3:452-60; http://dx.doi.org/10.4161/auto.4451
- Zhou C, Zhong W, Zhou J, Sheng F, Fang Z, Wei Y, Chen Y, Deng X, Xia B, Lin J. Monitoring autophagic flux by an improved tandem fluorescent-tagged LC3 (mTagRFP-mWasabi-LC3) reveals that high-dose rapamycin impairs autophagic flux in cancer cells. Autophagy 2012; 8:1215-26; http://dx.doi.org/10.4161/auto.20284
- Rosado CJ, Mijaljica D, Hatzinisiriou I, Prescott M, Devenish RJ. Rosella: a fluorescent pH-biosensor for reporting vacuolar turnover of cytosol and organelles in yeast. Autophagy 2008; 4:205-13; http://dx.doi.org/10.4161/auto.5331
- Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat Biotechnol 2002; 20:83-7; http://dx.doi.org/10.1038/nbt0102-83
- Katayama H, Kogure T, Mizushima N, Yoshimori T, Miyawaki A. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem Biol 2011; 18:1042-52; http://dx.doi.org/10.1016/j.chembiol.2011.05.013
- Strozyk E, Kulms D. The role of AKT/mTOR pathway in stress response to UV-irradiation: implication in skin carcinogenesis by regulation of apoptosis, autophagy and senescence. Int J Mol Sci 2013; 14:15260-85; http://dx.doi.org/10.3390/ijms140815260
- Drake KR, Kang M, Kenworthy AK. Nucleocytoplasmic distribution and dynamics of the autophagosome marker EGFP-LC3. PLoS One 2010; 5:e9806; http://dx.doi.org/10.1371/journal.pone.0009806
- Buktenica S, Frankfater A. Effect of subunit size and conformation on the rate of lysosomal degradation of extracellular proteins in cultured mouse peritoneal macrophages. J Biol Chem 1987; 262:11611-5..
- Kneen M, Farinas J, Li Y, Verkman AS. Green fluorescent protein as a noninvasive intracellular pH indicator. Biophys J 1998; 74:1591-9; http://dx.doi.org/10.1016/S0006-3495(98)77870-1
- Sankaranarayanan S, De Angelis D, Rothman JE, Ryan TA. The use of pHluorins for optical measurements of presynaptic activity. Biophys J 2000; 79:2199-208; http://dx.doi.org/10.1016/S0006-3495(00)76468-X
- Bizzarri R, Nifosì R, Abbruzzetti S, Rocchia W, Guidi S, Arosio D, Garau G, Campanini B, Grandi E, Ricci F, et al. Green fluorescent protein ground states: the influence of a second protonation site near the chromophore. Biochemistry 2007; 46:5494-504; http://dx.doi.org/10.1021/bi602646r
- Lam AJ, St-Pierre F, Gong Y, Marshall JD, Cranfill PJ, Baird MA, McKeown MR, Wiedenmann J, Davidson MW, Schnitzer MJ, et al. Improving FRET dynamic range with bright green and red fluorescent proteins. Nat Methods 2012; 9:1005-12; http://dx.doi.org/10.1038/nmeth.2171
- Peter M, Ameer-Beg SM, Hughes MKY, Keppler MD, Prag S, Marsh M, Vojnovic B, Ng T. Multiphoton-FLIM quantification of the EGFP-mRFP1 FRET pair for localization of membrane receptor-kinase interactions. Biophys J 2005; 88:1224-37; http://dx.doi.org/10.1529/biophysj.104.050153
- Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng Des Sel 1999; 12:1035-40; http://dx.doi.org/10.1093/protein/12.12.1035
- Subach OM, Patterson GH, Ting L-M, Wang Y, Condeelis JS, Verkhusha V V. A photoswitchable orange-to-far-red fluorescent protein, PSmOrange. Nat Methods 2011; 8:771-7; http://dx.doi.org/10.1038/nmeth.1664
- Bizzarri R, Serresi M, Luin S, Beltram F. Green fluorescent protein based pH indicators for in vivo use: a review. Anal Bioanal Chem 2009; 393:1107-22; http://dx.doi.org/10.1007/s00216-008-2515-9
- Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 2014; 15:155-62; http://dx.doi.org/10.1038/nrm3757
- Panieri E, Toietta G, Mele M, Labate V, Ranieri SC, Fusco S, Tesori V, Antonini A, Maulucci G, De Spirito M, et al. Nutrient withdrawal rescues growth factor-deprived cells from mTOR-dependent damage. Aging 2010; 2:487-503.
- Li M, Khambu B, Zhang H, Kang J-H, Chen X, Chen D, Vollmer L, Liu P-Q, Vogt A, Yin X-M. Suppression of lysosome function induces autophagy via a feedback down-regulation of MTOR complex 1 (MTORC1) activity. J Biol Chem 2013; 288:35769-80; http://dx.doi.org/10.1074/jbc.M113.511212
- Maulucci G, Pani G, Labate V, Mele M, Panieri E, Papi M, Arcovito G, Galeotti T, De Spirito M. Investigation of the spatial distribution of glutathione redox-balance in live cells by using Fluorescence Ratio Imaging Microscopy. Biosens Bioelectron 2009; 25:682-7; http://dx.doi.org/10.1016/j.bios.2009.07.038
- Maulucci G, Pani G, Fusco S, Papi M, Arcovito G, Galeotti T, Fraziano M, De Spirito M. Compartmentalization of the redox environment in PC-12 neuronal cells. Eur Biophys J 2010; 39:993-9; http://dx.doi.org/10.1007/s00249-009-0470-9
- Bernard A, Popelka H, Klionsky DJ. A unique hairpin-type tail-anchored SNARE starts to solve a long-time puzzle. Autophagy 2013; 9:813-4; http://dx.doi.org/10.4161/auto.24359
- Mauvezin C, Nagy P, Juhász G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun 2015; 6:7007; http://dx.doi.org/10.1038/ncomms8007
- Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: measurement, determinants, and regulation. Physiology 2004; 19:207-15; http://dx.doi.org/10.1152/physiol.00005.2004
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol 2010; 12:831-5; http://dx.doi.org/10.1038/ncb0910-831
- Mari M, Tooze SA, Reggiori F. The puzzling origin of the autophagosomal membrane. F1000 Biol Rep 2011; 3:25.
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 2012; 31:1095-108; http://dx.doi.org/10.1038/emboj.2012.32
- Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol 2013; 14:283-96; http://dx.doi.org/10.1038/nrm3565
- Yamada E, Singh R. Mapping autophagy on to your metabolic radar. Diabetes 2012; 61:272-80; http://dx.doi.org/10.2337/db11-1199
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp red fluorescent protein. Nat Biotechnol 2004; 22:1567-72; http://dx.doi.org/10.1038/nbt1037
- Awaji T, Hirasawa A, Shirakawa H, Tsujimoto G, Miyazaki S. Novel green fluorescent protein-based ratiometric indicators for monitoring pH in defined intracellular microdomains. Biochem Biophys Res Commun 2001; 289:457-62; http://dx.doi.org/10.1006/bbrc.2001.6004
- Maulucci G, Labate V, Mele M, Panieri E, Arcovito G, Galeotti T, Østergaard H, Winther JR, De Spirito M, Pani G. High-resolution imaging of redox signaling in live cells through an oxidation-sensitive yellow fluorescent protein. Sci Signal 2008; 1:pl3.
- Angelucci C, Maulucci G, Lama G, Proietti G, Colabianchi A, Papi M, Maiorana A, De Spirito M, Micera A, Balzamino OB, et al. Epithelial-stromal interactions in human breast cancer: effects on adhesion, plasma membrane fluidity and migration speed and directness. PLoS One 2012; 7:e50804; http://dx.doi.org/10.1371/journal.pone.0050804
- Maulucci G, Troiani D, Eramo SLM, Paciello F, Podda MV, Paludetti G, Papi M, Maiorana A, Palmieri V, De Spirito M, et al. Time evolution of noise induced oxidation in outer hair cells: Role of NAD(P)H and plasma membrane fluidity. Biochim Biophys Acta 2014; 1840(7):2192-202.
- Miesenböck G, De Angelis DA, Rothman JE. Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 1998; 394:192-5; http://dx.doi.org/10.1038/28190
- Hanson GT, McAnaney TB, Park ES, Rendell MEP, Yarbrough DK, Chu S, Xi L, Boxer SG, Montrose MH, Remington SJ. Green fluorescent protein variants as ratiometric dual emission pH sensors. One. Structural characterization and preliminary application. Biochemistry 2002; 41:15477-88; http://dx.doi.org/10.1021/bi026609p
- Bizzarri R, Arcangeli C, Arosio D, Ricci F, Faraci P, Cardarelli F, Beltram F. Development of a novel GFP-based ratiometric excitation and emission pH indicator for intracellular studies. Biophys J 2006; 90:3300-14; http://dx.doi.org/10.1529/biophysj.105.074708
