ABSTRACT
Overexpressed CCND1 (cyclin D1) is associated with hepatocellular carcinoma (HCC) and we used 147 tumor tissue samples from HCC patients and 3 murine models to reveal an inverse correlation between low autophagic activity and high CCND1 expression. These 2 phenomena in combination correlated with poor overall survival in HCC patients. Mechanistic analysis showed that activated autophagy triggered CCND1 ubiquitination followed by SQSTM1 (sequestosome 1)-mediated selective phagophore recruitment, autophagosome formation, fusion with a lysosome, and degradation. Functional studies revealed that autophagy-selective degradation of CCND1 suppresses DNA synthesis, cell proliferation, and colony, and liver tumor formation by arresting the cell cycle at the G1 phase. Most importantly, diverse pharmacological inducers (rapamycin and amiodarone) effectively suppress tumor growth in orthotopic liver tumor and subcutaneous tumor xenograft models. In conclusion, we have demonstrated a link between degradative autophagy and the cell cycle regulator CCND1, and have discovered the underlying mechanism by which the autophagic degradation machinery regulates the turnover of the cell-cycle regulator CCND1, which in turn affects HCC tumorigenesis.
Abbreviations: CCDN1: cyclin D1; HBV: hepatitis B virus; HCC: hepatocellular carcinoma; HCV: hepatitis C virus; SQSTM1: sequestosome 1
Hepatitis B virus (HBV) and hepatitis C virus (HCV) infection may lead to chronic liver diseases including hepatitis, fibrosis, cirrhosis, and HCC. Patients are often diagnosed at the late stages of HCC development due to lack of good diagnosis markers. Overexpression of CCND1 may trigger hepatocellular carcinogenesis through promoting liver cell proliferation and tumor formation. Autophagy and the proteasome are 2 major degradation systems regulating the physiological process of homeostasis in the cell. Emerging evidence shows that impaired autophagy or CCND1 overexpression are possible risk factors for HCC incidence.
In our recent report [Citation1], we analyzed 147 virus-associated HCC patients and found that low autophagic activity (low BECN1 and ATG5 expression and high SQSTM1 accumulation) was significantly correlated with high CCND1 expression, and in combination was associated with poor overall survival rate in HCC patients, implying that autophagy and CCND1 play critical roles in HCC development. We further found that the G1 phase cell cycle regulator CCND1 is an oncogenic factor regulated by both the proteasome and autophagy degradation systems. Intriguingly, only the latter selectively recruits and regulates CCND1 turnover to prevent HCC. Mechanistic analysis shows that activated autophagy triggers GSK3B-mediated CCND1 phosphorylation (Thr286) and ubiquitination followed by SQSTM1-mediated selective phagophore recruitment, subsequent autophagosome formation, fusion with a lysosome, and degradation.
We utilized CCND1 ubiquitination sites mutants and SQSTM1 gene knockout approaches to confirm that the ubiquitination sites of CCND1, as well as SQSTM1, are essential for selective recruitment and binding with LC3-II on the phagophore following induction of autophagy. Functional analyses showed that activated autophagy plays suppressive roles in cell proliferation, colony formation, and development of tumors through the degradation of ubiquitinated CCND1. Interestingly, we demonstrated that induction of autophagy by either off-label use of the drug ‘amiodarone’ or the traditional autophagy inducer ‘rapamycin’ effectively suppresses tumor growth in orthotopic rat liver tumor and subcutaneous tumor xenograft mouse models. Taken together, our data reveal the underlying mechanism by which the autophagic degradation machinery regulates the turnover of the cell-cycle regulator CCND1, which in turn affects HCC development.
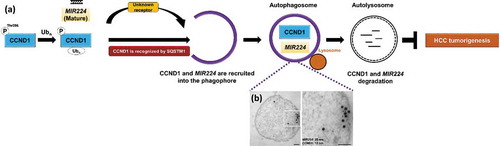
We previously reported that autophagy preferentially degrades the oncogenic microRNA MIR224 to prevent HCC occurrence. In a rat orthotopic model, liver tumors are significantly suppressed by amiodarone-induced autophagy, and levels of both MIR224 and CCND1 are decreased. Based on these findings, along with the results of the current study, we hypothesize that these 2 oncogenic factors (MIR224 and CCND1) are simultaneously regulated by the autophagic degradation machinery to maintain cellular homeostasis, thereby preventing HCC occurrence ()). Consistent with this hypothesis, we observe colocalization of MIR224 and CCND1 by confocal microscopy and coexistence of these 2 molecules in purified autophagosomes by transmission electron microscopy ()). Because the receptor for MIR224 recruitment has not been identified, it has yet to be determined whether these 2 molecules in the autophagosome use the same degradation pathway. This is the first study to demonstrate that autophagy affects HCC tumorigenesis through selective recruitment and degradation of 2 oncogenic factors, MIR224 and CCND1.
Figure 1. A schematic hypothetical diagram of degradative autophagy, selective regulation of MIR224 and CCND1 and evidence of MIR224 and CCND1 coexistence in the autophagosome. (a) CCND1 is selectively recruited to the phagophore through the specific receptor SQSTM1. MIR224 is preferentially recruited to the phagophore by unknown factors. Both of the oncogenic factors are present in autophagosomes after selective recruitment; subsequent autophagosome fusion with the lysosome is followed by degradation. (b) Human liver cancer Hep 3B cells were treated with the autophagy inducer amiodarone. Coexistence of immunogold-labeled CCND1 (12-nm bead) and MIR224 (20-nm bead) after miRNA in situ hybridization in the purified autophagosomes was investigated using TEM. Scale bar: 100 nm.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Wu S-Y, Lan S-H, Wu S-R, et al. Hepatocellular carcinoma-related cyclin D1 is selectively regulated by autophagy degradation system. Hepatology. 2018 July;68(1):141–154.
