ABSTRACT
Circadian rhythms help cells to organize complex processes, but how they shape the kinetics of protein catabolism is unclear. In a recent paper, we employed proteomics to map daily biological rhythms in autophagic flux in mouse liver, and correlated these rhythms with proteasome activity. We also explored the effect of inflammation caused by endotoxin on autophagy dynamics. Our data provide insight into how circadian rhythms serve as a framework for connecting the spatial, temporal, and metabolic aspects of autophagy at a system-wide level. Our observations also have implications for how to optimize autophagy-directed therapies in patients.
Circadian rhythms are daily oscillations in biological activity that allow organisms to anticipate environmental changes imposed by the solar day. These rhythms are ubiquitous in nature and can be found at every biological scale, from sleep-wake cycles to the rhythmic abundance of biomolecules. Research into circadian rhythms has accelerated in recent years with the discovery that they originate from a cell autonomous molecular circadian clock, consisting of transcription factors called ‘clock genes’ that impart daily rhythms to gene expression, cellular metabolism, and ultimately organism physiology. With large swaths of the transcriptome and metabolome subject to rhythmic expression, the circadian clock is regarded as part of the core operating system of the cell. In a recent paper, we explored how circadian rhythms shape autophagic activity during health and during severe inflammation [Citation1].
From the standpoint of cellular physiology, autophagy and circadian rhythms have much in common. Deletion of autophagy genes and circadian clock genes confer a similar spectrum of phenotypes in mice, including the accumulation of dysfunctional mitochondria, excess free radicals, and altered lipid metabolism. Moreover, interventions such as intermittent fasting that have beneficial effects on metabolism and are dependent on macroautophagy also reset the circadian clock. Direct evidence of a connection between circadian rhythms and macroautophagy first came from ultrastructural studies, which showed that autophagosome number varies cyclically across the day. However, fundamental questions remained: for example, is macroautophagy unique in displaying circadian rhythms or do other autophagy pathways oscillate as well? Are the temporal dynamics of selective autophagy and bulk autophagy similar or different? What is the relationship between rhythms in autophagic activity and the behavior of non-lysosomal catabolic pathways, such as the ubiquitin-proteasome system? Finally, what happens to autophagy dynamics when circadian rhythms are disrupted by inflammation, a common feature of many diseases?
To address these questions, we developed a proteomic approach to measuring autophagic flux in mouse liver and then applied this method to detect daily biological rhythms in autophagy at a proteome-wide scale. Our data yielded several insights into the dynamics of autophagy in vivo. First, we found that autophagic flux is globally rhythmic, encompassing not just macroautophagy targets but also clients of chaperone-mediated autophagy. Viewed from a temporal perspective, mediators of ‘selective’ macroautophagy such as Sequestesome-1 (SQSTM1/p62) and Next to BRCA gene 1 (NBR1) are not so selective: they are degraded with the same dynamics as targets of bulk macroautophagy. Instead, autophagy rhythms confer a different kind of substrate selectivity based on subcellular location. Proteins that localize to the cytosol and nucleus tend on a statistical basis to be degraded during the day while mitochondrial, ER, and peroxisomal proteins tend to be degraded at night (). The key lesson is that a connection exists between the spatial and temporal dynamics of autophagy, which we speculate reflects alternative regulation of autophagosome formation in different regions of the cell. Autophagy, it seems, occurs across a landscape of ‘space-time’.
Figure 1. Autophagy is regulated around a 24-h rhythm in mouse liver, catabolizing substrates during the day or night-time in distinct cellular regions.
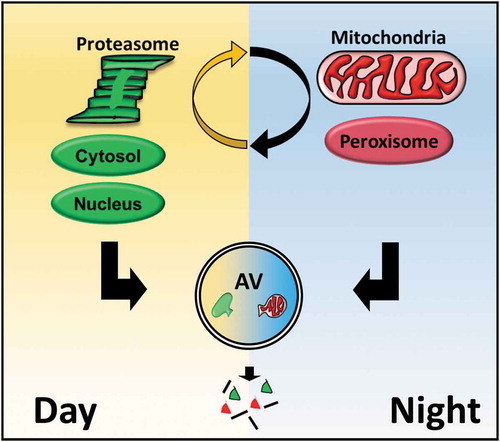
A second insight from our study is that daily oscillations in autophagy are part of a larger rhythmic coordination of protein catabolism that includes the proteasome. By adapting our turnover assay to employ bortezomib instead of leupeptin, we identified daily rhythms in liver proteasomal flux in parallel with those of autophagic flux. Given that proteasomes are degraded via autophagy (which our data confirmed), one might think rhythms in proteasomal flux would be reciprocal to those of autophagy. Instead, we found that the dynamics of basal autophagic and proteasomal flux are roughly aligned such that liver protein catabolism is concentrated during the daytime in mice.
Finally, we found that treating mice with endotoxin disrupts the temporal structure of liver autophagy just as it disturbs clock gene expression. However, rather than categorically suppressing or stimulating autophagic flux, endotoxin induces a shift in substrate preferences to favor mitochondrial proteins at the expense of cytosolic proteins. Such a shift would be predicted to reduce mitochondrial mass and to support a cellular shift towards greater dependence on glycolysis, both of which occur during sepsis. These results illustrate the role autophagy plays in metabolic reprogramming during inflammation. They also suggest that spatiotemporal regulation of autophagy persists during disease states.
Our proteome-wide profiling of autophagy rhythms raises new questions and has practical implications. The mechanism behind how autophagy targets subcellular compartments at certain times of day or under specific health states needs to be explored. Factors controlling the alignment between autophagic and proteasomal rhythms represents another topic for future research. From a translational standpoint, our data imply that pharmacological modulation of autophagy to treat various diseases could be optimized by identifying the best time of day to administer these drugs, a concept known a chronotherapy. This might limit the doses of medications needed to achieve results and potentially reduce unwanted side effects. To successfully deploy autophagy research in the clinic, we may discover that timing is everything.
Acknowledgments
MR, AE, and JH are supported by grants from the National Heart Lung and Blood Institute (NHLBI), and the Children’s Discovery Institute (CDI).
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
Reference
- Ryzhikov M, Ehlers A, Steinberg A, et al. Dirunal rhythms spatially and temporally organize autophagy. Cell Rep. 2019;26:1880–1892.
