ABSTRACT
Macroautophagy/autophagy is a cellular degradation process that sequesters organelles or proteins into a double-membrane structure called the phagophore; this transient compartment matures into an autophagosome, which then fuses with the lysosome or vacuole to allow hydrolysis of the cargo. Factors that control membrane traffic are also essential for each step of autophagy. Here we demonstrate that 2 monomeric GTP-binding proteins in Saccharomyces cerevisiae, Arl1 and Ypt6, which belong to the Arf/Arl/Sar protein family and the Rab family, respectively, and control endosome-trans-Golgi traffic, are also necessary for starvation-induced autophagy under high temperature stress. Using established autophagy-specific assays we found that cells lacking either ARL1 or YPT6, which exhibit synthetic lethality with one another, were unable to undergo autophagy at an elevated temperature, although autophagy proceeds normally at normal growth temperature; specifically, strains lacking one or the other of these genes are unable to construct the autophagosome because these 2 proteins are required for proper traffic of Atg9 to the phagophore assembly site (PAS) at the restrictive temperature. Using degron technology to construct an inducible arl1Δ ypt6Δ double mutant, we demonstrated that cells lacking both genes show defects in starvation-inducted autophagy at the permissive temperature. We also found Arl1 and Ypt6 participate in autophagy by targeting the Golgi-associated retrograde protein (GARP) complex to the PAS to regulate the anterograde trafficking of Atg9. Our data show that these 2 membrane traffic regulators have novel roles in autophagy.
Introduction
Autophagy is a lysosome/vacuole-dependent mechanism which is used to sequester and degrade cytoplasmic components by using specialized vesicular structures that will ultimately fuse with the degradative compartment.Citation1 Autophagy is essential for survival under stressful conditions; it can serve as a way for cells to overcome starvation and is responsible for the removal of protein aggregates, damaged organelles, and for developmental remodeling. Thus, it plays an important role in controlling intracellular homeostasis.Citation2 Autophagy can be roughly divided into selective and nonselective forms. In this paper, we will use the term “autophagy” to refer to the nonselective form, also called macroautophagy.
Generally, autophagy starts by sensing an upstream signal that is controlled by the target of rapamycin (TOR) protein complex. Then a phagophore forms at the phagophore assembly site (PAS) and encloses cellular components such as misfolded proteins or damaged organelles. The expansion of the phagophore leads to the formation of a double-membrane structure called the autophagosome. Subsequently, the autophagosome, which contains the cytoplasmic components to be degraded, fuses with the vacuole, transferring the cargo for subsequent hydrolysis. The inner membrane as well as the engulfed cargo are degraded and the resulting building blocks are released into the cytoplasm by vacuolar membrane permeases for reuse.Citation3 The autophagy machinery, encompassing more than 40 autophagy-related (ATG) genes in S. cerevisiae and other fungi, is complex and highly regulated.Citation4
Although most cytoplasmic contents can be turned over through nonselective autophagy, in some situations, substrates or organelles are degraded by specific forms of autophagy. For example, in yeast, 3 vacuolar proteases, the precursor forms of Ape1 (aminopeptidase I; prApe1), Ams1 (α-mannosidase; prAms1) and Ape4 (aspartyl aminopeptidase; prApe4) are synthesized in the cytoplasm and transported to the vacuole for further processing through a selective autophagy pathway named the cytoplasm-to-vacuole targeting (Cvt) pathway.Citation5-7 The Cvt pathway shares high similarity with autophagy and requires most of the ATG genes, except it is predominant during growing conditions, whereas autophagy is strongly induced by starvation.Citation8
Upon induction of autophagy, a complex consisting of Atg17-Atg31-Atg29 assembles with Atg1 kinase and Atg13 to serve as the platform for other Atg proteins to form the autophagosome.Citation9 Then Atg9, a transmembrane protein, brings additional membrane to establish the phagophore. The initiation step of vesicle nucleation and efficient elongation of the phagophore requires the covalent attachment of phosphatidylethanolamine (PE) to Atg8. Atg4 proteolyzes the C terminus of Atg8, exposing a glycine residue, which is followed by attachment of PE to Atg8, making it phagophore-associated Atg8–PE. At this point, Atg8 is fully activated and recruits membrane for the elongation of the phagophore and eventual establishment of the autophagosome.Citation10
Besides the Atg proteins, autophagy also requires membrane sources to form the autophagosome. There is ample evidence, particularly in S. cerevisiae, that many membrane-traffic regulators, including several monomeric GTP-binding proteins are indispensable for autophagy.Citation11 Examples include the Rab protein family members Ypt1, Ypt31-Ypt32 and Ypt7,Citation12-14 and the Arf/Arl/Sar family members Arf1-Arf2 and Sar1.Citation15,16 These monomeric GTP-binding proteins function in different stages of the secretory or endocytic pathway. When autophagy is triggered, these proteins participate in different steps of autophagy, from recruiting membrane components to the PAS to forming the autophagosome and regulating the fusion between the autophagosome and the vacuole.
Previous work has shown that Arl1 and Ypt6 are important regulators of membrane traffic. Arl1 belongs to the Arf/Arl/Sar family, while Ypt6 is a member of the Rab family. Both function at the trans-Golgi membranes, regulating vesicular traffic between the trans-Golgi network (TGN) and the early endosome. Like other GTP-binding proteins, when associated with GTP, Arl1 and Ypt6 are associated with membranes and control vesicle traffic; but when the GTP is hydrolyzed into GDP, they remain in the cytoplasm and are inactive.Citation17-19 Specifically, the GTP-bound form of Arl1 controls vesicle trafficking in 3 different pathways, recruiting the golgin protein Imh1 to the TGN, recruiting the clathrin adaptor protein Gga1,Citation20 and transporting the glycosylphosphatidylinositol-anchored protein Gas1 to the plasma membrane.Citation21 In addition, a multisubunit tethering complex called GARP (Golgi-associated retrograde protein)/VFT (Vps53) interacts with Arl1-GTP as determined by affinity chromatography.Citation22 Although deleting ARL1 seems to have no effect on the cellular localization of the GARP complex,Citation22 Arl1 shows synthetically lethality and physical interaction with Vps53, one of the subunits of the GARP complex,Citation23 indicating that at least some functions of the GARP complex overlap with Arl1.
Ypt6 has not been studied as thoroughly as Arl1 and many of the other monomeric guanine-nucleotide binding proteins. However, GTP-bound Ypt6 also shows the ability to bind to the GARP complex through interaction with the Vps52 subunit.Citation24 Because both Arl1 and Ypt6 interact with subunits of the GARP complex, the 2 proteins may have some overlapping functions. This hypothesis is in keeping with genetic evidence that demonstrates these 2 proteins show synthetic lethality with one another, meaning that cells lacking both ARL1 and YPT6 are nonviable.Citation23,25
In this paper, we show Arl1 and Ypt6 are required for autophagy under high-temperature stress. Yeast lacking either ARL1 or YPT6 are able to conduct autophagy normally at the permissive temperature of 30°C but have a complete defect at the restrictive temperature of 37°C. Furthermore, we found the defect at 37°C in arl1Δ and ypt6Δ strains is a result of a block in anterograde trafficking of Atg9 to the PAS at high temperature. Importantly, upon construction of a conditional double mutant, we observed that the double mutant was defective for autophagy at the permissive temperature. Finally, we found the targeting of the GARP complex to the PAS was defective at high temperature when either ARL1 or YPT6 was deleted. Therefore in this work, we demonstrate that these 2 monomeric GTP-binding proteins, Arl1 and Ypt6, in addition to their established roles as regulators of membrane trafficking, have novel roles in autophagy.
Results
Arl1 and Ypt6 are involved in autophagy at 37°C
Arl1 and Ypt6 are both involved in vesicle trafficking between the TGN and the early endosome.Citation17,19 To investigate whether Arl1 or Ypt6 is required in autophagy, we first examined growth in the presence of rapamycin, a TOR inhibitor that induces autophagy.Citation26 Previous studies have shown that cells with autophagy defects cannot grow in the presence of rapamycin.Citation27 A strain with the deletion of the gene ATG1, which is essential for autophagy,Citation28 was used as the negative control for this and subsequent experiments. We found arl1Δ and ypt6Δ strains were able to grow on plates containing 5 ng/ml rapamycin at 30°C but, surprisingly, not at 37°C, suggesting that autophagy in these 2 strains was inhibited at high temperature (). Because autophagy in yeast can also be induced by nitrogen starvation, we tested the ability of wild-type (WT), atg1Δ, arl1Δ, and ypt6Δ strains to survive in nitrogen-deficient (SD-N) liquid medium at both 30°C and 37°C, followed by plating on nitrogen-replete medium. We found results similar to the rapamycin growth experiment in that the arl1Δ and ypt6Δ strains had a significant defect (i.e., similar to the atg1Δ strain) in ability to form colonies after incubation in SD-N medium at 37°C. The arl1Δ and ypt6Δ strains had an intermediate phenotype at 30°C (Fig. S1).
Figure 1. Arl1 and Ypt6 are involved in autophagy at the restrictive temperature of 37°C. (A) arl1Δ and ypt6Δ strains are sensitive to rapamycin at 37°C. All strains were streaked on YPD + DMSO (4.57 nM) or YPD + 5 ng/ml rapamycin in DMSO and cultured either at 30°C or 37°C. Plates were photographed after 3 d of growth. (B) GFP-Atg8 degradation was decreased in the arl1Δ strain at 37°C. WT (BY4743), atg1Δ, and arl1Δ strains were cultured at 30°C in nonstarvation medium until log-phase; then all the strains were incubated at 37°C or 30°C for 30 min. The cells were then washed twice with SD-N medium, and cultured in SD-N for 3 h either at 37°C or at 30°C. An aliquot of cells at 37°C were transferred back to 30°C and cultured in SD-N for an additional 3 h (denoted “R”). Whole cell lysates were subjected to immunoblot with anti-GFP antibody. (C) GFP-Atg8 degradation was decreased in the ypt6Δ strain at 37°C. The procedure was the same as . (D) The arl1Δ (YSA003) and ypt6Δ (YSA004) strains show decreased Pho8Δ60 activity at 37°C. Error bars represent standard deviation from 3 biological replicates. The samples were done in 3 technical replicates for each biological replicate.
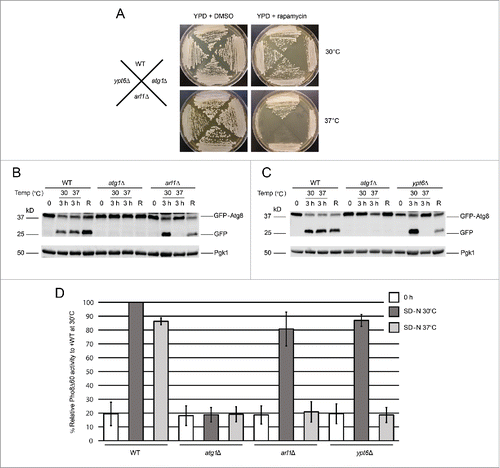
To confirm that Arl1 and Ypt6 have roles in autophagy at 37°C, an assay that follows cleavage of a modified version of Atg8, green fluorescent protein (GFP)-Atg8, was used to monitor the transport of Atg8 to the vacuole through autophagy (). Once autophagy is triggered, Atg8 is normally conjugated to the lipid PE on its C terminus, which helps expand the membranes of the autophagosome.Citation29 The protein GFP-Atg8 will thus be transferred to the vacuole through autophagy. While inside the vacuole, Atg8 will be degraded, but the GFP moiety is resistant to degradation and can therefore be detected as free GFP on western blots if autophagy proceeds normally. Under nonstarvation conditions (0 time in ), no free GFP was detected in any strain (WT, atg1Δ, arl1Δ, or ypt6Δ). Under starvation conditions, specifically incubation of cells for 3 h in SD-N medium (“3 h” in ) at normal growth temperature (30°C), GFP-Atg8 processing was detected in WT, arl1Δ, and ypt6Δ strains, suggesting normal autophagy; whereas in atg1Δ, the negative control, no processing was observed, meaning defective autophagy. In contrast, no free GFP was detected in arl1Δ and ypt6Δ strains incubated at 37°C. Importantly, when the arl1Δ and ypt6Δ strains initially incubated at 37°C were transferred back to 30°C and starved for an additional 3 h, autophagy activity was recovered (“R” in ), indicating that the defect in GFP-Atg8 processing was not due to cell death.
We previously found the arl1Δ strain had increased sensitivity to H2O2 (Fig. S2A). In order to rule out the possibility that the autophagy defect occurs in response to stress in general, we performed the GFP-Atg8 processing assay under oxidative stress conditions, namely in the presence of 1 mM hydrogen peroxide (Fig. S2B) and found arl1Δ and ypt6Δ strains were able to perform autophagy normally, suggesting that the autophagy defect is specifically related to heat stress.
In S. cerevisiae, the activity of a truncated form of the vacuolar alkaline phosphatase Pho8 (Pho8Δ60) is widely used to measure the magnitude of autophagy.Citation30 Normally, Pho8 is trafficked to the vacuole via part of the secretory pathway, where it is activated by proteolytic removal of a C-terminal propeptide. Upon removal of the N-terminal 60 amino acids, Pho8 can only be trafficked to the vacuole by autophagy. Thus, the amount of Pho8 enzymatic activity under these circumstances is a measure of autophagy. We therefore performed the Pho8Δ60 assay in arl1Δ (YSA003) and ypt6Δ (YSA004) strains at both 30 and 37°C (). The results show that the arl1Δ and ypt6Δ strains had a significant decrease in Pho8Δ60 activity compared with WT (YSA001) at 37°C but not at 30°C, consistent with the results of the GFP-Atg8 processing assay ().
GTP-bound Arl1 and Ypt6 are able to suppress the autophagy defect of the arl1Δ strain or the ypt6Δ strain at 37°C
As a GTP-binding proteinCitation17,18 Arl1 has several different conformations, which can be modeled using site-directed mutant alleles based on results from other members of the Ras superfamily. We investigated which alleles could complement the autophagy defect. The Arl1 mutants used include Arl1Q72L, a form of the protein that is predicted to be unable to hydrolyze GTP based on the comparisons with Ras and Arf1 proteins and thus is GTP-restricted;Citation31 Arl1G2A, predicted to be myristoylation-defective and thus cannot bind to membranes;Citation17 Arl1N127I, predicted to be unable to bind GTP compared to Ras and Arf proteins and thus is GDP-restricted;Citation32 Arl1D130N, predicted to bind xanthine nucleotides in vitro by homology to Rab and thus to be nucleotide-free in vivo.Citation33 We transformed these alleles of Arl1 into the arl1Δ strain and performed the GFP-Atg8 processing and Pho8Δ60 assays to determine which alleles suppressed the autophagy defect at high temperature (). From the results of the 2 assays we found that only the WT and the GTP-bound Arl1Q72L were able to complement the autophagy defect at 37°C. Overall, these data suggest GTP-bound Arl1, the version active for membrane traffic, is essential for autophagy at high temperature.
Figure 2. GTP-bound Arl1 or Ypt6 are able to suppress the autophagy defect of the arl1Δ or ypt6Δ strains at 37°C. (A) GFP-Atg8 degradation was recovered when the GTP-restricted form of Arl1 was expressed in the arl1Δ strain. WT Arl1, empty vector (YEp352), N-terminal myristoylation defective Arl1 (G2A), GTP-bound Arl1 (Q72L), GDP-bound Arl1 (N127I), or nucleotide-free Arl1 (D130N) were transformed into the arl1Δ strain. The GFP-Atg8 processing assay was performed. (B) Pho8Δ60 activity was recovered when the GTP-restricted form of Arl1 was expressed in the arl1Δ (YSA003) strain. Error bars represent standard deviation from 3 biological replicates. The samples were done in 3 technical replicates for each biological replicate. (C) GFP-Atg8 degradation was recovered when the GTP-bound form of Ypt6 was expressed. Empty vector (pRS316), WT, GTP-bound Ypt6 (Q69L) or GDP-bound Ypt6 (T24N) were transformed into the ypt6Δ strain. The GFP-Atg8 processing assay was performed. (D) Pho8Δ60 activity was recovered when the GTP-bound form of Ypt6 was expressed in the ypt6Δ strain (YSA004). Error bars represent standard deviation from 3 biological replicates. The samples were done in 3 technical replicates for each biological replicate. EV, empty vector.
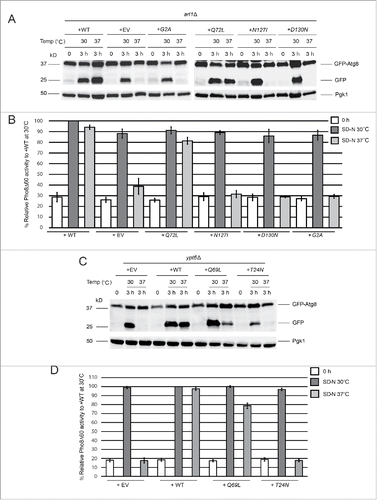
For Ypt6, we performed a similar analysis, first generating GTP-restricted and GDP-restricted alleles of Ypt6, Q69L and T24N, respectively,Citation34 via site-directed mutagenesis.Citation35 We transformed all 3 alleles of YPT6 into the ypt6Δ strain and performed the GFP-Atg8 processing and Pho8Δ60 assays. The results demonstrate that the WT and the GTP-restricted Q69L forms of the protein complemented the autophagy defect of the ypt6Δ strain at 37°C (), suggesting that the membrane-trafficking function of Ypt6 is important for autophagy at high temperature similar to what we observed with Arl1.
In additional to expression of different Arl1 alleles in the arl1Δ strain, we also overexpressed YPT6 in the arl1Δ strain to examine the epistatic relationship between Arl1 and Ypt6. As shown by the GFP-Atg8 processing assay in Figure S3A, the overexpression of YPT6 suppressed the autophagy phenotype to about the same extent as the overexpression of wild-type ARL1. However, when we did the converse experiment we found the overexpression of ARL1 in the ypt6Δ background could not suppress the autophagy defect of ypt6Δ cells at high temperature (Fig. S3B). This result may suggest that Arl1 functions upstream of Ypt6. Alternatively, this may suggest that Ypt6 plays a larger role in autophagy than Arl1.
Atg8 is mislocalized in the arl1Δ and ypt6Δ strains under starvation conditions at 37°C
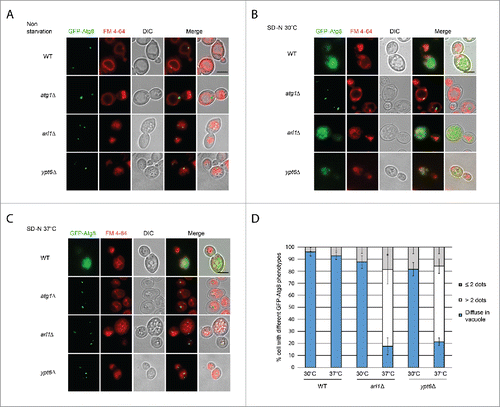
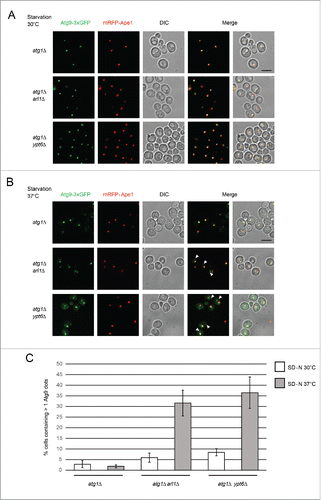
To directly monitor the autophagy phenotype, we visualized the transport of GFP-tagged Atg8 under starvation conditions at different temperatures by fluorescence microscopy (). FM 4–64 was used to mark the vacuolar membranes.Citation36 Under nonstarvation conditions, a single GFP-Atg8 dot, denoting the PAS,Citation37 was detected outside the vacuole in all 4 strains (WT, atg1Δ, arl1Δ and ypt6Δ). Under starvation conditions at 30°C (), diffuse GFP labeling was observed inside the vacuole in the WT strain, indicating the normal processing of GFP-Atg8, consistent with the findings in and . In arl1Δ and ypt6Δ cells, although the vacuoles were not intact even at the permissive temperature as previously described,Citation38,39 diffuse GFP was detected inside the small vacuolar fragments so that despite the fragmentation, these 2 strains were able to perform autophagy at 30°C, consistent with results shown in . Under starvation conditions at 37°C (), the WT strain showed diffuse green staining within the vacuole. However, in the arl1Δ and ypt6Δ strains multiple green GFP-Atg8 dots were found outside the vacuole fragments. This pattern was similar to atg1Δ, indicating defective autophagy in at 37°C. We also observed that the number of the GFP-Atg8 dots in the arl1Δ and ypt6Δ cells at 37°C was significantly higher than in the WT cells under the same conditions (). The mislocalization of GFP-Atg8 under these conditions indicates Arl1 and Ypt6 are required for the transport of GFP-Atg8 to the vacuole at high temperature.
Figure 3. Arl1 and Ypt6 are required for Atg8 transport to the vacuole at 37°C. Atg8 is mislocalized in arl1Δ and ypt6Δ strains at 37°C. Cells were grown, then starved for nitrogen as described. FM 4–64 was used to stain the vacuolar membrane. Experiments were repeated 3 times and the results shown are from a single experiment. (A) Fluorescence images for WT, atg1Δ, arl1Δ and ypt6Δ in nonstarvation conditions. (B) Fluorescence images for WT, atg1Δ, arl1Δ and ypt6Δ in starvation conditions at 30°C. (C) Fluorescence images for WT, atg1Δ, arl1Δ and ypt6Δ in starvation conditions at 37°C. (D) The percentage of cells with Atg8 dots in 3 categories: 0, ≤ 2 and multiple dots (> 2 dots per cell) was quantified. At least 150 cells were counted for each strain. Error bars represent standard deviation from 3 biological replicates. Scale bar: 3 µm.
Arl1 and Ypt6 are required for the formation of the autophagosome at 37°C
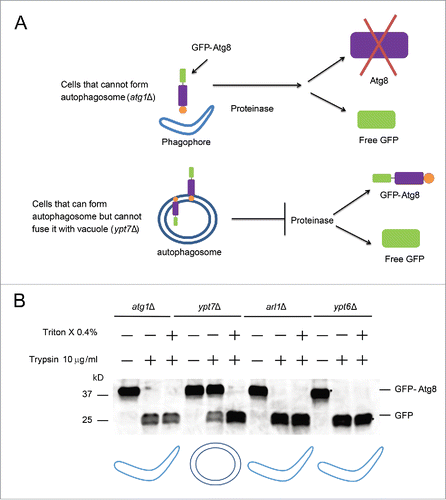
The multiple dots of GFP-Atg8 in the arl1Δ and ypt6Δ strains could either result from defective formation of the autophagosome so that Atg8 molecules accumulate in some abnormal location; or defective fusion between the autophagosome and the vacuole. In order to distinguish between these possibilities, we utilized the GFP-Atg8 proteinase protection assayCitation40 (). Two deletion mutants, atg1Δ and ypt7Δ, which block the formation of the autophagosome or the fusion of the autophagosome with the vacuole, respectively, were used as controls. As shown in , the GFP-Atg8 fusion protein was not detected in either the arl1Δ or the ypt6Δ strain upon treatment with trypsin, similar to the phenotype observed in atg1Δ, suggesting that Arl1 and Ypt6 are required for the formation of the autophagosome at high temperature ().
Figure 4. Arl1 and Ypt6 are required in the formation of the autophagosome at 37°C. (A) Illustration of the GFP-Atg8 proteinase protection assay. (B) GFP-Atg8 proteinase protection assay was done as indicated in Materials and Methods. The arl1Δ and ypt6Δ strains were cultured in SD-N medium at 37°C, while the atg1Δ and ypt7Δ strains were at 30°C. The cartoon underneath represents different structures for autophagy in the presence of trypsin. The boomerang represents the phagophore. The double circles represent the autophagosome.
The conditional YPT6 and ARL1 double-knockout strain shows an autophagy defect at 30°C
As mentioned previously, ARL1 and YPT6 show synthetic lethality. We therefore created a conditional double mutant to test the hypothesis that cells lacking both Arl1 and Ypt6 will be unable to perform autophagy at 30°C. In order to make the double mutant, we took advantage of the auxin-inducible degron (AID) system.Citation41 We first integrated the F-box protein gene TIR1 from Oryza sativa into the ypt6Δ strain, then subsequently tagged ARL1 by looping in a cassette containing 3 × mini AID and 5 × FLAG sequences on its 3′ end. To induce the degradation of the Arl1 protein, 1-naphthaleneacetic acid (NAA) was added to the medium to a final concentration of 1 mM and the culture was incubated for 30 min. Next, autophagy was induced by nitrogen starvation along with continued treatment with NAA to prevent the accumulation of Arl1. The GFP-Atg8 processing assay was performed as described previously. As the result in demonstrates, autophagy was completely inhibited at 30°C when this strain was cultured with NAA. As a control, Western blot analysis was performed to confirm that cells had undetectable levels of FLAG-tagged Arl1, the only version of Arl1 present. In the presence of the NAA carrier (ethanol), autophagy proceeded normally.
Figure 5. The YPT6 ARL1 conditional double knockout strain is defective for autophagy at 30°C. The YSA021 strain, containing the ARL1AID allele, or the ypt6Δ strain was transformed with the pRS316-GFP-Atg8 plasmid. Cells were cultured until OD600 = 0.6 and divided into 2 portions. One portion was cultured with 1 mM NAA for 30 min, another one was with 0.17% ethanol alone. Autophagy was induced as described. For the YSA021 strain, after 3 h in SD-N medium, an aliquot of the + NAA culture was washed 3 times by SD-N medium and cells were cultured in SD-N medium without NAA for an additional 3 h. All the samples were collected and subjected to either Western blot with the anti-GFP antibody and anti-FLAG antibody, or live-cell fluorescence microscopy. (A) GFP-Atg8 assay shows the degradation of Arl1 caused the autophagy defect in the ypt6Δ strain background (YSA021) at 30°C. This defect can be reversed by washing out NAA (“washed”). (B) GFP-Atg8 assay shows that adding NAA to the ypt6Δ strain did not affect autophagy. (C) Fluorescence microcopy shows the punctate phenotype of mislocalized GFP-Atg8 after Arl1 was degraded. The diffuse green phenotype of normal autophagy reappeared after NAA was removed. Scale bar: 3 µm.
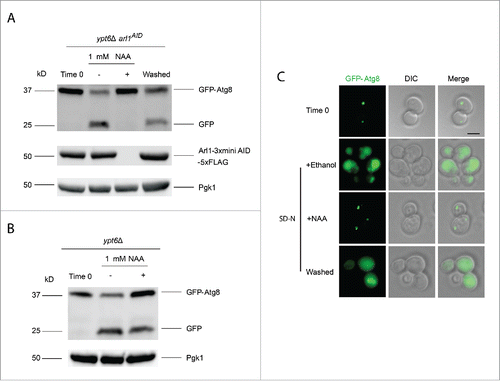
Because the effects of auxin are reversible, we also tested whether autophagy was recovered by removing the NAA. Cells were washed 3 times with SD-N liquid medium. After washing and further incubation in SD-N for 3 h we observed that autophagy was indeed restored, correlating with the reappearance of the Arl1 protein. We also confirmed that adding NAA alone to the ypt6Δ strain (with the normal ARL1 allele) had no effect on autophagy (). The autophagy phenotype was further confirmed by using fluorescence microcopy to detect the GFP-Atg8 signal (). After addition of NAA, GFP-Atg8 remained as multiple green dots at 30°C in the cell, indicative of defective/stalled autophagy. After washing out NAA, a diffuse green phenotype was observed, demonstrating restoration of autophagy. In conclusion, upon deletion of both ARL1 and YPT6, cells are unable to perform autophagy even at 30°C.
Arl1 and Ypt6 are required for the anterograde trafficking of Atg9
Atg9 is an essential component for creation of the autophagosome and is localized in several cytoplasmic sites including the Golgi apparatus and mitochondria; it normally cycles between these organelles and the PAS.Citation42-44 The recycling of Atg9 from the PAS depends on several other Atg proteins including Atg1-Atg13 and Atg2-Atg18.Citation45,46 Thus, if cells lack ATG1, the retrograde transport of Atg9 from the PAS is blocked. In this condition, the number of Atg9 dots per cell and the colocalization of Atg9 with the PAS marker prApe1 represent the level of the anterograde transport of Atg9 to the PAS.Citation47 Because Atg9 works upstream of Atg8 and we found that Atg8 is mislocalized in the arl1Δ and ypt6Δ mutants (), we examined whether the trafficking of Atg9 is affected in these mutant strains. As shown in , we found in starvation conditions at 30°C, the deletion of ARL1 or YPT6 in an atg1Δ deletion mutant background (YSA009, to make YSA010 and YSA011, respectively) had no effect on colocalization of Atg9-3×GFP with the PAS marker mRFP-Ape1, indicating that normal anterograde trafficking of Atg9 to the PAS occurred, consistent with the previous observations that autophagy occurred normally at this temperature. However, when we treated the cells under starvation conditions at 37°C (), we found both the atg1Δ arl1Δ and atg1Δ ypt6Δ strains showed multiple Atg9 dots in the cells while the atg1Δ strain had a single Atg9-3×GFP dot (). Thus, Arl1 and Ypt6 are required for the anterograde trafficking of Atg9 to the PAS at 37°C.
Figure 6. Arl1 and Ypt6 are required for the anterograde trafficking of Atg9. mRFP-Ape1 and Atg9-3×GFP were integrated into the yeast genome at the APE1 and ATG9 loci, respectively. Cells were grown in YPD then transferred to SD-N medium as described. (A) Fluorescence images for atg1Δ (YSA009), atg1Δ arl1Δ (YSA010) and atg1Δ ypt6Δ (YSA011) strains under starvation conditions at 30°C. (B) Fluorescence images for atg1Δ (YSA009), atg1Δ arl1Δ (YSA010) and atg1Δ ypt6Δ (YSA011) strains under starvation conditions at 37°C, Arrows point to the additional Atg9 dots. (C) The percentage of the cells with more than 1 Atg9 dot in and . At least 50 cells were counted for each strain. Error bars represent standard deviation from 3 biological replicates. Scale bar: 5 µm.
It has been suggested that the Golgi apparatus is one of the origins for the autophagosomal membrane.Citation48 To test this hypothesis, we integrated DsRed-tagged Sec7 into strains of interest as a marker for the TGN. We performed the Atg9 trafficking assay in atg1Δ (YSA012), atg1Δ arl1Δ (YSA013) and atg1Δ ypt6Δ (YSA014) strains to determine whether Atg9 was trapped in the TGN, as we found a fraction of the Atg9-3xGFP protein could not be transferred to the PAS in atg1Δ arl1Δ and atg1Δ ypt6Δ strains at 37°C. As the results in Fig. S4 show, approximately 25% of the atg1Δ arl1Δ and atg1Δ ypt6Δ cells showed colocalization between the Atg9-3xGFP and Sec7-DsRed at 37°C, suggesting the origin of some of the Atg9-containing membrane is the TGN.
Arl1 and Ypt6 are also required in the Cvt pathway
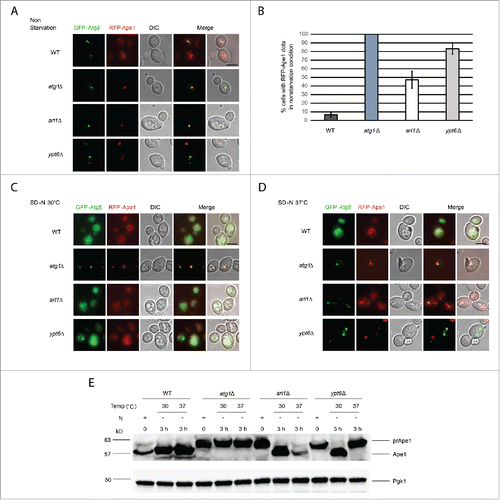
To investigate whether Arl1 and Ypt6 are required in one of the selective autophagy pathways, specifically the Cvt pathway, the fate of mRFP-tagged prApe1 was examined. As shown in , in nonstarvation conditions, WT cells (YSA005) showed a diffuse red phenotype indicating that the Cvt pathway was active. The negative control, the atg1Δ mutant (YSA006), showed a single red dot, demonstrating the Cvt pathway was incapable of transferring the mRFP-prApe1 into the vacuole. In contrast, arl1Δ (YSA007) and ypt6Δ (YSA008) cells had more mRFP-prApe1 dots than the WT strain (), suggestive of a partially defective Cvt pathway in the 2 deletion mutants under nonstarvation conditions (). In addition, we found both the arl1Δ and ypt6Δ strains were able to undergo transfer of mRFP-prApe1 to the vacuole with starvation at 30°C (; diffuse red phenotype). However, when we increased the temperature to 37°C along with starvation, the ypt6Δ strain showed single red dots, whereas the arl1Δ strain showed the normal diffuse red phenotype (). To confirm these results, Western blots to follow the proteolytic processing of prApe1 to the vacuolar mature form were performed. As shown in , neither the arl1Δ nor ypt6Δ strain could process prApe1 normally under nonstarvation conditions. Conversely, in the arl1Δ strain this defect could be reversed by nitrogen starvation at both 30°C and 37°C; in the cells lacking YPT6, the prApe1 processing defect could only be reversed by starvation at 30°C, again suggesting the autophagy defect in the ypt6Δ strain might be more severe than in the arl1Δ strain. In conclusion, Arl1 is required in the Cvt pathway only under nonstarvation conditions, but Ypt6 is required for this pathway in both nonstarvation and starvation conditions with high temperature stress.
Figure 7. Arl1 and Ypt6 are required in the Cvt pathway. mRFP-Ape1 was integrated into the yeast genome using the pRS305-mRFP-Ape1 plasmid. Cells were grown and treated under starvation conditions as described. (A) Fluorescence images for WT (YSA005), atg1Δ (YSA006), arl1Δ (YSA007) and ypt6Δ (YSA008) strains in nonstarvation conditions. (B) The percentage of cells counted from that contain a red prApe1 dot rather than the diffuse red phenotype. Error bars represent the standard deviation from 3 biological replicates. At least 80 cells were counted for each strain. (C) Fluorescence images for WT (YSA005), atg1Δ (YSA006), arl1Δ (YSA007) and ypt6Δ (YSA008) strains under starvation conditions at 30°C. (D) Fluorescence images for WT (YSA005), atg1Δ (YSA006), arl1Δ (YSA007) and ypt6Δ (YSA008) strains under starvation conditions at 37°C. (E) prApe1 processing assay for WT, atg1Δ, arl1Δ and ypt6Δ strains under nonstarvation conditions (time 0), starvation for 3 h at 30°C and starvation for 3 h at 37°C. N, nitrogen. Scale bar: 3 µm.
Arl1 and Ypt6 are required for translocating GARP to the PAS
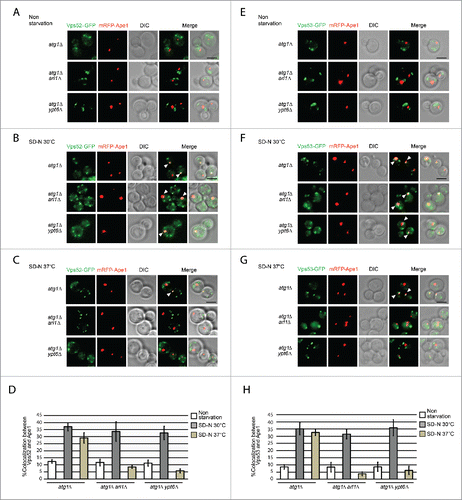
With respect to the results shown above, it seems feasible that a regulator of autophagy that interacts with both Arl1 and Ypt6 malfunctions at high temperature when either Arl1 or Ypt6 is missing, or at the permissive temperature when cells lack both Arl1 and Ypt6. We hypothesized that the Golgi-associated retrograde protein (GARP) complex is the effector that connects Arl1 and Ypt6 with autophagy. First, Arl1 and Ypt6 share the ability to bind to subunits of GARP, Vps53 and Vps52, respectively.Citation22 Second, GARP not only affects the successful delivery of cargo via the Cvt pathway and the sorting of Atg9 from the mitochondria (the deletion of VPS52 blocks Atg9 sorting from mitochondria),Citation49,50 but also recruits the syntaxin-like t-SNARE Tlg2 to the TGN.Citation24,51 Further, the deletion of TLG2 affects the magnitude of autophagy as well as the anterograde transport of Atg9.Citation52 As the autophagy defect in both the arl1Δ and ypt6Δ strains can only be detected at high temperature, it is possible that GARP can be properly localized at the normal temperature if cells lack one protein or the other, but not both. However, at the restrictive temperature, the increased membrane fluidity or some other property of the membranes causes GARP mislocalization because a single protein, Arl1 or Ypt6, is no longer sufficient to retain GARP at the proper location. To test this hypothesis, we tagged the Vps52 and Vps53 subunits with GFP to see whether they were translocated to the PAS during autophagy at the restrictive temperature when either Arl1 or Ypt6 was missing. As the results in demonstrate, Vps52-GFP colocalized with the PAS marker mRFP-Ape1 in the atg1Δ (YSA015), atg1Δ arl1Δ (YSA016) and atg1Δ ypt6Δ (YSA017) strains when autophagy was triggered ( compared to ) at the permissive temperature, suggesting that GARP was transferred to the PAS during autophagy. Similarly, Vps53-GFP colocalized with mRFP-Ape1 in atg1Δ (YSA018) atg1Δ arl1Δ (YSA019), and atg1Δ ypt6Δ (YSA020) strains under starvation conditions at the permissive temperature ( compared to ).
Figure 8. Arl1 and Ypt6 are required for GARP subunits Vps52 and Vps53 to be translocated to the PAS. ((A)– C) Fluorescence images of the yeast strains atg1Δ (YSA015), atg1Δ arl1Δ (YSA016) and atg1Δ ypt6Δ (YSA017) under nonstarvation conditions, starvation at 30°C and starvation at 37°C. Arrows point to the colocalization between Vps52-GFP and mRFP-Ape1. (D) The percentage of cells with colocalization between Vps52-GFP and mRFP-Ape1. At least 90 cells were counted for each strain at each condition. Error bars represent standard deviation from 3 biological replicates. ((E)– G) Fluorescence images of the yeast strain atg1Δ (YSA018), atg1Δ arl1Δ (YSA019) and atg1Δ ypt6Δ (YSA020) under nonstarvation conditions, starvation at 30°C and starvation at 37°C. Arrows point to the colocalization between Vps53-GFP and mRFP-Ape1. (H) The percentage of cells with colocalization between Vps53-GFP and mRFP-Ape1. At least 90 cells were counted for each strain under each condition. Error bars represent standard deviation from 3 biological replicates. Scale bar: 3 µm.
However, we found in both the atg1Δ arl1Δ and the atg1Δ ypt6Δ strains that there were significant decreases in the number of cells that had colocalization between Vps52 and prApe1 () or Vps53 and prApe1 () at high temperature compared with the permissive temperature. We also observed that Vps52 and Vps53 had both diffuse and punctate straining in both the atg1Δ arl1Δ and the atg1Δ ypt6Δ strains at both temperatures, indicating that some Vps52 and Vps53 subunits cannot be localized properly when cells lack Arl1 or Ypt6. Overall, these data suggest the transfer of GARP to the PAS is dependent on both Arl1 and Ypt6 at high temperature.
Discussion
Autophagy is an important aspect of a cell′s survival strategy against stressful conditions. This process relies intensively on the transport of membrane in order to form the autophagosome. One of the major questions is the sources of the membrane components that make up the autophagosome. To date, many membrane-bound organelles such as the ER,Citation53 the mitochondriaCitation54 and the Golgi apparatusCitation55 have been suggested to supply portions of the autophagosomal membrane. For example, many late secretory components localized in the TGN such as Sec4 and Arf1-Arf2 can be redirected and enable the network to send Golgi-derived vesicles to the PAS for the formation of the autophagosome.Citation13 These results demonstrate that components of the membrane traffic apparatus, such as the monomeric GTP-binding proteins and their effectors are important players in generating the membrane-bound compartments necessary for autophagy.
Previous work has elucidated roles for several monomeric GTP-binding proteins,Citation11 including some known to function in post-Golgi compartments, such as Ypt31-Ypt32.Citation13 In this study, we showed that 2 additional monomeric GTP-binding proteins that function in this area, Arl1 and Ypt6, also have roles in autophagy. However, their pattern in autophagy is significantly different from others that have been previously described. A simple knockout of one of these 2 genes did not impair autophagy at the permissive temperature, yet both arl1Δ and ypt6Δ strains showed completely defective autophagy at the restrictive temperature (). The defect appears to stem from an inability to form an intact autophagosome (). This temperature-sensitive phenotype indicates some essential machinery of the early stages of autophagy fails to function properly at high temperature when one of 2 genes, ARL1 or YPT6, has been deleted. Upon degradation of Arl1 in the ypt6Δ background, cells were unable to perform autophagy even at the permissive temperature.
The GTP-bound conformations of Arl1 and Ypt6 recruit their downstream effectors and regulate membrane trafficking in the cell. GTP-bound Arl1 interacts with Imh1, Gga1, and Vps53; whereas GTP-bound Ypt6 interacts with Vps52 and perhaps other proteins yet to be determined. In this study we confirmed that only the GTP-restricted alleles of Arl1 and Ypt6, Arl1Q72L and Ypt6Q69L, respectively, which are the ones known to function in membrane traffic, were able to rescue the autophagy defect at high temperature. These results also suggest the ability to recruit downstream effectors such as the GARP complex by GTP-bound Ypt6 and Arl1 is essential in autophagy.
In addition, we found the anterograde trafficking of Atg9 is impaired in both the arl1Δ and ypt6Δ strains at high temperature, the result of the defective formation of the autophagosome rather than defective fusion between the autophagosome and the vacuole, suggesting that Arl1 and Ypt6 participate in the recruitment of Atg9-containing vesicles to the PAS. In both the arl1Δ and ypt6Δ strains at high temperature, the anterograde trafficking of Atg9 to the PAS is adversely affected, leading to an overall block in autophagy because the autophagosomal membrane cannot be constructed. Moreover, using Sec7-DsRed as the TGN marker, we are able to confirm in either the arl1Δ or ypt6Δ strains, some fraction of Atg9 was trapped in the TGN, further suggesting that the TGN is one of the origins of the autophagosomal membrane.
For the most part, the arl1Δ and ypt6Δ strains showed similar phenotypes, although in some instances the phenotypes observed in the ypt6Δ strain were slightly more severe. One important difference we observed was with respect to the Cvt pathway. Although cells lacking either ARL1 or YPT6 have a block in the Cvt pathway under normal growth conditions (at the permissive temperature and in nitrogen-replete media), the defect in arl1Δ can be overcome through starvation treatment regardless of the temperature. In contrast, the defect in the ypt6Δ strain can only be recovered by starvation at 30°C not at 37°C (). Processing defects for prApe1 can be reversed by starvation,Citation50 where the autophagosomes supersede the ability of the Cvt vesicles to transport prApe1 to the vacuole. The fact that the ypt6Δ strain cannot transfer prApe1 to the vacuole under starvation conditions at the restrictive temperature is interesting because a previous study indicated prApe1 transits to the vacuole for processing even when autophagy is badly defective, such as in cells lacking Atg8,Citation56 so the precise role of Ypt6 in prApe1 traffic under starvation conditions remains to be resolved.
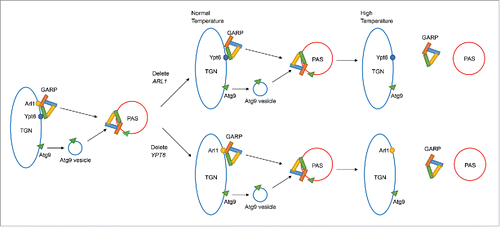
GARP is a multisubunit tethering complex that normally regulates the membrane trafficking between the endosome and the TGN by receiving cargo vesicles.Citation57 Some other tethering complexes in the secretory pathway have been shown to have roles in autophagy including the TRAPP (TRAnsport Protein Particle) complexCitation58 and the COG (conserved oligomeric Golgi) complex,Citation59 both of which help in the formation of the autophagosome. We tested whether the localization of the GARP complex is affected by loss of Arl1 or Ypt6 under conditions for autophagy. We found the localization of GARP subunits to the PAS required both Arl1 and Ypt6 at high temperature. Because GARP interacts with both Arl1 and Ypt6 and also functions in autophagy, this is consistent with our hypothesis that an important regulator of autophagy, which interacts with both Arl1 and Ypt6, fails to function properly at high temperature when one of the genes encoding these proteins has been deleted. Our model describing the roles of Arl1 and Ypt6 in autophagy is described in . Normally Arl1 and Ypt6 are localized at the TGN and assist in targeting the GARP complex to the PAS, which regulates the anterograde trafficking of the Atg9-containing vesicle for the formation of the autophagosome. At the permissive temperature, the loss of Arl1 or Ypt6 does not affect the targeting of the GARP complex; thus, Atg9 can still be transported to the PAS. However, at the restrictive temperature the transfer of the GARP complex to the PAS is inhibited upon the loss of either Arl1 or Ypt6 and the resulting defect of anterograde trafficking of Atg9 inhibits autophagosome formation. When both Arl1 and Ypt6 are missing, the cells are defective for autophagy even at the permissive temperature. In conclusion we demonstrate here that Arl1 and Ypt6, both monomeric GTP-binding proteins that function in TGN-endosome traffic, also play unique roles in autophagy. These roles are likely due to their interaction with and ability to target the GARP complex properly to the PAS to receive Atg9-containing vesicles to create the autophagosome.
Figure 9. Model for Arl1 and Ypt6 in autophagy. Arl1 and Ypt6 regulate the targeting of the GARP complex to the PAS and the anterograde trafficking of Atg9. At the permissive temperature, Arl1 or Ypt6 alone is sufficient for the transport of GARP and Atg9 to the PAS. However, at the restrictive temperature, a single protein, Arl1 or Ypt6, is insufficient for targeting GARP to the PAS. Therefore under increased temperature, the anterograde trafficking of Atg9 to the PAS is impaired.
Materials and methods
Strains, plasmids and reagents
Yeast strains and plasmids used in this study are summarized in Tables S1 and S2. All yeast transformations were done with the lithium acetate method.Citation60 The deletion mutant strains atg1Δ, ypt6Δ, arl1Δ, ypt7Δ, and rpe1/pos18Δ were obtained from the homozygous diploid deletion collection developed by the Saccharomyces Genome Deletion Project.Citation61 Strains for the Pho8Δ60 assay were constructed as previously described.Citation30 To integrate mRFP-Ape1 (prApe1 tagged by the monomeric red fluorescent protein), Atg9-3xGFP, and Sec7-DsRed into the yeast genome, the integration plasmid pRFP-Ape1 (pRS305-mRFP-APE1)Citation62 (a gift of Daniel Klionsky, University of Michigan), pAtg9-3×GFP (pRS306-ATG9-3×GFP)Citation63 (a gift of Daniel Klionsky) and YIplac204-TC-Sec7-6xDsred-M1Citation64 were linearized by restriction enzymes AvrII, BglII and EcoNI, respectively. Homologous recombination was performed for integration into the APE1, ATG9, or SEC7 locus, respectively. To tag Vps52 and Vps53 with GFP, the GFP fragment bounded by the TRP1 gene was amplified from the plasmid pFA6a-GFP(S65T)-TRP1Citation65 with the gene flanking regions of the C-terminal regions of VPS52 and VPS53. Homologous recombination was performed for integration into the genome. In each instance, correct recombination was verified by PCR using diagnostic primers for the appropriate integration. All the chromosomal gene deletions were accomplished through PCR amplification of the nutritional markers HIS3 or URA3 from the plasmid pRS313 or pRS316 with gene flanking regions for recombination. Marker swap experiments were done as described previously.Citation66
To clone YPT6 into the pRS316 plasmid, a 1600-base pair fragment containing the YPT6 gene with its 5′ and 3′ untranslated regions was amplified from the yeast genome by using oligonucleotides with either a SacI or ClaI restriction site. The PCR product and the pRS316 plasmid were digested with SacI and ClaI. The digested products were ligated with T4 DNA ligase and transformed into E. coli. The resulting pRS316-YPT6 plasmid was confirmed by sequencing. YPT6 (Q69L) and YPT6 (T24N) were made by PCR-based site-directed mutagenesis, using the Q5 Site-Directed Mutagenesis Kit from NEB (NEB, E0554S), following the manufacturer's protocol. The presence of the desired point mutations was confirmed by sequencing. Sequencing here and elsewhere was performed by Genewiz (South Plainfield, NJ).
For construction of the YPT6 ARL1 conditional double-knockout strain (ypt6Δ arl1AID), the TIR1 gene from Oryza sativa was integrated into the SSN6 locus of the ypt6Δ strain by cutting the plasmid pST1868Citation67 with BbvCI and performing homologous recombination. To tag Arl1 with AID, the fragment of 3xmini AID-5xFLAG along with the KanMX gene was amplified from the plasmid pST1933Citation67 with the primers containing gene flanking regions of the C-terminal region of ARL1. Homologous recombination was performed to integrate the fragment into the C terminus of ARL1.
Antibodies used were anti-GFP primary antibody from mouse (Roche Diagnostics, 11814460001); anti-Pgk1/phosphoglycerate kinase-1 antibody from mouse (Molecular Probes, A6457); anti-Ape1 antibody from rabbit (the gifts of Yoshinori Ohsumi, Tokyo Institute of Technology and Susan Ferro-Novick, University of California, San Diego), anti-FLAG antibody from mouse (Clontech, 635691), anti-mouse IgG horseradish peroxidase-linked secondary antibody from sheep (GE Healthcare, NA931) and anti-rabbit IgG horseradish peroxidase-linked secondary antibody from donkey (GE Healthcare, NA934). The enhanced chemiluminescence (ECL) prime kit was from GE Healthcare (RPN2236).
All chemical reagents were from Sigma-Aldrich, unless otherwise noted. FM 4–64 (N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide) (T3166) and geneticin (G418) (11811023) were from Thermo Scientific. Restriction enzymes were from New England Biolabs. Phusion High-Fidelity DNA Polymerase was from Thermo Scientific (F-534S). Zymolyase 20T was from Sunrise (320921). Molecular biology grade trypsin was from Roche (11047841001).
Yeast culture condition and the induction of autophagy
Yeast strains were grown in YPD (1% yeast extract, 2% peptone, and 2% glucose) or synthetic dropout (SD) media (2% glucose, 0.67% yeast nitrogen base without amino acids, supplemented with appropriate nutrients and magnesium sulfate).Citation68 Experiments to induce autophagy by nitrogen starvation were performed in SD medium lacking nitrogen (SD-N; 2% glucose, 0.17% yeast nitrogen base without amino acids, ammonium sulfate, or vitamins). All chemicals for media were from Fisher Scientific except yeast nitrogen base (Sunrise, 1500–500).
Yeast strains were normally grown at permissive temperature (30°C) in YPD medium or SD medium with appropriate supplements to maintain selection for the plasmids. To induce autophagy, yeast strains were grown until log phase (OD600 = 0.6), incubated at either 30°C or 37°C in nonstarvation conditions for 30 min before washing twice in SD-N medium and further incubation in SD-N at the same temperature for 3 h. In some experiments, cultures were returned to 30°C for 3 h after incubation at 37°C (see appropriate figure legends).
Western blots
Proteins were extracted from yeast cells by using trichloroacetic acid (TCA) precipitation. Cells were collected, washed with 20% TCA then the pellet fractions were resuspended in 150 μl 20% TCA and subjected to glass bead lysis. The supernatant fractions were collected, and glass beads were subsequently washed with 300 μl 5% TCA. Proteins were pelleted by centrifugation, washed once with 100% ethanol, and solubilized in 90 μl 3× SDS-PAGE sample buffer (60 mM Tris, pH 6.8, 10% glycerol, 100 mM DTT, 0.2% bromophenol blue, 2% SDS) plus 60 μl 1 M Tris, pH 8.0. After incubating at 95°C for 20 min, the insoluble components were removed by centrifugation (10 min, 10,000 × g) and the supernatant fractions were subjected to Western blot. The total proteins from 0.8 OD600 units of the cells (1 ml of cells at OD600 = 1.0 corresponds to 1 unit) were separated on 4–15% pre-cast polyacrylamide gels (Bio-Rad, 456–9036), transferred to nitrocellulose membranes (Thermo Scientific, 88018), and subjected to protein detection using specific primary antibodies, followed by HRP-conjugated secondary antibodies (GE Healthcare, NA931 or NA934. Protein bands were visualized by using ECL Prime kit (GE Healthcare, RPN2236) and detected with an ImageQuant LAS 4010 imager (GE Healthcare). Each set of western blot experiments was repeated at least twice. Representative examples are shown in each figure.
Live-cell fluorescence microscopy
Yeast strains with GFP-tagged, RFP-tagged, or DsRed-tagged proteins were collected from rich medium (YPD or SD medium lacking the appropriate nutrients) or SD-N medium at different growth temperatures and washed once with water before imaging. To stain vacuoles, FM 4–64 was added to a final concentration of 1.6 μM for one h before visualization at 30°C or 37°C. Cells were visualized with a Zeiss AxioImager M2 florescence microscopy system using a 63× oil lens. Images were captured and deconvolved using Volocity 6.3 (PerkinElmer) software. Each set of fluorescence microscopy experiments was repeated at least twice.
Pho8Δ60 assay
The Pho8Δ60 assay to quantify the magnitude of autophagy was performed as previously described.Citation30 Briefly, cell lysates from 0.5 OD600 units of cells were incubated with 5.5 mM α-naphthyl phosphate (disodium salt; Sigma-Aldrich, N7255) for 20 min at 30°C in reaction buffer (250 mM Tris-HCl, pH 9.0, 10 mM MgSO4, 10 µM ZnSO4). The reaction was stopped with an equal volume of 2 M glycine-NaOH, pH 11.0. Fluorescence emissions of the product 1-napthol were measured (λex = 330, λem = 472) by using a GloMax plate reader (Promega) with a UV filter. Protein concentrations were determined by the Bradford assay.Citation69 All experiments were repeated 3 times with triplicate samples.
Precursor Ape1 processing assay
Cells were grown at 30°C until OD600 = 0.6. Autophagy was induced as described. Five ODs of cells from time 0 and 3 h at 30°C, and 3 h at 37°C were collected. The cells were washed with water and 50 μl of the 2x protein sample buffer was added along with glass beads. All the samples were vortexed for 5 min and another 50 μl of the sample buffer was added. All the samples were boiled for 10 min and centrifuged. The supernatant fractions were collected and subsequently analyzed by western blot by using an anti-Ape1 antibody.
GFP-Atg8 proteinase protection assay
The GFP-Atg8 proteinase protection assay was modified from that previously described.Citation40 Briefly, after nitrogen starvation, cells were collected and washed once with DTT buffer (10 mM Tris-HCl, pH 9.4, 10 mM DTT). Then cells were incubated in DTT buffer for 15 min at 37°C, pelleted by centrifugation, then resuspended in SP buffer (1 M sorbitol [Sigma-Aldrich, S1876], 20 mM PIPES, pH 6.8). Zymolyase 20T was added to a final concentration of 0.4 mg/ml and cells were incubated for 25 min. The resulting spheroplasts were pelleted and hypotonically lysed in SP200 buffer (20 mM PIPES, pH 6.8, 200 mM sorbitol, 5 mM MgCl2.). Two centrifugation steps, the first at 5000 × g for 5 min and collecting the supernatant fraction, and the second at 10,000 × g for 10 min and collecting the pellet fraction, were performed to remove unbroken cells and debris. The supernatant fractions were divided into 3 parts: untreated; treated with 10 μg/ml trypsin; and treated with 10 μg/ml tryspin in the presence of 0.4% Triton X-100 (Sigma-Aldrich, T9284). Samples were incubated in 30°C for 25 min. The reactions were stopped by the addition of 100% TCA to the final concentration of 16.7%. Proteins were precipitated on ice for 10 min and collected by centrifugation (10 min, 10,000 × g). Protein pellets were washed twice with ice-cold 100% acetone. The pellets were dried and resuspended in 2× protein sample buffer. Proteins were separated on 4–15% Tris-HCl gels as described above for western blots. Free GFP and GFP-Atg8 were detected using the anti-GFP antibody as described above.
Auxin-induced degradation of Arl1
The YPT6 ARL1 conditional double-knockout strain (ypt6Δ arl1AID) was cultured until OD600 = 0.6 and divided into 2 portions. One portion was cultured with 1 mM NAA (Sigma, N0640) for 30 min, another one was with 0.17% ethanol alone. Autophagy was induced as described. After the 3 h in SD-N medium, the portion with 1 mM NAA was washed 3 times with SD-N medium to wash out the NAA and cells were cultured in SD-N medium for an additional 3 h in the absence of added NAA. All the samples were collected and subjected to either protein gel blot with the anti-GFP antibody and anti-FLAG antibody, or live-cell fluorescence microscopy.
Abbreviations
| AID | = | auxin-inducible degron |
| Arl | = | ADP-ribosylation factor-like |
| Atg | = | autophagy-related |
| CFUs | = | colony forming units |
| Cvt | = | cytoplasm-to-vacuole targeting |
| COG | = | conserved oligomeric Golgi |
| DMSO | = | dimethyl sulfoxide |
| DTT | = | dithiothreitol |
| ER | = | endoplasmic reticulum |
| FM 4–64 | = | N-(3-triethylammoniumpropyl)-4-(6-(4-(diethylamino) phenyl) hexatrienyl) pyridinium dibromide |
| G418 | = | geneticin |
| GARP | = | Golgi-associated retrograde protein complex |
| GDP | = | guanosine diphosphate |
| GFP | = | green fluorescent protein |
| GTP | = | guanosine triphosphate |
| mRFP | = | monomeric red fluorescent protein |
| NAA | = | 1-naphthaleneacetic acid |
| PAS | = | phagophore assembly site |
| PE | = | phosphatidylethanolamine |
| Pho | = | phosphate metabolism |
| prAms1 | = | precursor of α-mannosidase |
| prApe1 | = | precursor of aminopeptidase I |
| prApe4 | = | precursor of aspartyl aminopeptidase |
| Rab | = | Ras-related in brain |
| SNARE | = | soluble NSF attachment protein receptor |
| TCA | = | trichloroacetic acid |
| TGN | = | trans-Golgi network |
| Tlg | = | t-SNARE affecting a late Golgi compartment |
| TOR | = | target of rapamycin |
| TRAPP | = | TRAnsport Protein Particle |
| Vps | = | vacuolar protein sorting |
| WT | = | wild type |
| Ypt | = | yeast protein |
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
KAUP_1196316_Supplemental_Figures_and_Tables.zip
Download Zip (5 MB)Acknowledgments
We would like to thank Drs. Daniel Klionsky, Yoshinori Ohsumi and Susan Ferro-Novick for materials including plasmids and antibodies. We also thank Drs. Ronda Rolfes and Peter Williamson for advice about the work. We thank the members of Dr. Maria Donoghue's lab for help with microscopy. We thank the members of the Rolfes and Rosenwald laboratory for their critical review of the manuscript, especially Dr. Erica Gerace.
References
- Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol 2010; 221:3-12; PMID:20225336; http://dx.doi.org/10.1002/path.2697
- Yang Z, Klionsky DJ. Eaten alive: a history of macroautophagy. Nat Cell Biol 2010; 12:814-22; PMID:20811353; http://dx.doi.org/10.1038/ncb0910-814
- Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ 2009; 16:57-69; PMID:18772897; http://dx.doi.org/10.1038/cdd.2008.130
- Klionsky DJ, Cregg JM, Dunn WA, Jr., Emr SD, Sakai Y, Sandoval IV, et al. A unified nomenclature for yeast autophagy-related genes. Dev Cell 2003; 5:539-45; PMID:14536056; http://dx.doi.org/10.1016/S1534-5807(03)00296-X
- Klionsky DJ, Cueva R, Yaver DS. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J Cell Biol 1992; 119:287-99; PMID:1400574; http://dx.doi.org/10.1083/jcb.119.2.287
- Hutchins MU, Klionsky DJ. Vacuolar localization of oligomeric α-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J Biol Chem 2001; 276:20491-8; PMID:11264288; http://dx.doi.org/10.1074/jbc.M101150200
- Yuga M, Gomi K, Klionsky DJ, Shintani T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J Biol Chem 2011; 286:13704-13; PMID:21343297; http://dx.doi.org/10.1074/jbc.M110.173906
- Lynch-Day MA, Klionsky DJ. The Cvt pathway as a model for selective autophagy. FEBS Lett 2010; 584:1359-66; PMID:20146925; http://dx.doi.org/10.1016/j.febslet.2010.02.013
- Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell 2012; 151:1501-12; PMID:23219485; http://dx.doi.org/10.1016/j.cell.2012.11.028
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagy from yeast to human. Autophagy 2007; 3:181-206; PMID:17224625; http://dx.doi.org/10.4161/auto.3678
- Yang S, Rosenwald AG. The roles of monomeric GTP-binding proteins in macroautophagy in Saccharomyces cerevisiae. Int J Mol Sci 2014; 15:18084-101; PMID:25302616; http://dx.doi.org/10.3390/ijms151018084
- Wang J, Menon S, Yamasaki A, Chou HT, Walz T, Jiang Y, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci U S A 2013; 110:9800-5; PMID:23716696; http://dx.doi.org/10.1073/pnas.1302337110
- Geng J, Nair U, Yasumura-Yorimitsu K, Klionsky DJ. Post-Golgi Sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol Biol Cell 2010; 21:2257-69; PMID:20444978; http://dx.doi.org/10.1091/mbc.E09-11-0969
- Kirisako T, Baba M, Ishihara N, Miyazawa K, Ohsumi M, Yoshimori T, et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J Cell Biol 1999; 147:435-46; PMID:10525546; http://dx.doi.org/10.1083/jcb.147.2.435
- van der Vaart A, Griffith J, Reggiori F. Exit from the Golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol Biol Cell 2010; 21:2270-84; PMID:20444982; http://dx.doi.org/10.1091/mbc.E09-04-0345
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, et al. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell 2001; 12:3690-702; PMID:11694599; http://dx.doi.org/10.1091/mbc.12.11.3690
- Rosenwald AG, Rhodes MA, Van Valkenburgh H, Palanivel V, Chapman G, Boman A, et al. ARL1 and membrane traffic in Saccharomyces cerevisiae. Yeast 2002; 19:1039-56; PMID:12210899; http://dx.doi.org/10.1002/yea.897
- Lee FJ, Huang CF, Yu WL, Buu LM, Lin CY, Huang MC, et al. Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae. J Biol Chem 1997; 272:30998-1005; PMID:9388248; http://dx.doi.org/10.1074/jbc.272.49.30998
- Li B, Warner JR. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J Biol Chem 1996; 271:16813-9; PMID:8663225; http://dx.doi.org/10.1074/jbc.271.28.16813
- Singer-Kruger B, Lasic M, Burger AM, Hausser A, Pipkorn R, Wang Y. Yeast and human Ysl2p/hMon2 interact with Gga adaptors and mediate their subcellular distribution. EMBO J 2008; 27:1423-35; PMID:18418388
- Liu YW, Lee SW, Lee FJ. Arl1p is involved in transport of the GPI-anchored protein Gas1p from the late Golgi to the plasma membrane. J Cell Sci 2006; 119:3845-55; PMID:16926193; http://dx.doi.org/10.1242/jcs.03148
- Panic B, Whyte JR, Munro S. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr Biol 2003; 13:405-10; PMID:12620189; http://dx.doi.org/10.1016/S0960-9822(03)00091-5
- Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, et al. Global mapping of the yeast genetic interaction network. Science 2004; 303:808-13; PMID:14764870; http://dx.doi.org/10.1126/science.1091317
- Siniossoglou S, Pelham HR. An effector of Ypt6p binds the SNARE Tlg1p and mediates selective fusion of vesicles with late Golgi membranes. EMBO J 2001; 20:5991-8; PMID:11689439; http://dx.doi.org/10.1093/emboj/20.21.5991
- Lafourcade C, Galan JM, Gloor Y, Haguenauer-Tsapis R, Peter M. The GTPase-activating enzyme Gyp1p is required for recycling of internalized membrane material by inactivation of the Rab/Ypt GTPase Ypt1p. Mol Cell Biol 2004; 24:3815-26; PMID:15082776; http://dx.doi.org/10.1128/MCB.24.9.3815-3826.2004
- Brown EJ, Albers MW, Shin TB, Ichikawa K, Keith CT, Lane WS, et al. A mammalian protein targeted by G1-arresting rapamycin-receptor complex. Nature 1994; 369:756-8; PMID:8008069; http://dx.doi.org/10.1038/369756a0
- Bugnicourt A, Mari M, Reggiori F, Haguenauer-Tsapis R, Galan JM. Irs4p and Tax4p: two redundant EH domain proteins involved in autophagy. Traffic 2008; 9:755-69; PMID:18298591; http://dx.doi.org/10.1111/j.1600-0854.2008.00715.x
- Matsuura A, Tsukada M, Wada Y, Ohsumi Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 1997; 192:245-50; PMID:9224897; http://dx.doi.org/10.1016/S0378-1119(97)00084-X
- Xie Z, Nair U, Klionsky DJ. Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 2008; 19:3290-8; PMID:18508918; http://dx.doi.org/10.1091/mbc.E07-12-1292
- Noda T, Klionsky DJ. The quantitative Pho8Delta60 assay of nonspecific autophagy. Methods Enzymol 2008; 451:33-42; PMID:19185711; http://dx.doi.org/10.1016/S0076-6879(08)03203-5
- Kahn RA, Clark J, Rulka C, Stearns T, Zhang CJ, Randazzo PA, et al. Mutational analysis of Saccharomyces cerevisiae ARF1. J Biol Chem 1995; 270:143-50; PMID:7814365; http://dx.doi.org/10.1074/jbc.270.1.143
- Dascher C, Balch WE. Dominant inhibitory mutants of ARF1 block endoplasmic reticulum to Golgi transport and trigger disassembly of the Golgi apparatus. J Biol Chem 1994; 269:1437-48; PMID:8288610
- Jones S, Jedd G, Kahn RA, Franzusoff A, Bartolini F, Segev N. Genetic interactions in yeast between Ypt GTPases and Arf guanine nucleotide exchangers. Genetics 1999; 152:1543-56; PMID:10430582
- Lee MT, Mishra A, Lambright DG. Structural mechanisms for regulation of membrane traffic by rab GTPases. Traffic 2009; 10:1377-89; PMID:19522756; http://dx.doi.org/10.1111/j.1600-0854.2009.00942.x
- Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 1989; 77:51-9; PMID:2744487; http://dx.doi.org/10.1016/0378-1119(89)90358-2
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol 1995; 128:779-92; PMID:7533169; http://dx.doi.org/10.1083/jcb.128.5.779
- Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J 2001; 20:5971-81; PMID:11689437; http://dx.doi.org/10.1093/emboj/20.21.5971
- Bonangelino CJ, Chavez EM, Bonifacino JS. Genomic screen for vacuolar protein sorting genes in Saccharomyces cerevisiae. Mol Biol Cell 2002; 13:2486-501; PMID:12134085; http://dx.doi.org/10.1091/mbc.02-01-0005
- Bensen ES, Yeung BG, Payne GS. Ric1p and the Ypt6p GTPase function in a common pathway required for localization of trans-Golgi network membrane proteins. Mol Biol Cell 2001; 12:13-26; PMID:11160819; http://dx.doi.org/10.1091/mbc.12.1.13
- Nair U, Thumm M, Klionsky DJ, Krick R. GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy 2011; 7:1546-50; PMID:22108003; http://dx.doi.org/10.4161/auto.7.12.18424
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 2009; 6:917-22; PMID:19915560; http://dx.doi.org/10.1038/nmeth.1401
- Noda T, Kim J, Huang WP, Baba M, Tokunaga C, Ohsumi Y, et al. Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J Cell Biol 2000; 148:465-80; PMID:10662773; http://dx.doi.org/10.1083/jcb.148.3.465
- Reggiori F, Shintani T, Nair U, Klionsky DJ. Atg9 cycles between mitochondria and the pre-autophagosomal structure in yeasts. Autophagy 2005; 1:101-9; PMID:16874040; http://dx.doi.org/10.4161/auto.1.2.1840
- Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol 2010; 190:1005-22; PMID:20855505; http://dx.doi.org/10.1083/jcb.200912089
- Shintani T, Suzuki K, Kamada Y, Noda T, Ohsumi Y. Apg2p functions in autophagosome formation on the perivacuolar structure. J Biol Chem 2001; 276:30452-60; PMID:11382761; http://dx.doi.org/10.1074/jbc.M102346200
- Guan J, Stromhaug PE, George MD, Habibzadegah-Tari P, Bevan A, Dunn WA, Jr., et al. Cvt18/Gsa12 is required for cytoplasm-to-vacuole transport, pexophagy, and autophagy in Saccharomyces cerevisiae and Pichia pastoris. Mol Biol Cell 2001; 12:3821-38; PMID:11739783; http://dx.doi.org/10.1091/mbc.12.12.3821
- Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Dev Cell 2004; 6:79-90; PMID:14723849; http://dx.doi.org/10.1016/S1534-5807(03)00402-7
- Tooze SA, Yoshimori T. The origin of the autophagosomal membrane. Nat Cell Biol 2010; 12:831-5; PMID:20811355; http://dx.doi.org/10.1038/ncb0910-831
- Reggiori F, Klionsky DJ. Atg9 sorting from mitochondria is impaired in early secretion and VFT-complex mutants in Saccharomyces cerevisiae. J Cell Sci 2006; 119:2903-11; PMID:16787937; http://dx.doi.org/10.1242/jcs.03047
- Reggiori F, Wang CW, Stromhaug PE, Shintani T, Klionsky DJ. Vps51 is part of the yeast Vps fifty-three tethering complex essential for retrograde traffic from the early endosome and Cvt vesicle completion. J Biol Chem 2003; 278:5009-20; PMID:12446664; http://dx.doi.org/10.1074/jbc.M210436200
- Dulubova I, Yamaguchi T, Gao Y, Min SW, Huryeva I, Sudhof TC, et al. How Tlg2p/syntaxin 16 ‘snares’ Vps45. EMBO J 2002; 21:3620-31; PMID:12110575; http://dx.doi.org/10.1093/emboj/cdf381
- Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, et al. SNARE proteins are required for macroautophagy. Cell 2011; 146:290-302; PMID:21784249; http://dx.doi.org/10.1016/j.cell.2011.06.022
- Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy 2009; 5:1180-5; PMID:19855179; http://dx.doi.org/10.4161/auto.5.8.10274
- Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, et al. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell 2010; 141:656-67; PMID:20478256; http://dx.doi.org/10.1016/j.cell.2010.04.009
- Geng J, Klionsky DJ. The Golgi as a potential membrane source for autophagy. Autophagy 2010; 6:950-1; PMID:20729630; http://dx.doi.org/10.4161/auto.6.7.13009
- Abeliovich H, Dunn WA, Jr., Kim J, Klionsky DJ. Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J Cell Biol 2000; 151:1025-34; PMID:11086004; http://dx.doi.org/10.1083/jcb.151.5.1025
- Bonifacino JS, Hierro A. Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol 2011; 21:159-67; PMID:21183348; http://dx.doi.org/10.1016/j.tcb.2010.11.003
- Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A 2010; 107:7811-6; PMID:20375281; http://dx.doi.org/10.1073/pnas.1000063107
- Yen WL, Shintani T, Nair U, Cao Y, Richardson BC, Li Z, et al. The conserved oligomeric Golgi complex is involved in double-membrane vesicle formation during autophagy. J Cell Biol 2010; 188:101-14; PMID:20065092; http://dx.doi.org/10.1083/jcb.200904075
- Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res 1992; 20:1425; PMID:1561104; http://dx.doi.org/10.1093/nar/20.6.1425
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 1999; 285:901-6; PMID:10436161; http://dx.doi.org/10.1126/science.285.5429.901
- He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol 2006; 175:925-35; PMID:17178909; http://dx.doi.org/10.1083/jcb.200606084
- Monastyrska I, He C, Geng J, Hoppe AD, Li Z, Klionsky DJ. Arp2 links autophagic machinery with the actin cytoskeleton. Mol Biol Cell 2008; 19:1962-75; PMID:18287533; http://dx.doi.org/10.1091/mbc.E07-09-0892
- Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS. Golgi maturation visualized in living yeast. Nature 2006; 441:1002-6; PMID:16699524; http://dx.doi.org/10.1038/nature04717
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 1998; 14:953-61; PMID:9717241; http://dx.doi.org/10.1002/(SICI)1097-0061(199807)14:10%3c953::AID-YEA293%3e3.0.CO;2-U
- Cross FR. 'Marker swap' plasmids: convenient tools for budding yeast molecular genetics. Yeast 1997; 13:647-53; PMID:9200814; http://dx.doi.org/10.1002/(SICI)1097-0061(19970615)13:7%3c647::AID-YEA115%3e3.0.CO;2-
- Tanaka S, Miyazawa-Onami M, Iida T, Araki H. iAID: an improved auxin-inducible degron system for the construction of a ‘tight’ conditional mutant in the budding yeast Saccharomyces cerevisiae. Yeast 2015; 32:567-81; PMID:26081484; http://dx.doi.org/10.1002/yea.3080
- Green SR, Moehle CM. Media and culture of yeast. Curr Protoc Cell Biol 2001; Chapter 1:Unit 1 6; PMID:18228294
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 1976; 72:248-54; PMID:942051; http://dx.doi.org/10.1016/0003-2697(76)90527-3
- Juhnke H, Krems B, Kotter P, Entian KD. Mutants that show increased sensitivity to hydrogen peroxide reveal an important role for the pentose phosphate pathway in protection of yeast against oxidative stress. Mol Gen Genet 1996; 252:456-64; PMID:8879247; http://dx.doi.org/10.1007/BF02173011
