ABSTRACT
Macroautophagy/autophagy is a catabolic process that is essential for cellular homeostasis. How autophagosomal vesicle forms in a spatio-temporally regulated manner remains elusive. Our recent study revealed that small GTPase, RAB37 (RAB37, member RAS oncogene family), functions as a key organizer of autophagosomal membrane biogenesis. RAB37 interacts with ATG5 (autophagy related 5) and promotes autophagosome formation by modulating ATG12–ATG5-ATG16L1 complex assembly. These findings provide new insights into autophagy regulation.
Autophagy is a catabolic process that is essential to maintaining cellular homeostasis. Dysregulation of autophagy is implicated in multiple human diseases, including cancer, neurodegenerative diseases, metabolic disorders and immune diseases. A fundamental question pertaining to autophagy is how the phagophore matures into a double-membrane autophagosomal vesicle. While the core machinery of autophagy, including the complex ATG12–ATG5-ATG16L1, is key for autophagosome membrane biogenesis, the molecular mechanisms of autophagy regulation remain elusive.
Autophagosome formation is actually a membrane-trafficking process. Intracellular membrane-trafficking involves vesicle budding, growth, motility, tethering and fusion with the specific target membrane. Small GTPases regulate membrane functions through a switch between 2 distinct conformations: the GTP (guanosine triphosphate)-bound (active) and the GDP (guanosine diphosphate)-bound (inactive) forms. To explore regulatory mechanisms of the formation of the autophagosome membrane, we investigated ATG proteins interacting with a small GTPase, RAB37, a member of the RAB family. Co-immunoprecipitation and confocal fluorescence microscopy were first used to identify RAB37-interacting ATG proteins, which showed that RAB37 is associated with ATG5 and ATG12–ATG5. Yeast 2-hybrid and GST-affinity isolation assays confirmed that RAB37 directly interacts with ATG5. As GTP-γ-S is a nonhydrolyzable GTP analog, GTP-γ-S assays were used to test whether RAB37 interaction with ATG5 is dependent on its GTP-binding function. The GTP-γ-S assay revealed that the direct interaction is dependent on GTP-binding of RAB37.
Two RAB37 mutants, a GTP-hydrolysis-deficient, constitutively active mutant (RAB37Q89L) and a GDP-stabilized, constitutively negative mutant (RAB37T43N), were used to test whether RAB37 functions as a GTPase. The mutation analysis confirmed that RAB37 interacts with ATG5 and localizes on autophagosomes in a GTP-dependent manner. In addition, RAB37 colocalization with an omegasome marker—ZFYVE1/DFCP1 (zinc finger FYVE-type containing 1), a PtdIns3P-binding ER protein—and ATG5 knockdown assays indicated that RAB37 puncta are colocalized with phagophore membranes in an ATG5-dependent manner.
To test a role of RAB37 in formation of the ATG12–ATG5-ATG16L1 complex, we established RAB37 knockdown HeLa cell lines. Co-immunoprecipitation indicated that ATG5 interacts with RAB37 and ATG16L1 respectively, and RAB37 knockdown inhibits interaction of the ATG12–ATG5 conjugate with ATG16L1. Immunofluorescence analysis revealed that RAB37 knockdown inhibits ATG5 colocalization with ATG16L1, whereas RAB37 overexpression promotes the colocalization. These data suggested a role of RAB37 in formation of the ATG12–ATG5-ATG16L1 complex. Two biochemical approaches were adopted to further investigate the role of RAB37 in formation of the ATG12–ATG5-ATG16L1 complex. First, sucrose density gradient centrifugation of the cell lysates (RAB37 overexpression, RAB37 knockdown and wild-type HeLa cells) followed by western blots showed that RAB37 knockdown decreases the molecular weight of the ATG12–ATG5-ATG16L1 complex, whereas RAB37 overexpression promotes complex formation. Second, gel filtration was used to confirm the role of RAB37 in ATG12–ATG5-ATG16L1 complex assembly. Gel filtration from cell lysis of RAB37 knockdown and mutations showed that RAB37 participates in formation of an ∼800-kDa multimeric protein complex, including approximately 8 sets of ATG12–ATG5-ATG16L1. We then investigated whether the complex formation is dependent on GTP-binding of RAB37. The constitutively active mutant increases the molecular size of the complex, whereas the constitutively negative mutant interferes with complex formation. These data suggested that RAB37 promotes formation of the ATG12–ATG5-ATG16L1 complex in a GTP-dependent manner, thus facilitating lipidation of MAP1LC3B/LC3B (microtubule-associated protein 1 light chain 3 beta) during autophagosome formation.
Autophagy flux was further tested using RAB37 knockdown and forced expression. Western blot analysis showed that RAB37 knockdown inhibits LC3B-II formation, whereas overexpression promotes LC3B-II formation upon starvation induction, and LC3B-II accumulates during bafilomycin A1 treatment. Autophagy flux tests using a tandem fluorescent indicator, mCherry-GFP-LC3, in RAB37 overexpression and knockdown cell lines, indicated that RAB37 knockdown inhibits formation of autophagosomes, whereas overexpression promotes the formation. Bafilomycin A1 treatment leads to an accumulation of autophagosomes in RAB37 overexpression cells. These data suggested that RAB37 promotes autophagosome formation, but does not affect its degradation process via lysosomes.
Because autophagy suppression can promote tumorigenesis, roles of RAB37 in autophagy regulation can be verified through tumorigenesis studies in vivo. The cells stably expressing RAB37 and miRNA to knock down RAB37 were used to implant nude mice. As expected, RAB37 knockdown promotes tumor growth, whereas RAB37 overexpression inhibits tumor growth. In RAB37 knocked-down tumors, LC3B-II protein is downregulated, whereas the protein is upregulated in RAB37-overexpressing tumors. Taken together, these data demonstrate a role of RAB37 in promoting autophagy.
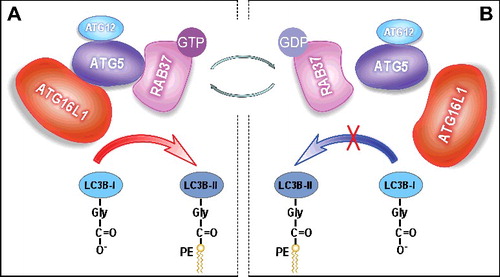
In summary, we demonstrate that RAB37 acts as a key organizer of autophagosomal membrane biogenesis. Upon autophagy induction, the active RAB37-GTP interacts directly with ATG5, and promotes formation of an ∼800-kDa multimeric ATG protein complex, which includes approximately 8 sets of ATG12–ATG5-ATG16L1. These protein complexes probably form scaffolds around the growing phagophore. Subsequently, the ATG12–ATG5-ATG16L1 complex recruits and lipidates LC3B-I to form active LC3B-II, which accelerates autophagosome formation (A). As a small GTPase, RAB37 can shift through a switch between 2 distinct conformations: the GTP-bound “on” and the GDP-bound “off” forms. RAB37-GTP can be transformed into the inactive RAB37-GDP through a GTPase activating protein. RAB37-GDP disassociates from ATG5, which destabilizes the ATG12–ATG5-ATG16L1 complex, and thus the autophagosome formation process is prevented (B). RAB37-GDP disassociates from the autophagosome membrane and spreads to the cytosol for recycling by a guanine nucleotide exchange factor to organize another initiation process of autophagosomal formation following autophagy induction. Further studies should uncover the regulatory mechanisms of recycling of RAB37 between 2 distinct conformations, GTP-bound “on” and GDP-bound “off”. Guanine nucleotide exchange factors probably activate RAB37 by stimulating GDP-release for GTP-binding. Conversely, GTPase-activating proteins could promote RAB37 inactivation via GTP-hydrolysis, which will lead to the inactive state RAB37-GDP. Further studies are therefore to identify these 2 types of proteins and uncover their regulatory mechanisms in autophagosomal formation. Implications of RAB37 in autophagy regulation should also be explored under various physiological and pathological conditions.
Figure 1. RAB37 regulates autophagosome formation by modulating ATG12–ATG5-ATG16L1 complex assembly. RAB37 as a key organizer participates in the process of ATG12–ATG5-ATG16L1 complex formation and promotes autophagy upon starvation induction in a GTP-dependent manner. RAB37, as a novel GTPase, regulates membrane functions through a switch between 2 distinct conformations: the GTP-bound “on” and the GDP-bound “off” forms. (A) RAB37-GTP interacts directly with ATG5 and promotes interaction of the ATG12–ATG5 conjugate with ATG16L1. The multimeric RAB37- ATG12–ATG5-ATG16L1 complex recruits and lipidates LC3B-I to form active LC3B-II, which accelerates autophagosomal formation. (B) RAB37-GTP can transform into the inactive RAB37-GDP probably through a GTPase activating protein. RAB37-GDP impairs its interaction with ATG5 and disassociates from ATG12–ATG5, then leads to separation of ATG12–ATG5 from ATG16L1. Finally, LC3B-I lipidation is prevented. Simultaneously, RAB37-GDP spreads to the cytosol for recycling via a guanine nucleotide exchange factor to organize another initiation process of autophagosomal formation upon autophagy induction.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
