ABSTRACT
In eukaryotes, most proteins are degraded through one of the 2 major proteolytic pathways: the ubiquitin-proteasome system (UPS) and macroautophagy/autophagy. Existing evidence suggests that these processes are critical to human physiology and pathology. Our study revealed a negative feedback system between proteasomal activity and autophagic flux in cells. We demonstrated that proteasome activation achieved by USP14 (ubiquitin specific peptidase 14) inhibition delays the fusion of autophagosomes with the lysosome. A new molecular circuit involving UVRAG (UV radiation resistance associated) was uncovered as a key linker between the systems, adding complexity to the regulatory crosstalk. These findings clearly demonstrate that the surveillance mechanisms for protein homeostasis and cell survival are not separate, but a coordinated system. We also found that proteasome activation promotes the clearance of MAPT (microtubule associated protein tau), while facilitating the aggregation of mutant HTT (huntingtin) in cells, indicating that the biochemical property of a protein might play a role in its response to degradation signals. Collectively, our results present novel mechanistic insights into the reciprocal communication between the UPS and autophagy, highlighting that while a strategy upregulating either the UPS or autophagy holds great potential, it may have caveats originating from the intrinsic feedback regulation between them.
KEYWORDS:
In eukaryotes, the UPS is the primary pathway for the degradation of most short-lived proteins in both the cytoplasm and nucleus. The UPS is also the key system for protein quality control in cells, which functions by eliminating misfolded proteins that escape the refolding machinery of molecular chaperones. Autophagy is biochemically analogous to the UPS, in terms of both processes consisting of enzymatic cascades, using ubiquitin-like modifiers, E1, E2, E3-like enzymes, and multiple proteases, but autophagy has a very distinct substrate pool from the UPS, such as dysfunctional organelles. When excessive misfolded proteins are produced, which the UPS alone can no longer clear, autophagy, otherwise operating at a basal level, is strongly induced and participates in the clearance of these potentially harmful proteins. The 2 systems are, therefore, generally considered compensatory and, as a whole, a cooperative protein surveillance mechanism. A number of bridging proteins or protein complexes have been identified. However, it is largely unknown how the upregulation of proteasomal activity or induction of autophagy affects the autophagic flux or the output of the UPS, respectively.
Upregulation of the UPS or autophagy has generally been considered a promising strategy to protect cells against proteotoxic stress. To elucidate the reciprocal consequences, we examined the change in autophagic flux after proteasome activation (via USP14 inhibition), and the stability of UPS substrates after autophagy induction (via nutrient starvation) [Citation1]. Our experiments revealed that the UPS and autophagy appear to operate under a negative feedback regulation system, with USP14 being the common denominator of the 2 proteolytic machineries. We also found that proteasome activation significantly delays the oligomerization process of MAPT, whereas the same condition facilitates the aggregation of HTT proteins with long polyglutamine repeats. These findings indicate that a protein’s responsiveness to a degradation system may be attributed to its intrinsic biochemical property, varying through post-translational modifications (PTMs). Therefore, how PTMs on a proteotoxic protein are changed during disease progression or the aging process is a substantive question to be answered.
Critical evidence on the negative feedback coordination between the UPS and autophagy was obtained from the experiments using IU1, a chemical inhibitor of USP14, that functions as an endogenous antagonist of the proteasome. Treatment with IU1 results in the elevation of LC3-II and SQSTM1 levels in a dose- and time-dependent manner. This effect is USP14-specific and is accompanied by proteasome activation. Inhibition of autophagy at the autophagosome-lysosome fusion step virtually abrogates the effects of IU1, whereas additive effects are observed by USP14 inhibition and autophagy induction. Consistent with the additional electron microscopy analysis and tandem mRFP-GFP assay, our results indicated that proteasome activation lessens cellular autophagic flux, mainly by delaying the fusion of the autophagosome and lysosome.
How does USP14, which mainly associates with the proteasome as a deubiquitinating enzyme, affect the autophagic flux? To address this question, we tested various autophagy-related proteins to determine whether they are USP14 substrates, and determined that USP14 directly interacts with UVRAG, a tumor suppressor and an autophagic maturation regulator. The in vivo half-life of UVRAG is prolonged by USP14-mediated deubiquitination or proteasome inhibition. A pathophysiological manifestation of UVRAG, linking the 2 proteolytic systems, was examined in HCT116 colon cancer cells that are deficient in both USP14 and UVRAG. The normal communication between the UPS and autophagy appears to be perturbed in HCT116 cells, suggesting that proper coordination between the UPS and autophagy may have a critical role in blocking tumorigenesis or tumor progression.
To further dissect the compensatory crosstalk between the systems, we monitored the proteasomal activity and the stability of UPS substrates upon autophagy induction. To induce overall autophagy in the cell, we used either amino acid deprivation or glucose starvation, and found that although these conditions have different effects on the magnitude and kinetics of the autophagic flux, they attenuate cellular proteasomal activity and stabilize the otherwise highly short-lived proteins. These results suggest that the cellular fluxes through the UPS and autophagy are dynamically modulated to compensate and counterbalance the other, which may be required to maintain overall protein homeostasis. It is important to determine their reciprocal influence at the translational level, and the possible regulation of the biogenesis of the UPS and autophagy components.
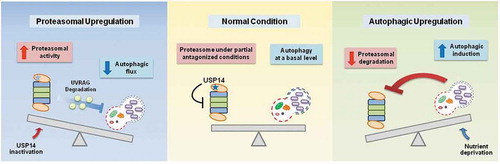
We report a relatively simple compensatory mechanism that may have multiple implications. First, upregulation of the UPS or autophagy is generally considered a promising strategy for the treatment of proteopathies. Our results suggest that elevating the overall UPS or autophagic flux in the cell may have a narrow therapeutic window if a disease has multiple pathological proteins, or if a proteotoxic protein’s degradation susceptibility is altered during disease progression. Second, our findings support the view that the UPS and autophagy are not 2 independent systems, but are closely coordinated processes that protect cells from a variety of stressed conditions. Third, the UPS and autophagy appear to primarily utilize their substrates for communication; for example, the cellular level of UVRAG is maintained under normal circumstances via the catalytic and noncatalytic actions of USP14. However, when USP14 is inhibited, UVRAG is degraded by the activated proteasome, limiting the cellular autophagic flux (). Therefore, this study highlights that USP14, by controlling UVRAG deubiquitination and degradation, has a dual function in cellular proteolytic flux, by negatively regulating proteasomal activity and positively regulating the autophagic flux.
Figure 1. Crosstalk between the UPS and autophagy. The proposed molecular circuit in which USP14 is a common denominator of the UPS and autophagy in the compensatory negative feedback connection. When USP14 is inhibited, activated proteasomes result in the degradation of UVRAG, which subsequently leads to a reduction in autophagic flux. Inversely, when cellular autophagy is induced via nutrient deprivation, proteasomal activity is reduced and levels of UPS substrates are elevated. The underlying mechanism mediating proteasome inactivation upon autophagy induction needs to be identified.
We anticipate that numerous proteins are involved in the UPS-autophagy communication and that they are specialized for various stress responses. We found that UVRAG senses proteasomal activity. Components of unfolded protein responses effectively recognize de novo misfolded proteins arising from abnormal translation and defective ribosomes. Some of the autophagic receptors and ubiquitin shuttling receptors are likely to affect the output of the other proteolytic system. Autophagic degradation of proteasomes, i.e., proteaphagy, might be the last resort to stop UPS-mediated proteolysis and, simultaneously, to eliminate inefficient proteasomes. Therefore, it appears essential to understand the interplay between the UPS and autophagy as a composite protein surveillance mechanism. The mechanistic clues from this study may contribute to resolve a part of this complex circuit.
Disclosure statement
No potential conflicts of interest were disclosed.
Additional information
Funding
Reference
- Kim E, Park S, Lee JH, et al. Dual function of USP14 deubiquitinase in cellular proteasomal activity and autophagic flux. Cell Rep. 2018;24:732–743.
