ABSTRACT
Different rabies virus (RABV) strains have their own biological characteristics, but little is known about their respective impact on autophagy. Therefore, we evaluated whether attenuated RABV HEP-Flury and wild-type RABV GD-SH-01 strains triggered autophagy. We found that GD-SH-01 infection significantly increased the number of autophagy-like vesicles, the accumulation of enhanced green fluorescent protein (EGFP)-LC3 fluorescence puncta and the conversion of LC3-I to LC3-II, while HEP-Flury was not able to induce this phenomenon. When evaluating autophagic flux, we found that GD-SH-01 infection triggers a complete autophagic response in the human neuroblastoma cell line (SK), while autophagosome fusion with lysosomes was inhibited in a mouse neuroblastoma cell line (NA). In these cells, GD-SH-01 led to apoptosis and mitochondrial dysfunction while triggering autophagy, and apoptosis could be decreased by enhancing autophagy. To further identify the virus constituent causing autophagy, 5 chimeric recombinant viruses carrying single genes of HEP-Flury instead of those of GD-SH-01 were rescued. While the HEP-Flury virus carrying the wild-type matrix protein (M) gene of RABV triggered LC3-I to LC3-II conversion in SK and NA cells, replacement of genes of nucleoprotein (N), phosphoprotein (P) and glycoprotein (G) produced only minor autophagy. But no one single structural protein of GD-SH-01 induced autophagy. Moreover, the AMPK signaling pathway was activated by GD-SH-01 in SK. Therefore, our data provide strong evidence that autophagy is induced by GD-SH-01 and can decrease apoptosis in vitro. Furthermore, the M gene of GD-SH-01 may cooperatively induce autophagy.
Introduction
The rabies virus (RABV) belongs to the genus Lyssavirus of the Rhabdoviridae family. It is a virus that invades the central nervous system and causes encephalitis, with fatal consequences, in humans and other warm-blooded vertebrates.Citation1 The genome of the RABV is a single negative-stranded RNA of approximately 12 kb and encodes 5 structural proteins in the order of 3′-leader, the nucleoprotein (N), the phosphoprotein (P), the matrix protein (M), the glycoprotein (G), the RNA-dependent RNA polymerase (L), trailer-5′.Citation2 Although rabies is one of the oldest known infectious diseases, it still poses a major challenge and cannot be cured after symptoms have developed. Human infection with the classical rabies virus has an extremely poor prognosis with almost 100% mortality.Citation3 Rabies can be prevented by vaccination and increasing research is focused on the development of new vaccines for rabies.Citation4-6 GD-SH-01 is a wild-type RABV strain, which was isolated from the brain tissue of a rabid pig in our laboratory;Citation7,8 HEP-Flury is an attenuated rabies virus strain that is a high-egg-passage strain.Citation9 With regard to the 5 viral proteins, the matrix protein plays a particularly indispensable role in the process of rabies infection. It has been shown that the M protein is primarily responsible for the assembly and budding of the rabies virus and the G protein contributes to these processes.Citation10 While the M protein also regulates viral RNA synthesis, this function is temporally and spatially separated from the previously mentioned one.Citation11 It has also been reported that the M protein activates host cell caspases and induces apoptosis.Citation12,13 Despite these findings regarding the pathogenicity of the RABV, the relationship between a viral infection and autophagy remains as unexplored as the potential role of the M gene regarding autophagy in the neurocyte, the preferential target of the RABV.
In eukaryotic cells, autophagy is a highly conserved process that can break down long-lived cytoplasmic proteins and damaged organelles by means of exposing them to various hydrolases present within lysosomes to maintain cellular homeostasis.Citation14,15 Additionally, autophagy can function in innate immunity to prevent infection from intracellular microbial pathogens. Autophagy can be triggered by a diversity of factors, including nutrient depletion, endoplasmic reticulum stress, oxidative stress, mitochondrial damage and microbial infection.Citation15 MAP1LC3/LC3 (microtubule-associated protein 1 light chain 3), is the most widely used molecular marker for estimating autophagy.Citation16-18 While the ubiquitin-like proteins Atg (autophagy-related) 12 and Atg8 are part of the conjugation systems in autophagy in yeast,Citation19 LC3-I to LC3-II conversion and the Atg12–Atg5 complex carry out a similar function in higher eukaryotes. Some viruses exploit mechanisms to escape autophagy of host cells,Citation20-22 or may even utilize autophagy to benefit their replication.Citation23-27 Several signaling pathways can be activated to regulate autophagy; one of them is the MTOR (mechanistic target of rapamycin [serine/threonine kinase]) pathway. AMP-activated protein kinase (AMPK) activates autophagy mainly through the inhibition of MTOR or by directly inhibiting the phosphorylation of its downstream target, RPS6KB (ribosomal protein S6 kinase).Citation28,29 Although it has been demonstrated previously that RABV may induce apoptosis,Citation12,13 little is known concerning RABVinduced autophagy and pathways relating it to apoptosis. Autophagy can inhibit apoptosis by maintaining cellular homeostasis, but autophagy and apoptosis may also cooperatively induce cell death.Citation30 In this study, we evaluated the relationship between those 2 machineries by enhancing autophagy with rapamycin, then detecting the change in the apoptosis rate.
To our knowledge, this is the first study providing evidence that autophagy is induced by RABV in the human neuroblastoma cell line (SK) and the mouse neuroblastoma cell line (NA); we conducted research in order to identify the autophagy signaling pathway that is activated by RABV. We conclude from our results that the M gene of GD-SH-01 plays a potential role in promoting RABV-induced autophagy and that autophagy caused by GD-SH-01 may serve as a protective mechanism in resistance to apoptosis.
Results
RABV infection triggers autophagy in NA and SK cells
To determine if RABV can induce autophagy, SK and NA cells were infected with virulent GD-SH-01 and attenuated HEP-Flury RABV. The autophagosome was observed under transmission electron microscopy (TEM) and the conversion from LC3-I to LC3-II was detected in western blot assays, while we observed EGFP-LC3 dot formation with a confocal microscope.
Massive double-membrane vesicles and single-membrane vesicles were observed in GD-SH-01-infected SK and NA cells in TEM assays. Many double-membrane vesicles containing organelles and debris were seen, but since the majority of the organelles had been degraded in autolysosomes, their morphology did not allow their identification. Single-membrane vesicles containing organelles were found in a portion of cells, and mitochondria were enclosed within an autophagosome (). The content of other single-membrane vesicles was considerably degraded and it was not possible to determine their origin (). Autophagosome-like vesicles were rarely observed in mock () and HEP-Flury-infected cells ().
Figure 1. GD-SH-01 infection increases the formation of autophagosome-like vesicles. (A) NA (i-iii) and SK (iv-vi) cells were mock-infected (iii and vi) or infected with HEP-Flury (ii and v) or GD-SH-01 (i and iv) at an MOI of 1 for 48 h and observed under an electron microscope. White arrows indicate single- and double-membrane vesicles of autophagosomes (i and iv). (B) Quantification of the number of autophagosome-like vesicles per cell in NA and SK cells. The average number of the vesicles in each cell was obtained from at least 15 cells undergoing each treatment. Mean ± SD of 3 independent experiments. Two-way ANOVA: ***, P < 0.001; #, P > 0.05.
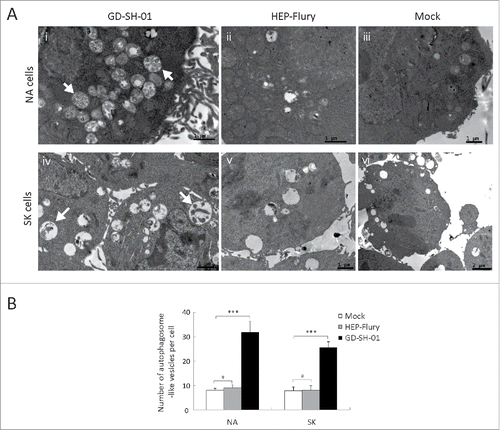
To further analyze if our observations were indeed provoked by an increase in autophagy, SK and NA cells were transfected with EGFP-LC3 plasmids, which can form detectable dots upon proteolytic processing and lipidation during autophagy.Citation18 EGFP-LC3 dots were visible in GD-SH-01-infected cells and after rapamycin treatment ( and ). This result indicates that GD-SH-01 infection can induce the formation of autophagosomes in SK and NA cells.
Figure 2. GD-SH-01 infection induces redistribution of the autophagy marker LC3 in SK cells. (A) SK cells treated with rapamycin (i) or infected with GD-SH-01 (ii), HEP-Flury (iii), mock (iv). Cells transfected with EGFP-LC3B plasmids are shown in green (i-iv); scale bar: 20 µm. The average number of LC3 puncta in each cell was determined from at least 50 cells in each group. Mean ± SD of 3 independent experiments. Two-way ANOVA: ***, P < 0.001 (B). Expression levels of LC3, BECN1 and ACTB at the end of the infection experiment in which SK cells were used as negative controls, infected with HEP-Flury or GD-SH-01. Mean ± SD of 3 independent experiments. Two-way ANOVA: *, P < 0.05; ***, P < 0.001. (Grouping of images of western blottting from different parts of the same gel.)
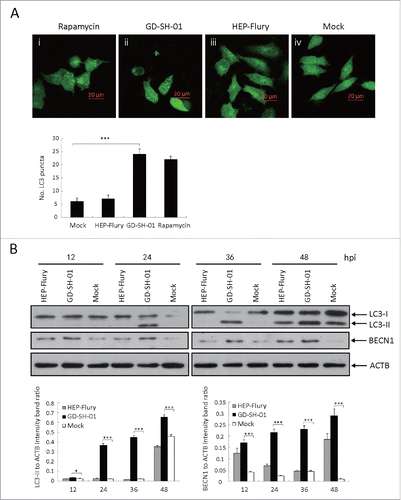
Figure 3. GD-SH-01 infection induces the redistribution of the autophagy marker LC3 in NA cells. (A) NA cells treated with rapamycin (i) or infected with GD-SH-01 (ii), HEP-Flury (iii), mock (iv). Cells transfected with EGFP-LC3B plasmids are shown in green (i-iv); scale bar: 20 µm. The average number of LC3 puncta in each cell was determined from at least 50 cells in each group. Mean ± SD of 3 independent experiments. Two-way ANOVA: ***, P < 0.001 (B). Expression levels of LC3, BECN1 and ACTB at the end of the infection experiment in which SK cells were used as negative controls, infected with HEP-Flury or GD-SH-01. Mean ± SD of 3 independent experiments. Two-way ANOVA: *, P < 0.05; ***, P < 0.001; #, P > 0.05. (Grouping of images of western blotting from different parts of the same gel.)
To further investigate whether autophagy is caused by viral infection, expression levels of proteins involved in autophagy pathways. LC3B and BECN1, were examined with immunoblots; ACTB served as a control protein. LC3-II specifically binds to the autophagosome membrane and is thus used as a marker for autophagy.Citation18 The LC3-II to ACTB ratio of GD-SH-01-infected cells was significantly increased when compared with HEP-Flury-infected cells or negative controls, particularly at later stages of the infection. BECN1, an early indicator for the formation of the autophagosome, was also upregulated in GD-SH-01-infected cells ( and ). Our data demonstrate that an RABV infection can activate the autophagy pathway and can induce the formation of autophagosomes.
Wild-type RABV strain infection is required for autophagosome accumulation
To further analyze whether all strains of RABV can trigger autophagy, the cells were infected with attenuated HEP-Flury or wild-type GD-SH-01. As shown in and , the ratio of LC3-II to ACTB in SK and NA cells inoculated with HEP-Flury were similar to those observed in negative controls. In contrast, a significant conversion of LC3-I to LC3-II was detectable in GD-SH-01-infected SK and NA cells ( and ), a fact that led us to conclude that the characteristic properties of RABV were required for the induction of autophagy. Furthermore, in cells that were transfected with EGFP-LC3B plasmids and subsequently infected with GD-SH-01, the amount of EGFP dots increased significantly. The same trend was observed in cells treated with rapamycin, but could not be seen in HEP-Flury-infected cells or negative controls ( and ). These observations regarding the EGFP dots further corroborate the hypothesis that features of RABV are necessary for the induction of autophagy.
Wild-type RABV enhances autophagic flux in SK cells and blocks autophagosome degradation in NA cells
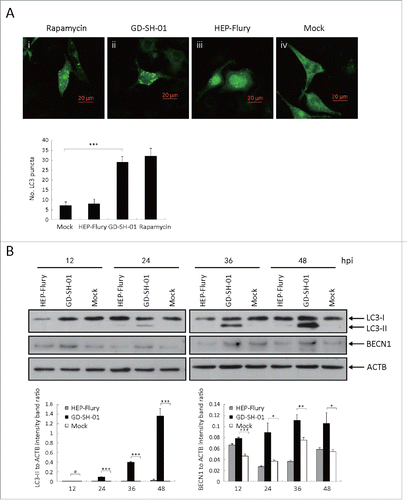
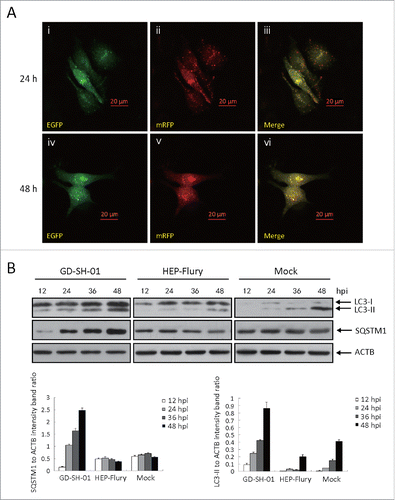
In order to examine the autophagic flux induced by GD-SH-01, we analyzed SQSTM1 protein expression in infected SK and NA cells 12, 24, 36, and 48 h after infection. We observed different effects in both cell lines: Whereas the expression of SQSTM1 (sequestosome 1) decreased in SK cells (), it increased in NA cells (). We also traced the autophagic flux morphologically with a monomeric red fluorescent protein (mRFP)-EGFP-LC3B plasmid tandem construct ( and ).Citation31 Upon observation 24 and 48 h postinfection (hpi), the amount of red dots increased in SK cells (), while only yellow puncta augmented in NA cells (). To further understand the molecular basis of the suppressed autophagic flux in NA cells, we conducted immunoblot analyses and thus determined if the upstream change is regular. As illustrated in and , the expression of BECN1 was significantly upregulated in SK and NA cells. This suggests that although the upstream change of autophagy is regular, autophagic flux is significantly suppressed in NA cells.
Figure 4. GD-SH-01 infection enhances autophagic flux in SK cells. (A) Colocalization of the EGFP and mRFP fluorescence in GD-SH-01-infected SK cells 24 and 48 h after transfection with mRFP-EGFP-LC3B plasmids. (B) Expression levels of LC3, SQSTM1 and ACTB at the end of the infection experiment in which SK cells were used as negative controls, infected with HEP-Flury or GD-SH-01. (Grouping of images of western blotting from different parts of the same gel.)
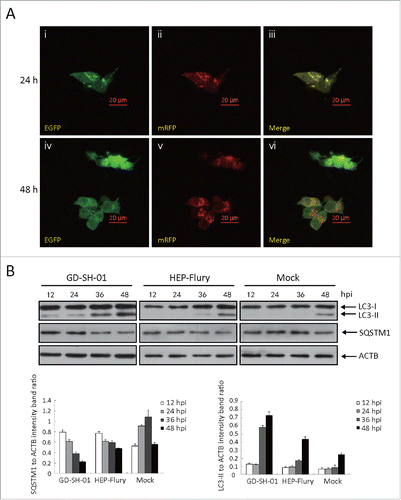
Figure 5. GD-SH-01 blocks autophagosome degradation in NA cells. (A) Colocalization of the EGFP and mRFP fluorescence in GD-SH-01-infected NA cells 24 and 48 h after transfection with mRFP-EGFP-LC3B plasmids. (B) Expression levels of LC3, SQSTM1 and ACTB at the end of the infection experiment in which NA cells were used as negative controls, infected with HEP-Flury or GD-SH-01. (Grouping of images of western blotting from different parts of the same gel.)
RABV induces apoptosis and mitochondrial dysfunction in SK and NA cells
GD-SH-01 induced autophagy as well as cytotoxicity. We thus aimed at clarifying if apoptotic pathways were activated and if these may explain the differences in cytotoxicity shown by GD-SH-01 and HEP-Flury. We used an Apoptosis Detection Kit and a flow cytometer to do so. As shown in , remarkable apoptosis was observed, and the percentage of early stage apoptotic cells as well as late stage apoptotic or even necrotic cells was significantly higher in GD-SH-01-infected cells when compared with HEP-Flury-infected cells and negative controls. Similar results were obtained with the cell viability assay (see below). This confirms the difference between the 2 virus strains regarding cell death activation and cytotoxicity.
Figure 6. RABV induces apoptosis and mitochondrial dysfunction. (A) GD-SH-01 induces apoptosis in SK and NA cells. Flow cytometry of mock-, HEP-Flury- and GD-SH-01-infected SK and NA cells after ANXA5-FITC and PI staining. Cells in early apoptosis and dead cells are represented as the percentage of ANXA5-FITC and PtdIns cells of total cells. Mean ± SD of 3 independent experiments. Two-way ANOVA: ***, P < 0.001. (B) Analysis of ΔΨm with flow cytometry using JC-1 dye. A representative density plot is shown for each condition. Expression of ΔΨm expressed as the JC-1 monomer to polymer fluorescence ratio. Mean ± SD of 3 independent experiments. Mean ± SD of 3 independent experiments. Two-way ANOVA: **, P < 0.01; ***, P < 0.001;#, P > 0.05.
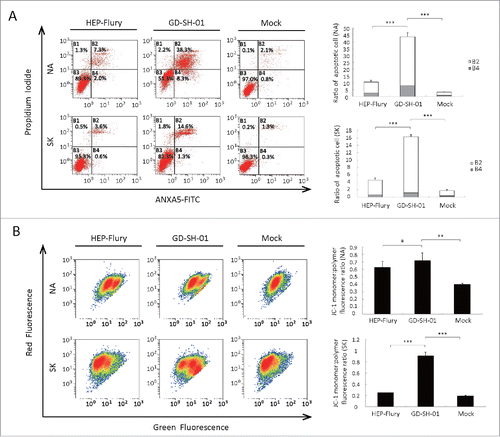
Since mitochondria depolarization is a mark of programmed cell death,Citation32 we examined the mitochondrial membrane potential (MMP) 48 hpi. MMP was inhibited by HEP-Flury and GD-SH-01 infection, but more severe mitochondria depolarization was seen in GD-SH-01-infected cells. This result is consistent with that of the apoptosis assay.
Enhancing autophagy with rapamycin decreases the apoptosis rate caused by GD-SH-01 infection
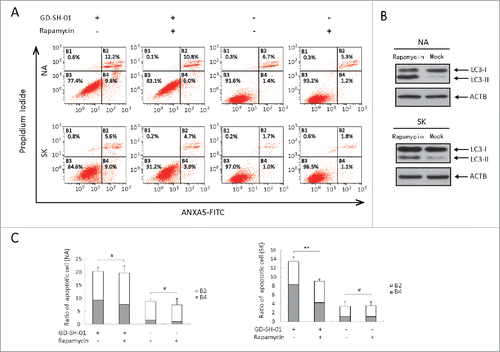
As shown previously, we observed that GD-SH-01 can trigger apoptosis and autophagy simultaneously. To reveal the interplay between autophagy and apoptosis, autophagy was enhanced with rapamycin, which can directly induce autophagy by specifically inhibiting the action of MTORCitation33,34 and the apoptosis rate was then measured with flow cytometry. The effective drug concentration in SK and NA cells turned out to be 50 nM and 10 nM (). As shown in , the apoptosis rate in GD-SH-01-infected cells decreased after pretreatment with rapamycin. In mock-infected cells, no difference could be detected regarding the apoptosis rate with or without the pretreatment with rapamycin. We concluded that autophagy may act as a protective mechanism against apoptosis induced by infection stress.
Figure 7. Apoptosis can be decreased by enhancing autophagy in GD-SH-01-infected cells. Cells pretreated with rapamycin or the corresponding solvent were infected with GD-SH-01, negative controls were not infected. (A) Flow cytometry of ANXA5-FITC and PI staining as a measure for apoptosis of SK and NA cells. (B) LC3-I to LC3-II conversion in SK (50 nM) and NA (10 nM) cells treated with rapamycin. (C) Cells in early apoptosis and dead cells. Mean ± SD of 3 independent experiments. Mean ± SD of 3 independent experiments. Two-way ANOVA: **, P < 0.01; #, P > 0.05. (Grouping of images of western blotting from different parts of the same gel.)
M protein of GD-SH-01 significantly promotes RABV-induced autophagy and is associated with cell apoptosis
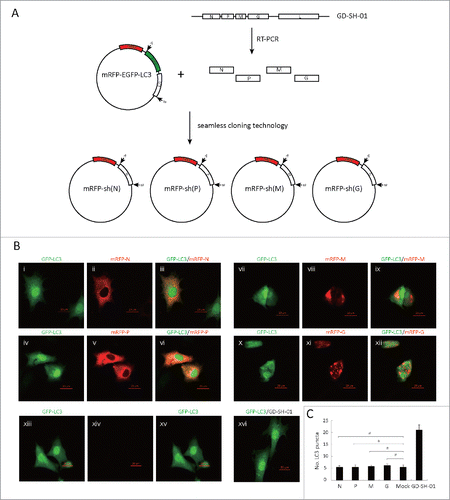
In order to identify the protein responsible for the induction of autophagy by GD-SH-01, the N, P, M, G, and L genes of HEP-Flury were replaced by the corresponding genes of GD-SH-01 (). Recombinant RABV were rescued and designated as rHEP-shN, rHEP-shP, rHEP-shM, rHEP-shG, and rHEP-shL by our laboratory (unpublished data). The recombinant viruses, HEP-Flury, and GD-SH-01 were inoculated into SK and NA cells, respectively, and the cell lysates were analyzed in a western blot assay 48 hpi. We found cells infected with GD-SH-01 or rHEP-shM to present a significant upregulation of LC3-II (). Since rHEP-shN, rHEP-shP, and rHEP-shG also provoked some LC3-I to LC3-II conversion, plasmids encoding N, P, M or G proteins were constructed and used, together with the EGFP-LC3B plasmid, to cotransfect SK cells. We did not find any differences regarding the number of LC3 dots in mock control cells and those cotransfected with mRFP-sh(N), mRFP-sh(P), mRFP-sh(M), or mRFP-sh(G) with EGFP-LC3B ().
Figure 8. M of GD-SH-01 is the main gene promoting RABV-induced autophagy and is associated with apoptosis. (A) Virus genomic structure and titers 48 hpi. White and black boxes represent open reading frames derived from HEP-Flury and GD-SH-01 strains, respectively. rHEP-shN, rHEP-shP, rHEP-shM, rHEP-shG, and rHEP-shL were rescued using reverse genetics technique. (B) M protein of GD-SH-01 promotes the conversion of from LC3-I to LC3-II. Expression levels of LC3, N and ACTB in SK and NA cells inoculated with HEP-Flury, GD-SH-01, rHEP-shN, rHEP-shP, rHEP-shM, rHEP-shG, and rHEP-shL 48 hpi. Mean ± SD of 3 independent experiments. Two-way ANOVA: ***, P < 0.001. (C) M protein of GD-SH-01 increases the apoptosis rate. ANXA5-FITC and PtdIns staining of cells infected with HEP-Flury, GD-SH-01 and rHEP-shM as well as mock controls to detect cells in early apoptosis and dead cells. Mean ± SD of 3 independent experiments. Two-way ANOVA: **, P < 0.01; ***, P < 0.001; #, P > 0.05. (D) Poor cell viability caused by M protein of GD-SH-01. Cell viability of SK and NA cells after infection with HEP-Flury, GD-SH-01, rHEP-shM and mock controls 24 and 48 hpi, normalized to cell viability of NA-Mock-24. Mean ± SD of 3 independent experiments. (Grouping of images of western blotting from different parts of the same gel.)
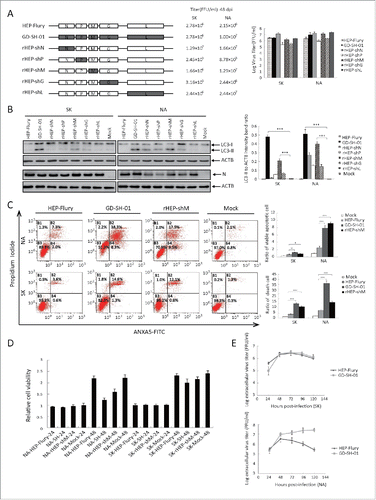
Figure 9. No one single protein of GD-SH-01 induces autophagy. (A) Graphic illustration of constructed plasmids of mRFP-sh(N), mRFP-sh(P), mRFP-sh(M) and mRFP-sh(G) (B) EGFP (green) and mRFP (red) expression in SK cells infected with mRFP-sh(N) (i, ii, iii), mRFP-sh(P) (iv, v, vi), mRFP-sh(M) (vii, viii, ix) and mRFP-sh(G) (x, xi, xii) or mock control (xiii, xiv, xv) cotransfected with EGFP-LC3B 24 h after transfection, or SK cells were infected with GD-SH-01 at an MOI of 1 for 48 h after being transfected with EGFP-LC3B-encoding plasmids (xvi). Scale bar: 20 µm. (C) Quantification of the number of LC3 dot per cell in SK cells. The average number of vesicles per cell was obtained from at least 15 cells undergoing each treatment. Mean ± SD of 3 independent experiments. Two-way ANOVA: #, P > 0.05.
We also found that the M protein is associated with cell apoptosis, as shown in , because the fraction of viable apoptotic cells increased from 0.2% to 0.8% in SK and from 2.0% to 9.5% in NA cells once the M gene of HEP-Flury was replaced by the M gene of GD-SH-01. The fraction of dead cells showed a similar development. The cell viability assay results further corroborated the role of the M protein, and the cytotoxicity of rHEP-shM was similar to cells infected with GD-SH-01 and at 48 hpi (). Dead cells showed a characteristic shrinking and floating in both cases. However, cells infected with HEP-Flury and negative controls presented a higher rate of cell survival.
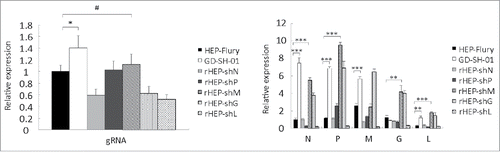
M of GD-SH-01 increases the transcription and replication abilities of rHEP-shM in SK cells
Since the M protein cannot induce obvious autophagy, we detected the levels of replication and transcription in SK cells that were induced to undergo complete autophagic flux. Primers for N, P, M, G, and L of GD-SH-01 and HEP-Flury were designed by our laboratory, and exhibited similar amplification efficiencies as the same structural gene. The real-time quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) results showed that the quantity of gRNA and almost all of the structural gene mRNA of GD-SH-01 and rHEP-shM were higher than those of HEP-Flury and other chimeric viruses, with the exception of the M mRNA of rHEP-shM and G mRNA of GD-SH-01 (). This indicates better transcription (mRNA) and replication (gRNA) abilities of GD-SH-01 and rHEP-shM than other RABVs, suggesting that M of GD-SH-01 increases the transcription and replication abilities of rHEP-shM and may contribute to an immune response that aids in inducing autophagy.
Figure 10. M of GD-SH-01 increases the transcription and replication abilities of rHEP-shM in SK cells. Cells were infected with HEP-Flury, GD-SH-01, and chimeric RABV for 48 h before the gRNA and the transcript abundances of N, P, M, G, and L were analyzed by qRT-PCR and calculated with the 2−ΔΔCt method. Mean ± SD of 3 independent experiments. Two-way ANOVA: *, P< 0.05; **, P < 0.01; ***, P < 0.001;#, P > 0.05.
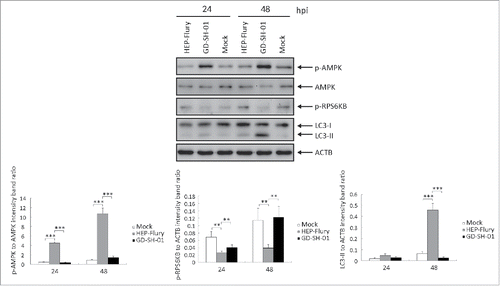
GD-SH-01 activates the AMPK signaling pathway in SK cells
SK cells presenting complete autophagic flux were used to study which signaling pathway triggered autophagy in GD-SH-01-infected cells. In order to identify the autophagy pathways implied in RABV infection, we analyzed the expression levels of proteins involved in autophagy and found the AMPK pathway to be activated. The AMPK signaling pathway was explored by analyzing the expression of phosphorylated (p)-AMPK (Thr172), AMPK, and p-RPS6KB. When compared with the HEP-Flury-infected cells, the cells infected with GD-SH-01 showed a significantly higher expression of the p-AMPK to AMPK ratio and a significantly lower expression of the p-RPS6KB to ACTB ratio 24 and 48 hpi (), suggesting that GD-SH-01 activates the expression of AMPK and inhibits the MTOR pathway in SK cells.
Figure 11. GD-SH-01 activates the AMPK signaling pathway in SK cells. p-AMPK, AMPK, p-RPS6KB and ACTB expression levels were analyzed in mock-, HEP-Flury- and GD-SH-01-infected SK cells 24 and 48 hpi and are represented here as p-AMPK to AMPK and p-RPS6KB to ACTB ratios. Mean ± SD of 3 independent experiments. Two-way ANOVA: **, P < 0.01; ***, P < 0.001
Discussion
Our research was performed in order to provide a better understanding of the interaction between a RABV infection and cellular autophagy. Our results show that autophagy was induced in RABV-infected SK and NA cells. Regarding the autophagic flux in these different neuroblastoma cell lines, contrary effects were observed upon infection with GD-SH-01. To our knowledge, this is the first demonstration that the M protein of RABV plays a crucial role in the activation of autophagy.
An increasing number of viruses have been shown to affect autophagy.Citation24,35,36 A recent study reports that vesicular stomatitis virus, viral hemorrhagic septicemia virus and spring viremia carp virus, members of the Rhabdoviridae family, can trigger autophagy.Citation37 However, to date there has been no report about RABV inducing autophagy. To answer this question, we focused on 2 RABV strains, the virulent RABV strain GD-SH-01 and the attenuated rabies virus strain HEP-Flury. In the present study, we demonstrated for the first time that a GD-SH-01 infection significantly increases the number of autophagosome-like vesicles in SK and NA cells. Accumulation of EGFP-LC3 dots in RABV-infected cells further confirmed that GD-SH-01 can induce autophagy. When examining the LC3 conversion in infected cells, we found that the ability of RABV to induce autophagosomes is limited to the virulent RABV strain GD-SH-01. The fact that HEP-Flury could not induce LC3 conversion suggests that a virulent RABV strain is required to induce autophagy. But additional research is required to confirm this finding applies for other virus strains.
Infection of GD-SH-01 induced a complete autophagic flux in SK cells, but inhibited autophagosome fusion with lysosomes in NA cells. Accumulating evidence suggests that virus infection can either enhance autophagic fluxCitation24,38–40 or does not enhance autophagic protein degradation,Citation41-45 but a virus triggering different changes regarding the autophagic flux in distinct cell types has not been described before. While both SK and NA cells are neuroblastoma cell lines, SK originates from the human and NA from the mouse. There are 2 mechanisms that may promote autophagosome accumulation: an increase in autophagic flux with an increase in the rate of autophagosome formation or a decrease in autophagic flux when any step of autophagosome fusion with lysosomes is blocked.Citation46 Apoptosis can be induced with activated caspase-3 by inhibiting autophagic flux.Citation47 Thus, we suppose that the incomplete autophagy process is correlated with cell apoptosis in NA cells. Regarding the rate of apoptosis in SK and NA cells, flow cytometry demonstrated that membrane disruption occurred in NA cells following infection with GD-SH-01, while HEP-Flury-infected cells did not show discernible differences to negative controls. GD-SH-01 as a wild-type RABV induced more apoptosis than Hep-Flury which is an attenuated RABV, this result is not consistent with previous studies indicating that virulent viruses are not able to stimulate apoptosis.Citation48,49 However, there are reports that support the notion that RABV does indeed induce apoptosis.Citation50-52 This difference may perhaps be due to the fact that induction of apoptosis is virus specific.
Mitochondrial membrane depolarization is a prelude to apoptosis.Citation32 Mitochondrial dysfunction activates caspases and promotes apoptosis.Citation53 Here, we have shown that HEP-Flury and particularly GD-SH-01 activate apoptosis and provoke a decrease of the MMP simultaneously. Autophagy as well as apoptosis constitutes 2 intracellular processes through which superfluous, damaged or aged cells or organelles are eliminated and several reports have provided evidence for a connection between these 2 pathways.Citation54-56 Autophagy and apoptosis often occur in the same cell and autophagy frequently precedes apoptosis.Citation57 While the autophagic response is stimulated by nonlethal stress, apoptosis is activated when stress levels exceed a certain threshold. Our results show that apoptosis caused by GD-SH-01 infection decreased when cells were pretreated with rapamycin, a fact that indicates that autophagy might be a self-protection mechanism of SK cells under GD-SH-01 infection stress. This hypothesis is supported by the fact that an increasing number of papers have shown a protective role of autophagy under stress.Citation58 In addition, autophagy is a mechanism controlling cellular homeostasis by self-digestion using lysosome enzymes while GD-SH-01 inhibited autophagosome fusion with lysosomes in NA cells, which might be the reason why there was no obvious decrease in the rate of apoptosis between rapamycin-treated and control groups upon GD-SH-01 infection. The relation between autophagy and apoptosis might serve as a starting point for the development of future preventive and therapeutic measures against rabies.
The genome of the RABV encodes 5 virus proteins, the N, P, M, G, and L proteins. Virulent and attenuated RABV differ greatly in their neuroinvasiveness. The trailer sequence, the RABV polymerase, and the pseudogene have been proven to contribute to neuroinvasiveness.Citation59 However, other reports have revealed that the M protein of Lyssavirus leads to the intracellular accumulation of virus particles and affects the cell death pathway.Citation12,60 We aimed at identifying the key protein in the induction of autophagy and found cells infected with GD-SH-01 and rHEP-shM to present a significant conversion of LC3-I to LC3-II, while rHEP-shN, rHEP-shP, and rHEP-shG induce autophagy to a lesser extent than rHEP-shM. These findings reveal that the M protein of the RABV plays a main role in the induction of autophagy. Moreover, we've also shown that rHEP-shM closely resembles GD-SH-01 regarding apoptosis rate and cell viability after infection. We also tried to figure out if the single M protein may trigger autophagy, but could not confirm this hypothesis, which seems to indicate that a single structure protein is not sufficient to trigger autophagy. The fact that rHEP-shM causes a strong increase in autophagy may be due to a more complex change of replication behavior of HEP-Flury by the M gene of GD-SH-01.5959 Although we observed a small amount of LC3-I to LC3-II conversion in cells infected with rHEP-shN, rHEP-shP and rHEP-shG, none of these single proteins induced autophagy. We found N protein levels to vary between viruses because of differences in replication efficiency between GD-SH-01, HEP-Flury, and the recombinant virus strains. Our results are consistent with a previous study which reports that the rabies M protein may indeed induce apoptosis.Citation12
Viral proteins and RNA are recognized through receptors such as the evolutionarily conserved toll-like receptors (TLRs).Citation61 TLR7, which binds single-stranded RNA,Citation62 is possibly targeted to endosomes by the autophagy pathway.Citation63 One report has indicated that SK cells express TLR7.Citation64 Although the M protein of GD-SH-01 alone cannot induce autophagy in vitro, the transcription and replication levels of GD-SH-01 and rHEP-shM were significantly higher than those of HEP-Flury and other chimeric RABVs. Therefore, we speculate that a stronger TLR7 was stimulated by higher expression of genomic RNA and mRNA in GD-SH-01 and rHEP-shM. This may be the reason that GD-SH-01 and rHEP-shM induced obvious autophagy.
AMPK is crucial to regulate cellular responses to metabolic stresses,Citation65 and activates autophagy mainly through the inhibition of MTOR or the phosphorylation of RPS6KB, its downstream target.Citation28,29 We found that infection stress activated AMPK and inhibited RPS6KB. GD-SH-01 was able to increase the ratio of p-AMPK to AMPK and decrease the phosphorylation of RPS6KB, implying that GD-SH-01 is involved in the activation of the AMPK-MTOR pathway.
Here, we are providing data that broadens our understanding of the pathogenicity of the rabies virus at the cellular level. Our results demonstrate for the first time that the M gene of the GD-SH-01 strain plays a potential role in promoting RABV-induced autophagy. Wild-type RABV GD-SH-01 induced opposite autophagy flux trends in 2 distinct cell types, and apoptosis also seems to be involved in RABV infection. The AMPK-MTOR pathway is activated upon GD-SH-01 infection. Moreover, we speculated that when stress caused by GD-SH-01 infection surpasses a certain limit, cells are not able to protect themselves by activating autophagy mechanisms and this may lead to cell death via apoptosis.
Materials and methods
Antibodies, reagents and plasmids
The primary antibodies against LC3B (2775), BECN1 (3495), p-AMPK (Thr172; 2535), AMPK (2603), and p-RPS6KB (Thr389; 9234) were purchased from Cell Signaling Technology, and other primary antibodies are as follows: SQSTM1 polyclonal antibody (ABclonal, A0682), mouse anti-rabies virus N monoclonal antibodies (Tongdian Biotechnology, Ab-0056) and ACTB (Beyotime, AA128). Horseradish peroxidase (HRP) affinipure goat anti-rabbit IgG (H+L) (E030120-01) and anti-mouse (E030110-01) secondary antibodies were purchased from EarthOx. Rapamycin (9904) was obtained from Cell Signaling Technology. mRFP-EGFP-LC3B (21074, deposited by Tamotsu Yoshimori Lab) was obtained from Addgene.
Cells and virus
The human neuroblastoma cell line (SK-N-SH, abbreviated as SK) (purchased from ATCC®, HTB-11™) and the mouse neuroblastoma cell line (NA) (obtained from the Wuhan Institute of Biological Products, China) were cultured in RPMI 1640 medium (Gibco, C22400500BT) containing 10% fetal bovine serum (FBS) at 37°C and 5% CO2. The wild-type RABV strain GD-SH-01 was isolated by our laboratory from the brain tissue of a rabid pig in China;Citation7 the attenuated rabies virus strain HEP-Flury was preserved in our laboratory.
Virus infection and drug treatment
SK and NA cell lines were grown to approximately 80% confluence and were inoculated with GD-SH-01 and HEP-Flury at a multiplicity of infection (MOI) of 1 in RPMI 1640 medium. One h later, the virus solution was removed and replaced by fresh medium complemented with 2% FBS and 100 U/mL of penicillin, 100 mg/mL of streptomycin and the cells were incubated at 37°C and 5% CO2 for 24 and 48 h. Since rapamycin triggers autophagy, we used it as a positive control. 50 nM and 10 nM were found to be the optimal concentrations of rapamycin to induce autophagy in SK and NA cells; cell viability is not affected by this concentration. SK and NA cells were pretreated with rapamycin for 24 h prior to virus infection. At the same time, the corresponding level of solvent of rapamycin, dimethyl sulfoxide (DMSO, 196055), served as a negative control and was applied to cells that were not inoculated with any virus.
DNA plasmids
The GD-SH-01 cDNA sequences of N, P, M, G were sequenced in our laboratory and have been uploaded to NCBI (GenBank: JX088694.1). In order to construct a plasmid capable of expressing the single RABV structure protein fused with mRFP, we obtained the corresponding sequences of GD-SH-01 by RT-PCR. We replaced the EGFP and LC3B genes of the mRFP-EGFP-LC3B plasmid using seamless cloning technology (Vazyme, C112) (). We obtained mRFP-sh(N), mRFP-sh(P), mRFP-sh(M) and mRFP-sh(G) plasmids that were used together with the EGFP-LC3B plasmid to cotransfect SK.
Cell transfection assays
After SK cells had grown to 80% confluence, cells were transfected with plasmids using the SuperFect Transfection Reagent (QIAGEN, 301307). Briefly, 2 μg of plasmids were dissolved in 75 μl of ddH2O, and 10 μl of SuperFect Transfection Reagent was directly pipetted into the DNA solution before 400 μl cell growth medium were added. Immediately, the total volume was transferred to the cells and an incubation period of 3 h at 37°C followed. Then, the transfection reagent medium was replaced with 2 ml of fresh RPMI 1640 containing 10% FBS and cells were further cultured at 37°C for 24 h.
Cell growth curve assay and measurements of virus titers
Culture supernatants of SK and NA cells infected with HEP-Flury and GD-SH-01 at a MOI of 1 were taken every 24 h for 5 d. Virus titers were calculated by using the Reed-Muench methodCitation66 and results are given as 50% tissue culture infectious doses per milliliter. In brief, cells cultivated in 96-well microtiter plates were inoculated with 10-fold serial dilutions of virus and the plates were kept at 37°C and 5% CO2 for 96 h. Subsequently, cells were fixed with 100 µL cold 80% acetone and kept at −20°C for 30 min. After washing twice with phosphate-buffered saline (PBS; Beyotime, C0221A, pH 7.4), the cells were treated with FITC-conjugated anti-rabies monoclonal globulin (Fujirabio, 800-092) for 1 h at 37°C. Virus titers were determined according to the presence of fluorescence with a fluorescence microscope (AMG, Mill Creek, Washington, USA).
Confocal fluorescence microscopy
Confocal fluorescence microscopy was used for analysis of LC3 expression after RABV infection or rapamycin treatment and the relationship between N, P, M or G protein expression and LC3 dot formation. Since measurement of the autophagic flux allows for more reliable conclusions regarding the induction of autophagy than the autophagosome-like vesicle accumulation,Citation17 we decided to use this parameter. In order to monitor autophagic flux, SK and NA cells transfected with the mRFP-EGFP-LC3B plasmid were evaluated under a fluorescence microscope. The autophagic flux was measured by spectral regions (488 nm and 558 nm) at 24 and 48 hpi.
To study if any one of the N, P, M or G proteins of GD-SH-01 could trigger an increase in LC3 dots, fluorescence signals in cells cotransfected with mRFP-sh(N), mRFP-sh(P), mRFP-sh(M) or mRFP-sh(G) and EGFP-LC3B were visualized with a LSM780 laser system using a LSCM 7DUO confocal fluorescence microscope (ZEISS, Oberkochen, Baden-Württemberg, Germany).
Transmission electron microscopy
TEM was used for observation of autophagy. The SK and NA cells infected with HEP-Flury or GD-SH-01 RABV as well as negative controls were collected in a 1.5-mL microcentrifuge tube. Then, the cells were centrifuged at 1,000 g for 10 min and the cell pellets were fixed with 2.5% glutaraldehyde and stored at 4°C overnight. The samples were fixed in 1% osmium tetroxide, dehydrated stepwise with graded ethanol, and embedded in epoxy resin (Sigma, 45347). Next, 60-nm to 80-nm sized ultrathin sections were obtained and then stained with uranyl acetate and lead citrate. Images of thin sections were acquired using a Tecnai G2 spirit twin transmission electron microscope (FEI, Hillsboro, Oregon, USA).
Western blotting
Treated cells were washed with cold PBS 3 times and lysed in radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, P0013B) containing 1 mM phenylmethanesulfonyl fluoride (Beyotime, ST506) on ice for 30 min. Cell lysates were centrifuged at 10,000g for 10 min at 4°C. Equal amounts of protein samples were boiled for 5 min in 6 × SDS-PAGE loading buffer, and were resolved by SDS-PAGE and electrotransferred to Hybond-polyvinylidene difluoride membranes (Milipore, IPVH00010) and then blocked with 5% (w:v) nonfat dry milk in phosphate buffered saline with 0.05% (w:v) Tween 20 (Sigma, V900548). Membranes were probed with primary antibodies overnight at 4°C and then exposed to the corresponding secondary antibodies conjugated to horseradish peroxidase. The protein bands were detected with the ECL Plus kit (Beyotime, P0018).
Flow cytometry
In order to examine apoptosis, we seeded 2 × 105 NA cells in each well of a 6-well plate and had them grow for 12 h to 80% confluence. HEP-Flury and GD-SH-01 RABV were then inoculated at an MOI of 1 and cells were incubated for 24 h and 48 h with each of the 2 virus strains. In order to evaluate a possible relationship between apoptosis and RABV-induced autophagy, SK and NA cells were treated with rapamycin or DMSO for 24 h and then incubated with GD-SH-01 or solvent for 48 h at an MOI of 0.5. Cells were subsequently centrifuged at 1,000 g for 5 min. After washing 2 times with PBS, cell pellets were resuspended in 500 μL binding buffer and incubated with 5 μL of ANXA5/annexin V-FITC and 5 μL of a propidium iodide (PI) solution at room temperature for 15 min, according to the manufacturer's instructions for the ANXA5-FITC Apoptosis Detection Kit (Keygentec, KGA108). ANXA5-FITC and PtdIns staining were measured with a Beckman FC 500 flow cytometer (Beckman Coulter, Fullerton, California, USA), followed by data analysis with the corresponding CXP Software.
To determine the ΔΨm of SK and NA cells, we used the mitochondrial membrane potential assay kit with JC-1 (5,5′,6,6′-tetrachloro-1,1′,3,3′-tetraethyl-imidacarbocyanine iodide) (Beyotime, C2006). Whether JC-1 forms monomers or polymers depends on the MMP. Since monomers and polymers emit fluorescence at different wavelengths upon excitation at 514 and 585 nm, the ratio of fluorescence emission at 529 nm to 590 nm allows conclusions regarding mitochondrial depolarization. Briefly, cells were incubated for 20 min at 37°C with JC-1 staining fluid and then analyzed by flow cytometry. In each sample, 30,000 events were measured and mean fluorescence emission after excitation at 514 nm and 585 nm were used for further analysis. Results are expressed as the JC-1 monomer to polymer fluorescence ratio.
Quantification of viral RNA
qRT-PCR was used to detect viral replication and transcription. Total RNA was extracted using the HiPure Universal RNA Mini Kit with gDNA Remover (Magen, R4130-02), and cDNA synthesis was performed using the HiScript II 1st Strand cDNA Synthesis Kit (vazyme, R211-02) with Oligo(dT) and RABV genomic RNA forward primer. For detection of the mRNA of the structural gene and gRNA, primer pairs were designed by our laboratory. The real-time qRT-PCR was performed using AceQ qPCR SYBR Green Master Mix (vazyme, Q111-02) on an iQ5 iCycler detection system (Bio-Rad) with the following reaction conditions: 95°C for 5 min, followed by 40 cycles of 95°C for 10 sec and 60°C for 32 sec. The relative expression of mRNA was normalized to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) and calculated with the traditional 2−ΔΔCt method.
Cell viability assay
Viability of SK and NA cells infected with GD-SH-01 or HEP-Flury for 24 and 48 h as well as viability of negative controls were evaluated with the Cell Counting Kit-8 (Dojindo, CK04) according to the manufacturer's instructions. Here, the optical density was used as a measure indicating cell viability and was determined at 570 nm with a microplate spectrophotometer (Bio-Rad; Hercules, California, USA; 1681002).
Statistical analysis
All results are expressed as the mean ± standard deviation (SD) and all statistical analyses were performed with 2-way ANOVA using IBM SPSS statistics software (version 19.0), multiple comparison between the groups was performed using S-N-K method. The intensities of western blot bands were analyzed with Image-Pro Plus (version 6.0). A value of P < 0.05 was considered to be statistically significant. For all tests, the notations used to indicate significance between groups are as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Abbreviations
| DMSO | = | dimethyl sulfoxide |
| EGFP | = | enhanced green fluorescent protein |
| ER | = | endoplasmic reticulum |
| FBS | = | fetal bovine serum |
| G | = | the glycoprotein |
| GAPDH | = | glyceraldehyde-3-phosphate dehydrogenase |
| gRNA | = | geneome RNA |
| hpi | = | hours postinfection |
| HRP | = | horseradish peroxidase |
| L | = | the RNA-dependent RNA polymerase |
| MAP1LC3/LC3 | = | microtubule-associated protein 1 light chain 3 |
| M | = | matrix protein |
| MMP | = | mitochondrial membrane potential |
| MOI | = | multiplicity of infection |
| mRFP | = | monomeric red fluorescent protein |
| MTOR | = | mechanistic target of rapamycin (serine/threonine kinase) |
| N | = | he nucleoprotein |
| NA | = | mouse neuroblastoma cell line |
| P | = | phosphoprotein |
| PBS | = | phosphate-buffered saline |
| qRT-PCR | = | quantitative reverse transcriptase polymerase chain reaction |
| RABV | = | rabies virus |
| RIPA | = | radio-immunoprecipitation assay |
| SD | = | standard deviation |
| SK | = | human neuroblastoma cell line |
| TEM | = | transmission electron microscopy |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Shile Huang (Department of Biochemistry and Molecular Biology, Louisiana State University Health Sciences Center, USA) for invaluable experimental help and discussion.
Funding
This study was partially supported by the National Program on Key Research Project of China (No. 2016YFD0500400), National Nature Science Foundation of China (No. 31172322), Nature Science Foundation of Guangdong (No.2015A03031103), and Special Fund for Agro-Scientific Research in the Public Interest (No. 201103032).
References
- Jackson AC. Rabies Virus Infection: An Update. J Neurovirol 2003; 9(2):253-8; PMID:12707856; http://dx.doi.org/10.1080/13550280390193975
- Marston DA, McElhinney LM, Johnson N, Muller T, Conzelmann KK, Tordo N, Fooks AR. Comparative analysis of the full genome sequence of European bat lyssavirus type 1 and type 2 with other lyssaviruses and evidence for a conserved transcription termination and polyadenylation motif in the G-L 3′ non-translated region. J Gen Virol 2007; 88(4):1302-14; PMID:17374776; http://dx.doi.org/10.1099/vir.0.82692-0
- WHO. Frequently asked questions on rabies. India: World Health Organization, 2013; 28
- Shin J, Sakoda Y, Yano S, Ochiai K, Kida H, Umemura T. Effective prevention against rabies by intracerebral immunization in mice. J Vet Med Sci 2009; 71(10):1331-6; PMID:19887739; http://dx.doi.org/10.1292/jvms.001331
- Faber M, Li J, Kean RB, Hooper DC, Alugupalli KR, Dietzschold B. Effective preexposure and postexposure prophylaxis of Rabies with a highly attenuated recombinant rabies virus. Proc Natl Acad Sci U S A 2009; 106(27):11300-5; http://dx.doi.org/10.1073/pnas.0905640106
- McGettigan JP. Experimental rabies vaccines for humans. Expert Rev Vaccines 2010; 9(10):1177-86; PMID:20923268; http://dx.doi.org/10.1586/erv.10.105
- Luo Y, Zhang Y, Liu X, Yang Y, Yang X, Zhang D, Deng X, Wu X, Guo X. Complete genome sequence of a highly virulent Rabies virus isolated from a rabid pig in South China. J Virol 2012; 86(22):12454-5; PMID:23087116; http://dx.doi.org/10.1128/JVI.02234-12
- Luo Y, Zhang Y, Liu X, Yang Y, Yang X, Zheng Z, Deng X, Wu X, Guo X. Characterization of a wild rabies virus isolate of porcine origin in China. Infect Genet Evol 2013; 17:147-52; http://dx.doi.org/10.1016/j.meegid.2013.03.046
- Koprowski H, Black J, Nelsen DJ. Studies on chick-embryo-adapted-rabies virus. VI. Further changes in pathogenic properties following prolonged cultivation in the developing chick embryo. J Immunol 1954; 72(1):94-106; PMID:13118196
- Mebatsion T, Weiland F, Conzelmann K. Matrix protein of rabies virus is responsible for the assembly and budding of bullet-shaped particles and interacts with the transmembrane spike glycoprotein G. J Virol 1999; 1(73):242
- Finke S, Conzelmann K. Dissociation of rabies virus matrix protein functions in regulation of viral RNA synthesis and virus assembly. J Virol 2003; 22(77):12074-82; http://dx.doi.org/10.1128/JVI.77.22.12074-12082.2003
- Larrous F, Gholami A, Mouhamad S, Estaquier J, Bourhy H. Two overlapping domains of a Lyssavirus matrix protein that acts on different cell death pathways. J Virol 2010; 84(19):9897-906; PMID:20631119; http://dx.doi.org/10.1128/JVI.00761-10
- Kassis R, Larrous F, Estaquier J, Bourhy H. Lyssavirus Matrix Protein Induces Apoptosis by a TRAIL-Dependent Mechanism Involving Caspase-8 Activation. J Virol 2004; 78(12):6543-55; PMID:15163747; http://dx.doi.org/10.1128/JVI.78.12.6543-6555.2004
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science 2000; 290(5497):1717-21; PMID:11099404; http://dx.doi.org/10.1126/science.290.5497.1717
- Kroemer G, Mariño G, Levine B. Autophagy and the Integrated Stress Response. Mol Cell 2010; 40(2):280-93; PMID:20965422; http://dx.doi.org/10.1016/j.molcel.2010.09.023
- Kirkegaard K, Taylor MP, Jackson WT. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat Rev Microbiol 2004; 2(4):301-14; PMID:15031729; http://dx.doi.org/10.1038/nrmicro865
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, Agholme L, Agnello M, Agostinis P, Aguirre-Ghiso JA, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy 2012; 4(8):445-544; http://dx.doi.org/10.4161/auto.19496
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 2000; 19(21):5720-8; PMID:11060023; http://dx.doi.org/10.1093/emboj/19.21.5720
- Ohsumi Y. Molecular dissection of autohagy:two ubiquitin-like systems. Nat Rev Mol Cell Biol 2001; 2(3):211-6; PMID:11265251
- Roizman B, Chou J. Herpes simplex virus 1 γ134.5 gene function, which blocks the host response to infection, maps in the homologous domain of the genes expressed during growth arrest and DNA damage. Biochemistry-US 1994; 91(12):5247-51
- Harrow S, Papanastassiou V, Harland J, Mabbs R, Petty R, Fraser M, Hadley D, Patterson J, Brown SM, Rampling R. HSV1716 injection into the brain adjacent to tumour following surgical resection of high-grade glioma: safety data and long-term survival. Gene Ther 2004; 11(22):1648-58; PMID:15334111; http://dx.doi.org/10.1038/sj.gt.3302289
- Suhy DA, Giddings TJ, Kirkegaard K. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J Virol 2000; 74(19):8953-65; PMID:10982339; http://dx.doi.org/10.1128/JVI.74.19.8953-8965.2000
- Sun M, Huang L, Wang R, Yu Y, Li C, Li P, Hu X, Hao H, Ishag HA, Mao X. Porcine reproductive and respiratory syndrome virus induces autophagy to promote virus replication. Autophagy 2014; 8(10):1434-47; http://dx.doi.org/10.4161/auto.21159
- Pei J, Zhao M, Ye Z, Gou H, Wang J, Yi L, Dong X, Liu W, Luo Y, Liao M, et al. Autophagy enhances the replication of classical swine fever virus in vitro. Autophagy 2013; 10(1):93-110; PMID:24262968; http://dx.doi.org/10.4161/auto.26843
- Rodriguez-Rocha H, Gomez-Gutierrez JG, Garcia-Garcia A, Rao X, Chen L, McMasters KM, Zhou HS. Adenoviruses induce autophagy to promote virus replication and oncolysis. Virology 2011; 416(1-2):9-15; PMID:21575980; http://dx.doi.org/10.1016/j.virol.2011.04.017
- Zhang Y, Li Z, Ge X, Guo X, Yang H. Autophagy promotes the replication of encephalomyocarditis virus in host cells. Autophagy 2014; 7(6):613-28; http://dx.doi.org/10.4161/auto.7.6.15267
- Zhu B, Xu F, Li J, Shuai J, Li X, Fang W. Porcine circovirus type 2 explores the autophagic machinery for replication in PK-15 cells. Virus Res 2012; 163(2):476-85; PMID:22134092; http://dx.doi.org/10.1016/j.virusres.2011.11.012
- Kimura N, Tokunaga C, Dalal S, Richardson C, Yoshino K, Hara K, Kemp BE, Witters LA, Mimura O, Yonezawa K. A possible linkage between AMP-activated protein kinase (AMPK) and mammalian target of rapamycin (mTOR) signalling pathway. Genes Cells 2003; 8(1):65-79; PMID:12558800; http://dx.doi.org/10.1046/j.1365-2443.2003.00615.x
- Cheng SWY, Fryer LGD, Carling D, Shepherd PR. Thr2446 is a novel mammalian target of rapamycin (mTOR) phosphorylation site regulated by nutrient status. J Biol Chem 2004; 279(16):15719-22; PMID:14970221; http://dx.doi.org/10.1074/jbc.C300534200
- Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ 2009; 16(7):966-75; PMID:19325568; http://dx.doi.org/10.1038/cdd.2009.33
- Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter Protein, tandem fluorescent-tagged LC3. Autophagy 2007; 3(5):452-60; PMID:17534139; http://dx.doi.org/10.4161/auto.4451
- Zamzami N, Marchetti P, Castedo M, Zanin C, Vayssière J, Petit PX, Kroemer G. Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J Exp Med 1995; 181(5):1661-72; PMID:7722446; http://dx.doi.org/10.1084/jem.181.5.1661
- Noda T, Ohsumi Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J Biol Chem 1998; 273(7):3963-6; PMID:9461583; http://dx.doi.org/10.1074/jbc.273.7.3963
- Klionsky DJ, Meijer AJ, Codogno P, Neufeld TP, Scott RC. Autophagy and p70S6 Kinase. Autophagy 2005; 1(1):59-61; PMID:16874035; http://dx.doi.org/10.4161/auto.1.1.1536
- Lee Y, Lei H, Liu M, Wang J, Chen S, Jiang-Shieh Y, Lin Y, Yeh T, Liu C, Liu H. Autophagic machinery activated by dengue virus enhances virus replication. Virology 2008; 374(2):240-8; PMID:18353420; http://dx.doi.org/10.1016/j.virol.2008.02.016
- Sir D, Chen W, Choi J, Wakita T, Yen TSB, Ou JJ. Induction of incomplete autophagic response by hepatitis C virus via the unfolded protein response. Hepatology 2008; 48(4):1054-61; PMID:18688877; http://dx.doi.org/10.1002/hep.22464
- García-Valtanen P, Ortega-Villaizán MDM, Martínez-López A, Medina-Gali R, Pérez L, Mackenzie S, Figueras A, Coll JM, Estepa A. Autophagy-inducing peptides from mammalian VSV and fish VHSV rhabdoviral G glycoproteins (G) as models for the development of new therapeutic molecules. Autophagy 2014; 10(9):1666-80; http://dx.doi.org/10.4161/auto.29557
- Sun Y, Yu S, Ding N, Meng C, Meng S, Zhang S, Zhan Y, Qiu X, Tan L, Chen H, et al. Autophagy benefits the replication of Newcastle disease virus in chicken cells and tissues. J Virol 2013; 88(1):525-37; PMID:24173218; http://dx.doi.org/10.1128/JVI.01849-13
- Liu Q, Qin Y, Zhou L, Kou Q, Guo X, Ge X, Yang H, Hu H. Autophagy sustains the replication of porcine reproductive and respiratory virus in host cells. Virology 2012; 429(2):136-47; PMID:22564420; http://dx.doi.org/10.1016/j.virol.2012.03.022
- Jin R, Zhu W, Cao S, Chen R, Jin H, Liu Y, Wang S, Wang W, Xiao G. Japanese Encephalitis virus activates autophagy as a viral immune evasion strategy. PLoS ONE 2013; 8(1):e52909; PMID:23320079; http://dx.doi.org/10.1371/journal.pone.0052909
- Wirblich C, Tan GS, Papaneri A, Godlewski PJ, Orenstein JM, Harty RN, Schnell MJ. PPEY motif within the rabies virus (RV) matrix protein is essential for efficient virion release and RV pathogenicity. J Virol 2008; 82(19):9730-8; PMID:18667490; http://dx.doi.org/10.1128/JVI.00889-08
- Vandergaast R, Fredericksen BL. West Nile virus (WNV) replication is independent of autophagy in mammalian cells. PLoS ONE 2012; 7(9):e45800; PMID:23029249; http://dx.doi.org/10.1371/journal.pone.0045800
- Beatman E, Oyer R, Shives KD, Hedman K, Brault AC, Tyler KL, David Beckham J. West Nile virus growth is independent of autophagy activation. Virology 2012; 433(1):262-72; PMID:22939285; http://dx.doi.org/10.1016/j.virol.2012.08.016
- Li J, Liu Y, Wang Z, Liu K, Wang Y, Liu J, Ding H, Yuan Z. Subversion of cellular autophagy machinery by Hepatitis B virus for viral envelopment. J Virol 2011; 85(13):6319-33; PMID:21507968; http://dx.doi.org/10.1128/JVI.02627-10
- Xiao Y, Ma C, Yi J, Wu S, Luo G, Xu X, Lin PH, Sun J, Zhou J. Suppressed autophagy flux in skeletal muscle of an amyotrophic lateral sclerosis mouse model during disease progression. Physiol Rep 2015; 3(1):e12271; PMID:25602021; http://dx.doi.org/10.14814/phy2.12271
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell 2010; 140(3):313-26; PMID:20144757; http://dx.doi.org/10.1016/j.cell.2010.01.028
- González-Polo R, Boya P, Pauleau A, Jalil A, Larochette N, Souquère S, Eskelinen E, Pierron G, Saftig P, Kroemer G. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 2005; 118(14):3091-102; http://dx.doi.org/10.1242/jcs.02447
- Thoulouze M, Lafage M, Montano-Htrose JA, Lafon M. Rabies virus infects mouse and human lymphocytes and induces apoptosis. J Virol 1997; 71(10):7372-80; PMID:9311815
- Yan X, Prosniak M, Curtis MT, Weiss ML, Faber M, Dietzschold B, Fu ZF. Silver-haired bat rabies virus variant does not induce apoptosis in the brain of experimentally infected mice. J Neurovirol 2001; 7(6):518-27; PMID:11704884; http://dx.doi.org/10.1080/135502801753248105
- Ubol S, Kasisith J, Pitidhammabhorn D, Tepsumethanol V. Screening of pro-apoptotic genes upregulated in an experimental street rabies virus-infected neonatal mouse brain. Microbiol Immunol 2005; 49(5):423-31; PMID:15905604; http://dx.doi.org/10.1111/j.1348-0421.2005.tb03746.x
- Ubol S, Kasisith J. Reactivation of Nedd-2, a developmentally down-regulated apoptotic gene, in apoptosis induced by a street strain of rabies virus. J Med Microbiol 2000; 49(11):1043-6; PMID:11073159; http://dx.doi.org/10.1099/0022-1317-49-11-1043
- Yin J, Ding Y, Huang Y, Tao X, Li H, Yu P, Shen X, Jiao W, Liang G, Tang Q, et al. Comparative Analysis of the Pathogenic Mechanisms of Street Rabies Virus Strains with Different Virulence Levels. Biomed Environ Sci 2014; 27(10):749-62; PMID:25341810
- Ravindran J, Gupta N, Agrawal M, Bala Bhaskar AS, Lakshmana Rao PV. Modulation of ROS/MAPK signaling pathways by okadaic acid leads to cell death via, mitochondrial mediated caspase-dependent mechanism. Apoptosis 2011; 16(2):145-61; PMID:21082355; http://dx.doi.org/10.1007/s10495-010-0554-0
- Maiuri MC, Criollo A, Kroemer G. Crosstalk between apoptosis and autophagy within the Beclin 1 interactome. EMBO J 2010; 29(3):515-6; PMID:20125189; http://dx.doi.org/10.1038/emboj.2009.377
- Mariño G, Niso-Santano M, Baehrecke EH, Kroemer G. Self-consumption: the interplay of autophagy and apoptosis. Nat Rev Mol Cell Bio 2014; 15(2):81-94; http://dx.doi.org/10.1038/nrm3735
- Mukhopadhyay S, Panda PK, Sinha N, Das DN, Bhutia SK. Autophagy and apoptosis: where do they meet? Apoptosis 2014; 19(4):555-66; PMID:24415198; http://dx.doi.org/10.1007/s10495-014-0967-2
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Bio 2007; 8(9):741-52; http://dx.doi.org/10.1038/nrm2239
- López-Larrea J, Martínez-Borra C. Autophagy and self-defense. Adv Exp Med Biol 2012; 738:169-84; http://dx.doi.org/10.1007/978-1-4614-1680-7_11
- Faber M, Pulmanausahakul R, Nagao K, Prosniak M, Rice AB, Koprowski H, Schnell MJ, Dietzschold B. Identification of viral genomic elements responsible for rabies virus neuroinvasiveness. Proc Natl Acad Sci U S A 2004; 101(46):16328-32; PMID:15520387; http://dx.doi.org/10.1073/pnas.0407289101
- Finke S, Granzow H, Hurst J, Pollin R, Mettenleiter TC. Intergenotypic replacement of Lyssavirus matrix proteins demonstrates the role of Lyssavirus M proteins in intracellular virus accumulation. J Virol 2010; 84(4):1816-27; PMID:19955305; http://dx.doi.org/10.1128/JVI.01665-09
- Finberg RW, Kurt-Jones EA. Viruses and Toll-like receptors. Microbes Infect 2004; 6(15):1356-60; PMID:15596120; http://dx.doi.org/10.1016/j.micinf.2004.08.013
- Diebold SS. Innate Antiviral Responses by Means of TLR7-Mediated Recognition of Single-Stranded RNA. Science 2004; 303(5663):1529-31; PMID:14976261; http://dx.doi.org/10.1126/science.1093616
- Delgado M, Singh S, De Haro S, Master S, Ponpuak M, Dinkins C, Ornatowski W, Vergne I, Deretic V. Autophagy and pattern recognition receptors in innate immunity. Immunol Rev 2009; 227:189-202; PMID:19120485; http://dx.doi.org/10.1111/j.1600-065X.2008.00725.x
- Mohanty MC, Deshpande JM. Differential induction of Toll-like receptors & type 1 interferons by Sabin attenuated & wild type 1 polioviruses in human neuronal cells. Indian J Med Res 2013; 138:209-18; PMID:24056597
- Stapleton D, Guang KIM, Widmer J, Michell BJ, Teh T, House CM, Fernandez CS, Cox T, Witters LA, Kemp BE, et al. Mammalian AMP-activated protein kinase subfamily. J Biol Chem 1996; 271(2):611-4; PMID:8557660; http://dx.doi.org/10.1074/jbc.271.2.611
- Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 1938; 27(3):493-7
